On the Cutting Edge of Oral Cancer Prevention: Finding Risk-Predictive Markers in Precancerous Lesions by Longitudinal Studies
Abstract
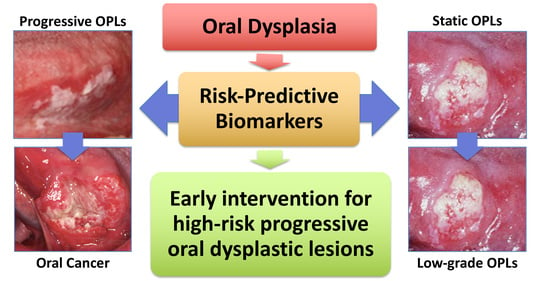
:1. Introduction
2. Risk-Predictive Markers Based on a Longitudinally Followed Study Design
3. Molecular Markers
