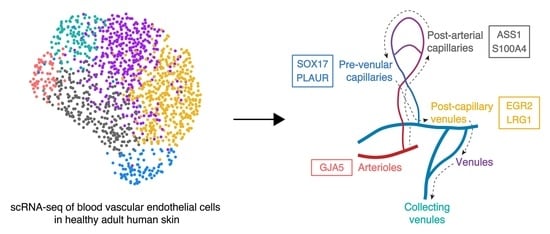
Novel Blood Vascular Endothelial Subtype-Specific Markers in Human Skin Unearthed by Single-Cell Transcriptomic Profiling
Abstract
:1. Introduction
2. Materials and Methods
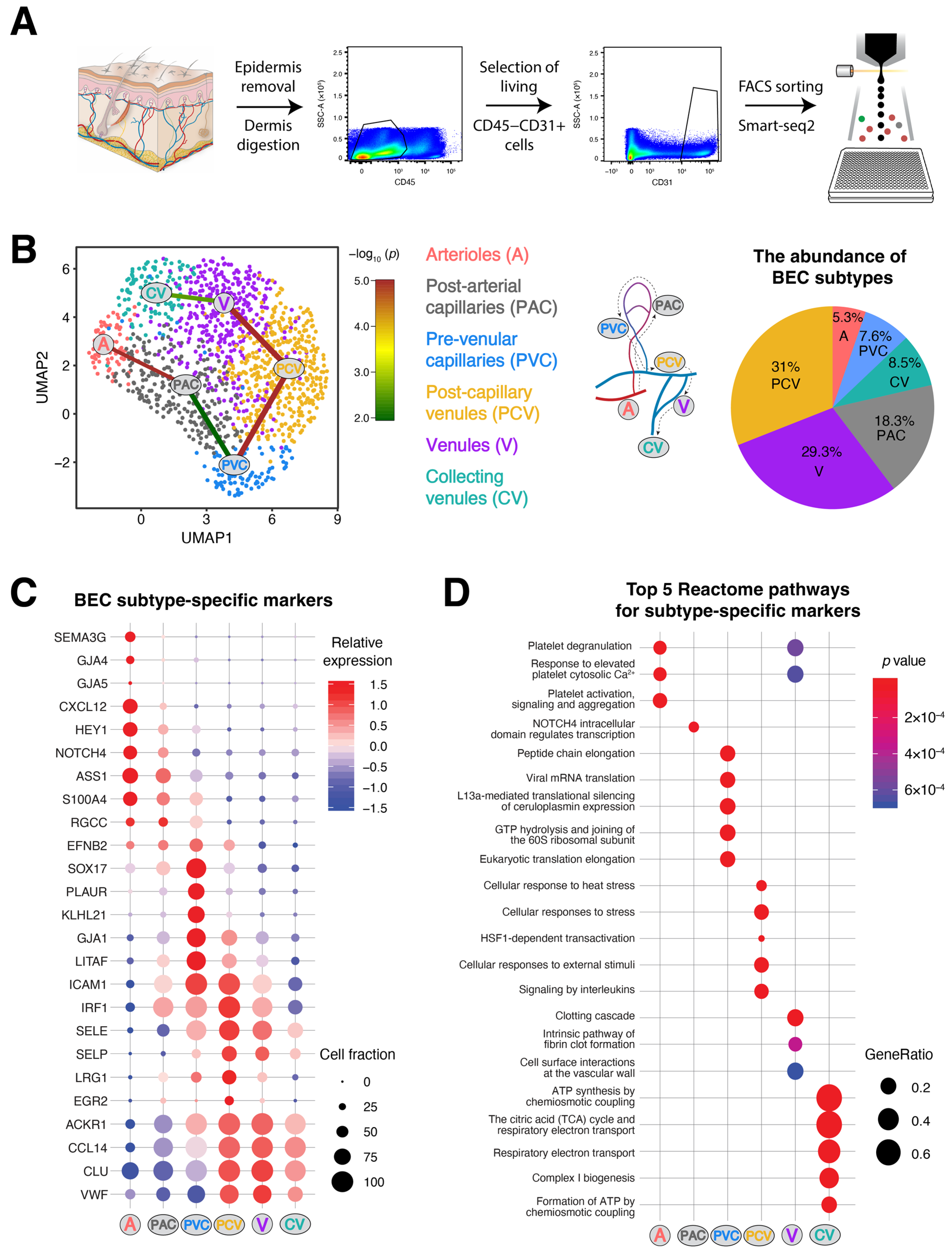
2.1. Isolation of Human Dermal Endothelial Cells
2.2. Full-Length Single-Cell RNA Sequencing
2.3. Preprocessing of scRNA-Seq Data
2.4. Clustering, Differential Expression and Pseudotime Trajectory Analyses
2.5. Immunofluorescence Staining of Skin Sections
2.6. Whole-Mount Immunostaining
3. Results
3.1. Identification of Six Distinct Human Dermal Blood Vascular Endothelial Cell Subtypes
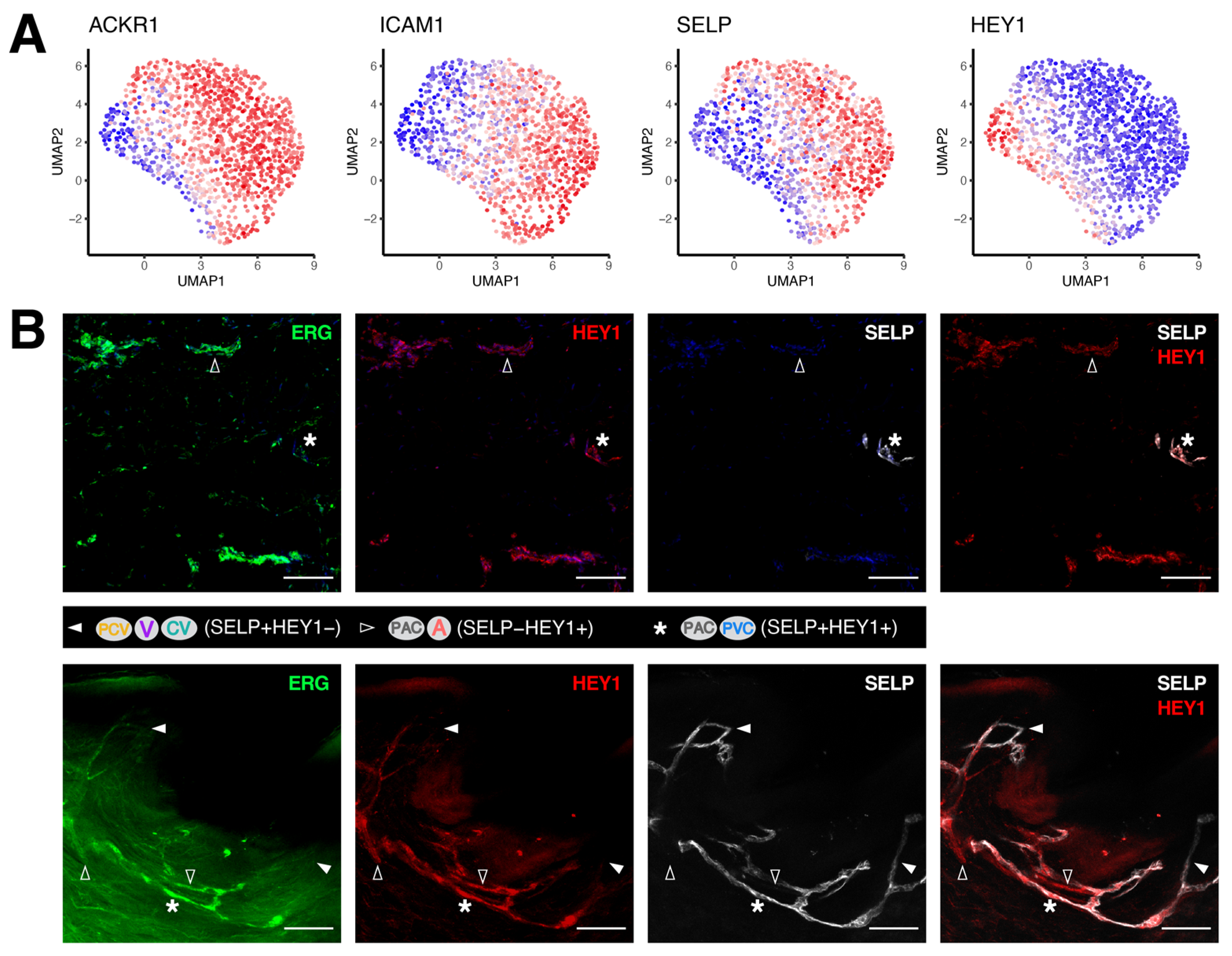
3.2. Discrimination between Venular and Non-Venular Endothelial Cells via the Expression of Established Markers
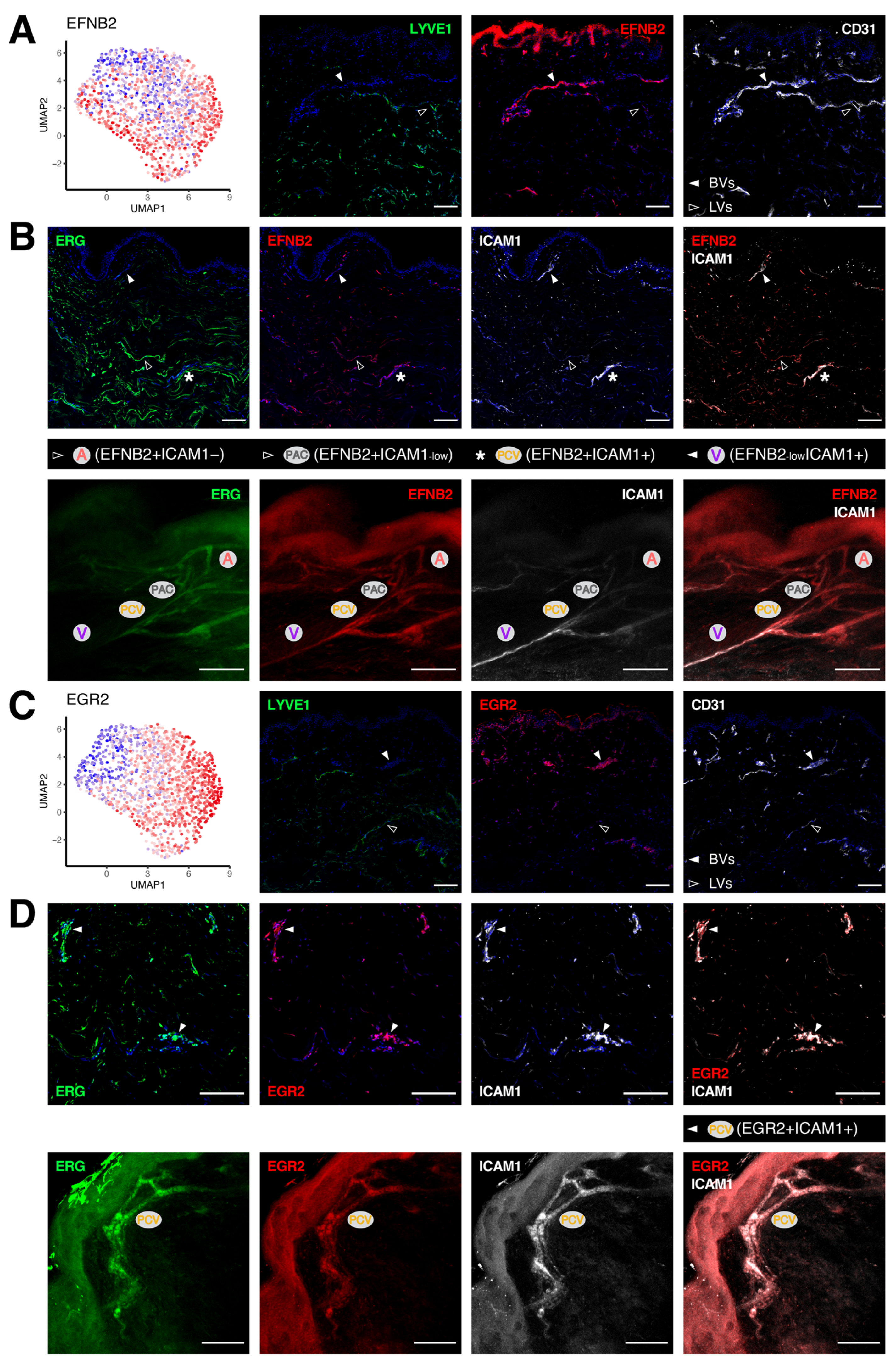
3.3. Unique Marker Combinations Delineate Venular Endothelial Cell Subpopulations
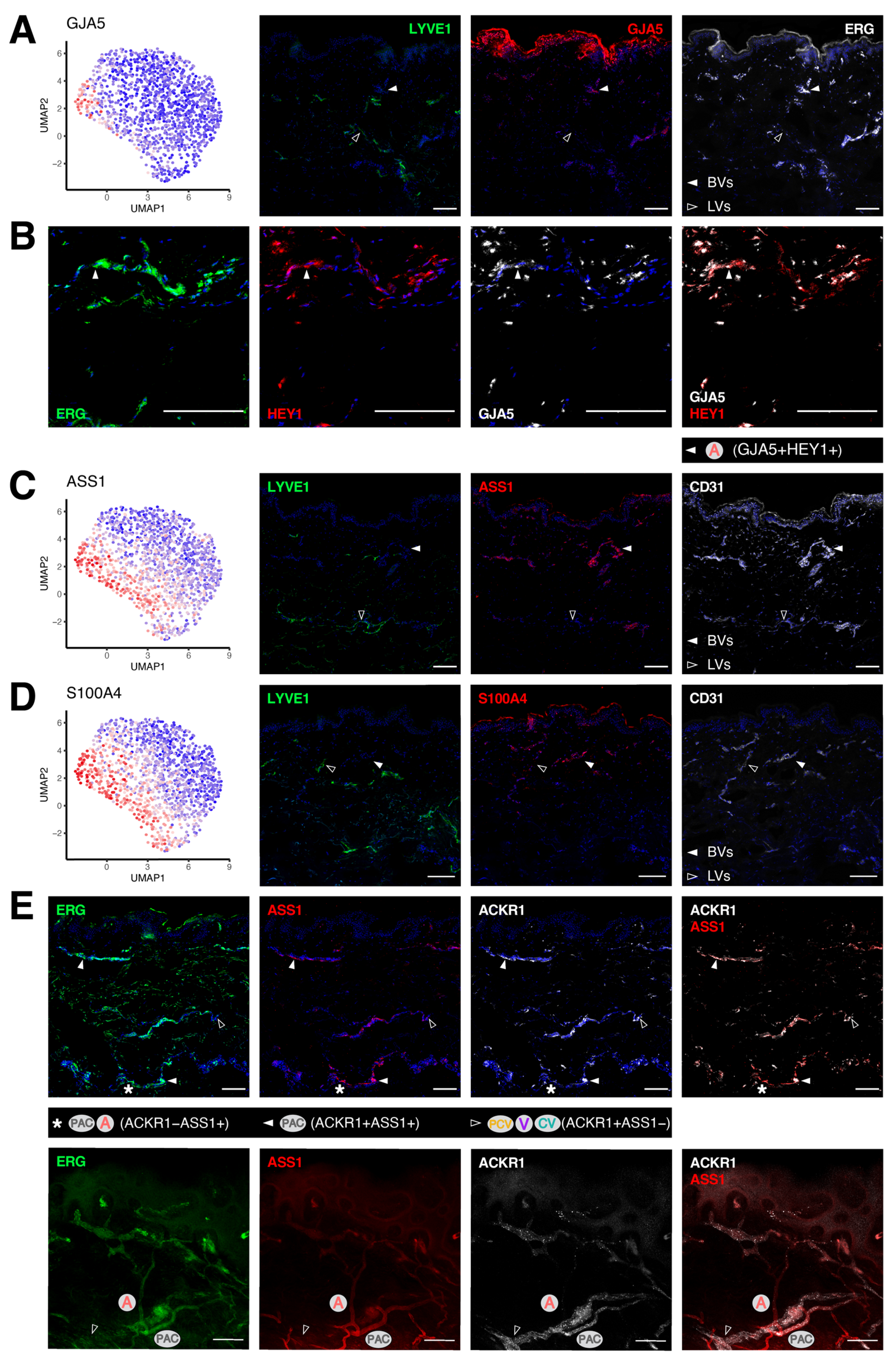
3.4. Transcriptomic Signatures of Blood Vascular Endothelial Cells Constituting Arterioles and Capillaries
3.5. Molecular Characterization of Pre-Venular Capillary-Lining Endothelial Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aird, W.C. Phenotypic heterogeneity of the endothelium. Circ. Res. 2007, 100, 174–190. [Google Scholar] [CrossRef] [Green Version]
- Potente, M.; Mäkinen, T. Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 477–494. [Google Scholar] [CrossRef]
- Pober, J.S.; Sessa, W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007, 7, 803–815. [Google Scholar] [CrossRef]
- Pober, J.S.; Sessa, W.C. Inflammation and the blood microvascular system. Cold Spring Harb. Perspect. Biol. 2015, 7, a016345. [Google Scholar] [CrossRef]
- Thurston, G.; Murphy, T.J.; Baluk, P.; Lindsey, J.R.; McDonald, D.M. Angiogenesis in mice with chronic airway inflammation: Strain-dependent differences. Am. J. Pathol. 1998, 153, 1099–1112. [Google Scholar] [CrossRef]
- Krausgruber, T.; Fortelny, N.; Fife-Gernedl, V.; Senekowitsch, M.; Schuster, L.C.; Lercher, A.; Nemc, A.; Schmidl, C.; Rendeiro, A.F.; Bergthaler, A.; et al. Structural cells are key regulators of organ-specific immune responses. Nature 2020, 583, 296–302. [Google Scholar] [CrossRef]
- Nakato, R.; Wada, Y.; Nakaki, R.; Nagae, G.; Katou, Y.; Tsutsumi, S.; Nakajima, N.; Fukuhara, H.; Iguchi, A.; Kohro, T.; et al. Comprehensive epigenome characterization reveals diverse transcriptional regulation across human vascular endothelial cells. Epigenet. Chromatin 2019, 12, 77. [Google Scholar] [CrossRef] [Green Version]
- Tacconi, C.; He, Y.; Ducoli, L.; Detmar, M. Epigenetic regulation of the lineage specificity of primary human dermal lymphatic and blood vascular endothelial cells. Angiogenesis 2021, 24, 67–82. [Google Scholar] [CrossRef]
- Feng, W.; Chen, L.; Nguyen, P.K.; Wu, S.M.; Li, G. Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front. Cardiovasc. Med. 2019, 6, 165. [Google Scholar] [CrossRef]
- Kalucka, J.; de Rooij, L.P.M.H.; Goveia, J.; Rohlenova, K.; Dumas, S.J.; Meta, E.; Conchinha, N.; Taverna, F.; Teuwen, L.-A.; Veys, K.; et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 2020, 180, 764–779.e20. [Google Scholar] [CrossRef]
- Kalluri, A.; Vellarikkal, S.K.; Edelman, E.; Nguyen, L.; Subramanian, A.; Ellinor, P.T.; Regev, A.; Kathiresan, S.; Gupta, R.M. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation 2019, 140, 147–163. [Google Scholar] [CrossRef]
- Paik, D.T.; Tian, L.; Williams, I.M.; Rhee, S.; Zhang, H.; Liu, C.; Mishra, R.; Wu, S.; Red-Horse, K.; Wu, J.C. Single-cell RNA-Seq unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation 2020, 91, 3527–3531. [Google Scholar] [CrossRef]
- Vanlandewijck, M.; He, L.; Mäe, M.A.; Andrae, J.; Ando, K.; Del Gaudio, F.; Nahar, K.; Lebouvier, T.; Laviña, B.; Gouveia, L.; et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018, 554, 475–480. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Suryawanshi, H.; Morozov, P.; Gay-Mimbrera, J.; Del Duca, E.; Kim, H.J.; Kameyama, N.; Estrada, Y.; Der, E.; Krueger, J.G.; et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J. Allergy Clin. Immunol. 2020, 145, 1615–1628. [Google Scholar] [CrossRef] [Green Version]
- Tabib, T.; Morse, C.; Wang, T.; Chen, W.; Lafyatis, R. SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. J. Investig. Dermatol. 2018, 138, 802–810. [Google Scholar] [CrossRef] [Green Version]
- Philippeos, C.; Telerman, S.B.; Oules, B.; Pisco, A.O.; Shaw, T.J.; Elgueta, R.; Lombardi, G.; Driskell, R.R.; Soldin, M.; Lynch, M.D.; et al. Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations. J. Investig. Dermatol. 2018, 138, 811–825. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.B.; Sedgewick, A.J.; Finnegan, A.I.; Harirchian, P.; Lee, J.; Kwon, S.; Fassett, M.S.; Golovato, J.; Gray, M.; Ghadially, R.; et al. Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep. 2018, 25, 871–883. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, N.; He, Y.; D’Addio, M.; Tacconi, C.; Detmar, M.; Dieterich, L.C. Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes. PLoS Biol. 2020, 18, e3000704. [Google Scholar] [CrossRef] [Green Version]
- Picelli, S.; Faridani, O.R.; Björklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [Green Version]
- Lun, A. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016, 17, 75. [Google Scholar] [CrossRef]
- Bacher, R.; Chu, L.-F.; Leng, N.; Gasch, A.P.; Thomson, J.A.; Stewart, R.M.; Newton, M.; Kendziorski, C. SCnorm: Robust normalization of single-cell RNA-seq data. Nat. Methods 2017, 14, 584–586. [Google Scholar] [CrossRef] [Green Version]
- Haghverdi, L.; Lun, A.T.L.; Morgan, M.D.; Marioni, J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018, 36, 421–427. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- Finak, G.; McDavid, A.; Yajima, M.; Deng, J.; Gersuk, V.; Shalek, A.K.; Slichter, C.K.; Miller, H.W.; McElrath, M.J.; Prlic, M.; et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015, 16, 278. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. Clusterprofiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Herman, J.S.; Sagar; Grün, D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods 2018, 15, 379–386. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Commerford, C.D.; Dieterich, L.C.; He, Y.; Hell, T.; Montoya, J.; Noerrelykke, S.F.; Russo, E.; Röcken, M.; Detmar, M. Mechanisms of Tumor-Induced Lymphovascular Niche Formation in Draining Lymph Nodes. Cell Rep. 2018, 25, 3554–3563.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz, N.G.D.; D’Amore, P.A. Arterial versus venous endothelial cells. Cell Tissue Res. 2008, 335, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Thiriot, A.; Perdomo, C.; Cheng, G.; Novitzky-Basso, I.; McArdle, S.; Kishimoto, J.K.; Barreiro, O.; Mazo, I.; Triboulet, R.; Ley, K.; et al. Differential DARC/ACKR1 expression distinguishes venular from non-venular endothelial cells in murine tissues. BMC Biol. 2017, 15, 45. [Google Scholar] [CrossRef]
- Kalna, V.; Yang, Y.; Peghaire, C.R.; Frudd, K.; Hannah, R.; Shah, A.V.; Almagro, L.O.; Boyle, J.J.; Göttgens, B.; Ferrer, J.; et al. The Transcription Factor ERG Regulates Super-Enhancers Associated with an Endothelial-Specific Gene Expression Program. Circ. Res. 2019, 124, 1337–1349. [Google Scholar] [CrossRef]
- Rocha, S.F.; Adams, R.H. Molecular differentiation and specialization of vascular beds. Angiogenesis 2009, 12, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Girbl, T.; Lenn, T.; Perez-Gutierrez, L.; Rolas, L.; Barkaway, A.; Thiriot, A.; del Fresno, C.; Lynam, E.; Hub, E.; Thelen, M.; et al. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis. Immunity 2018, 49, 1062–1076.e6. [Google Scholar] [CrossRef] [Green Version]
- Pusztaszeri, M.P.; Seelentag, W.; Bosman, F.T. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand Factor, and Fli-1 in normal human tissues. J. Histochem. Cytochem. 2006, 54, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Shu, B.; Zhang, Y.; Wang, M. Endothelial response to pathophysiological stress. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 11. [Google Scholar] [CrossRef]
- To, W.K.L.; Kumar, P.; Marshall, J.M. Hypoxia is an effective stimulus for vesicular release of ATP from human umbilical vein endothelial cells. Placenta 2015, 36, 759–766. [Google Scholar] [CrossRef] [Green Version]
- Taefehshokr, S.; Key, Y.A.; Khakpour, M.; Dadebighlu, P.; Oveisi, A. Early growth response 2 and Egr3 are unique regulators in immune system. Cent. Eur. J. Immunol. 2017, 2, 205–209. [Google Scholar] [CrossRef]
- Fang, F.; Ooka, K.; Bhattachyya, S.; Wei, J.; Wu, M.; Du, P.; Lin, S.; del Galdo, F.; Feghali-Bostwick, C.A.; Varga, J. The early growth response gene Egr2 (Alias Krox20) is a novel transcriptional target of transforming growth factor-β that is up-regulated in systemic sclerosis and mediates profibrotic responses. Am. J. Pathol. 2011, 178, 2077–2090. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Abraham, S.; McKenzie, J.A.G.; Jeffs, N.; Swire, M.; Tripathi, V.B.; Luhmann, U.F.O.; Lange, C.A.K.; Zhai, Z.; Arthur, H.M.; et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature 2013, 499, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhou, J.; Xie, Z.; Wang, J.; Ho, C.-K.; Zhang, Y.; Li, Q. Mechanical strain promotes skin fibrosis through LRG-1 induction mediated by ELK1 and ERK signalling. Commun. Biol. 2019, 2, 359. [Google Scholar] [CrossRef]
- Mun, G.I.; Kim, I.-S.; Lee, B.-H.; Boo, Y.C. Endothelial argininosuccinate synthetase 1 regulates nitric oxide production and monocyte adhesion under static and laminar shear stress conditions. J. Biol. Chem. 2011, 286, 2536–2542. [Google Scholar] [CrossRef] [Green Version]
- Pepper, M.S. Role of the matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1104–1117. [Google Scholar] [CrossRef] [Green Version]
- Semov, A.; Moreno, M.J.; Onichtchenko, A.; Abulrob, A.; Ball, M.; Ekiel, I.; Pietrzynski, G.; Stanimirovic, D.; Alakhov, V. Metastasis-associated Protein S100A4 Induces Angiogenesis through Interaction with Annexin II and Accelerated Plasmin Formation. J. Biol. Chem. 2005, 280, 20833–20841. [Google Scholar] [CrossRef] [Green Version]
- Schmidt-Hansen, B.; Örnås, D.; Grigorian, M.; Klingelhöfer, J.; Tulchinsky, E.; Lukanidin, E.; Ambartsumian, N. Extracellular S100A4(mts1) stimulates invasive growth of mouse endothelial cells and modulates MMP-13 matrix metalloproteinase activity. Oncogene 2004, 23, 5487–5495. [Google Scholar] [CrossRef] [Green Version]
- Rakic, J.M.; Maillard, C.; Jost, M.; Bajou, K.; Masson, V.; Devy, L.; Lambert, V.; Foidart, J.M. Role of plasminogen activator-plasmin system in tumor angiogenesis. Cell. Mol. Life Sci. 2003, 60, 463–473. [Google Scholar] [CrossRef]
- Poettler, M.; Unseld, M.; Mihaly-Bison, J.; Uhrin, P.; Koban, F.; Binder, B.R.; Zielinski, C.C.; Prager, G.W. The urokinase receptor (CD87) represents a central mediator of growth factor-induced endothelial cell migration. Thromb. Haemost. 2012, 108, 357–366. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, L.; Marsboom, G.; Jambusaria, A.; Xiong, S.; Toth, P.; Benevolenskaya, E.V.; Rehman, J.; Malik, A.B. Sox17 is required for endothelial regeneration following inflammation-induced vascular injury. Nat. Commun. 2019, 10, 2126. [Google Scholar] [CrossRef] [Green Version]
- Corada, M.; Orsenigo, F.; Morini, M.F.; Pitulescu, M.E.; Bhat, G.; Nyqvist, D.; Breviario, F.; Conti, V.; Briot, A.; Iruela-Arispe, M.L.; et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat. Commun. 2013, 4, 2609. [Google Scholar] [CrossRef] [Green Version]
- Goveia, J.; Zecchin, A.; Rodriguez, F.M.; Moens, S.; Stapor, P.; Carmeliet, P. Endothelial cell differentiation by SOX17. Circ. Res. 2014, 115, 205–207. [Google Scholar] [CrossRef] [Green Version]
- Ha, E.H.; Choi, J.-P.; Kwon, H.-S.; Park, H.J.; Lah, S.J.; Moon, K.-A.; Lee, S.-H.; Kim, I.; Cho, Y.S. Endothelial Sox17 promotes allergic airway inflammation. J. Allergy Clin. Immunol. 2019, 144, 561–573.e6. [Google Scholar] [CrossRef] [Green Version]
- van Kempen, M.J.; Jongsma, H.J. Distribution of connexin37, connexin40 and connexin43 in the aorta and coronary artery of several mammals. Histochem. Cell Biol. 1999, 112, 479–486. [Google Scholar] [CrossRef]
- Su, T.; Stanley, G.; Sinha, R.; D’Amato, G.; Das, S.; Rhee, S.; Chang, A.H.; Poduri, A.; Raftrey, B.; Dinh, T.T.; et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature 2018, 559, 356–362. [Google Scholar] [CrossRef]
- Duarte, A. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004, 18, 2474–2478. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Coon, B.G.; Gillis, N.; Chen, Z.; Qiu, J.; Chittenden, T.W.; Burt, J.M.; Schwartz, M.A.; Hirschi, K.K. Addendum: Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat. Commun. 2018, 9, 720. [Google Scholar] [CrossRef]
- Yan, R.; van Meurs, M.; Popa, E.R.; Jongman, R.M.; Zwiers, P.J.; Niemarkt, A.E.; Kuiper, T.; Kamps, J.A.; Heeringa, P.; Zijlstra, J.; et al. Endothelial interferon regulatory factor 1 regulates lipopolysaccharide-induced VCAM-1 expression independent of NFκB. J. Innate Immun. 2017, 9, 546–560. [Google Scholar] [CrossRef]
- Bijli, K.M.; Fazal, F.; Rahman, A. Regulation of Rela/p65 and endothelial cell inflammation by proline-rich tyrosine kinase 2. Am. J. Respir. Cell Mol. Biol. 2012, 47, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Potente, M.; Urbich, C.; Sasaki, K.; Hofmann, W.K.; Heeschen, C.; Aicher, A.; Kollipara, R.; Depinho, R.A.; Zeiher, A.M.; Dimmeler, S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Investig. 2005, 115, 2382–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Tempel, D.; Dekker, W.K.D.; Haasdijk, R.; Chrifi, I.; Bos, F.L.; Wagtmans, K.; van de Kamp, E.H.; Blonden, L.; Biessen, E.A.; et al. Ets2 Determines the Inflammatory State of Endothelial Cells in Advanced Atherosclerotic Lesions. Circ. Res. 2011, 109, 382–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funk, S.D.; Finney, A.C.; Yurdagul, A.; Pattillo, C.B.; Orr, A.W. EphA2 stimulates VCAM-1 expression through calcium-dependent NFAT1 activity. Cell Signal. 2018, 49, 30–38. [Google Scholar] [CrossRef]
- Hneino, M.; Blirando, K.; Buard, V.; Tarlet, G.; Benderitter, M.; Hoodless, P.; François, A.; Milliat, F. The TG-interacting factor TGIF1 regulates stress-induced proinflammatory phenotype of endothelial cells. J. Biol. Chem. 2012, 287, 38913–38921. [Google Scholar] [CrossRef] [Green Version]
- Craig, M.P.; Sumanas, S. ETS transcription factors in embryonic vascular development. Angiogenesis 2016, 19, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Inverso, D.; Shi, J.; Lee, K.H.; Jakab, M.; Ben-Moshe, S.; Kulkarni, S.R.; Schneider, M.; Wang, G.; Komeili, M.; Vélez, P.A.; et al. A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie-Wnt signaling axis in the liver. Dev. Cell 2021, 56, 1677–1693.e10. [Google Scholar] [CrossRef]
- Reynolds, G.; Vegh, P.; Fletcher, J.; Poyner, E.F.M.; Stephenson, E.; Goh, I.; Botting, R.A.; Huang, N.; Olabi, B.; Dubois, A.; et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 2021, 371. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, Z.; Wang, L.; Lin, Y.; Fang, H.; Lei, J.; Cao, T.; Wang, G.; Dang, E. Single-cell transcriptome profiling reveals vascular endothelial cell heterogeneity in human skin. Theranostics 2021, 11, 6461–6476. [Google Scholar] [CrossRef]
- Goveia, J.; Rohlenova, K.; Taverna, F.; Treps, L.; Conradi, L.-C.; Pircher, A.; Geldhof, V.; de Rooij, L.P.; Kalucka, J.; Sokol, L.; et al. An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell 2020, 37, 421. [Google Scholar] [CrossRef]

| Clusters | GJA5 | ASS1 | HEY1 | S100A4 | SOX17 | PLAUR | EFNB2 | ACKR1 | ICAM1 | EGR2 | LRG1 | SELP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arterioles (A) | + | + | + | + | + | − | + | − | − | − | − | − |
| Post-arterial capillaries (PAC) | − | + | + | + | + | low | + | low | low | low | low | low |
| Pre-venular capillaries (PVC) | − | low | + | + | high | high | + | + | + | low | low | low |
| Post-capillary venules (PCV) | − | − | − | − | low | low | + | + | + | high | high | + |
| Venules (V) | − | − | − | − | − | − | low | + | + | low | low | + |
| Collecting venules (CV) | − | − | − | − | − | − | − | + | low | − | − | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Tacconi, C.; Dieterich, L.C.; Kim, J.; Restivo, G.; Gousopoulos, E.; Lindenblatt, N.; Levesque, M.P.; Claassen, M.; Detmar, M. Novel Blood Vascular Endothelial Subtype-Specific Markers in Human Skin Unearthed by Single-Cell Transcriptomic Profiling. Cells 2022, 11, 1111. https://doi.org/10.3390/cells11071111
He Y, Tacconi C, Dieterich LC, Kim J, Restivo G, Gousopoulos E, Lindenblatt N, Levesque MP, Claassen M, Detmar M. Novel Blood Vascular Endothelial Subtype-Specific Markers in Human Skin Unearthed by Single-Cell Transcriptomic Profiling. Cells. 2022; 11(7):1111. https://doi.org/10.3390/cells11071111
Chicago/Turabian StyleHe, Yuliang, Carlotta Tacconi, Lothar C. Dieterich, Jihye Kim, Gaetana Restivo, Epameinondas Gousopoulos, Nicole Lindenblatt, Mitchell P. Levesque, Manfred Claassen, and Michael Detmar. 2022. "Novel Blood Vascular Endothelial Subtype-Specific Markers in Human Skin Unearthed by Single-Cell Transcriptomic Profiling" Cells 11, no. 7: 1111. https://doi.org/10.3390/cells11071111
APA StyleHe, Y., Tacconi, C., Dieterich, L. C., Kim, J., Restivo, G., Gousopoulos, E., Lindenblatt, N., Levesque, M. P., Claassen, M., & Detmar, M. (2022). Novel Blood Vascular Endothelial Subtype-Specific Markers in Human Skin Unearthed by Single-Cell Transcriptomic Profiling. Cells, 11(7), 1111. https://doi.org/10.3390/cells11071111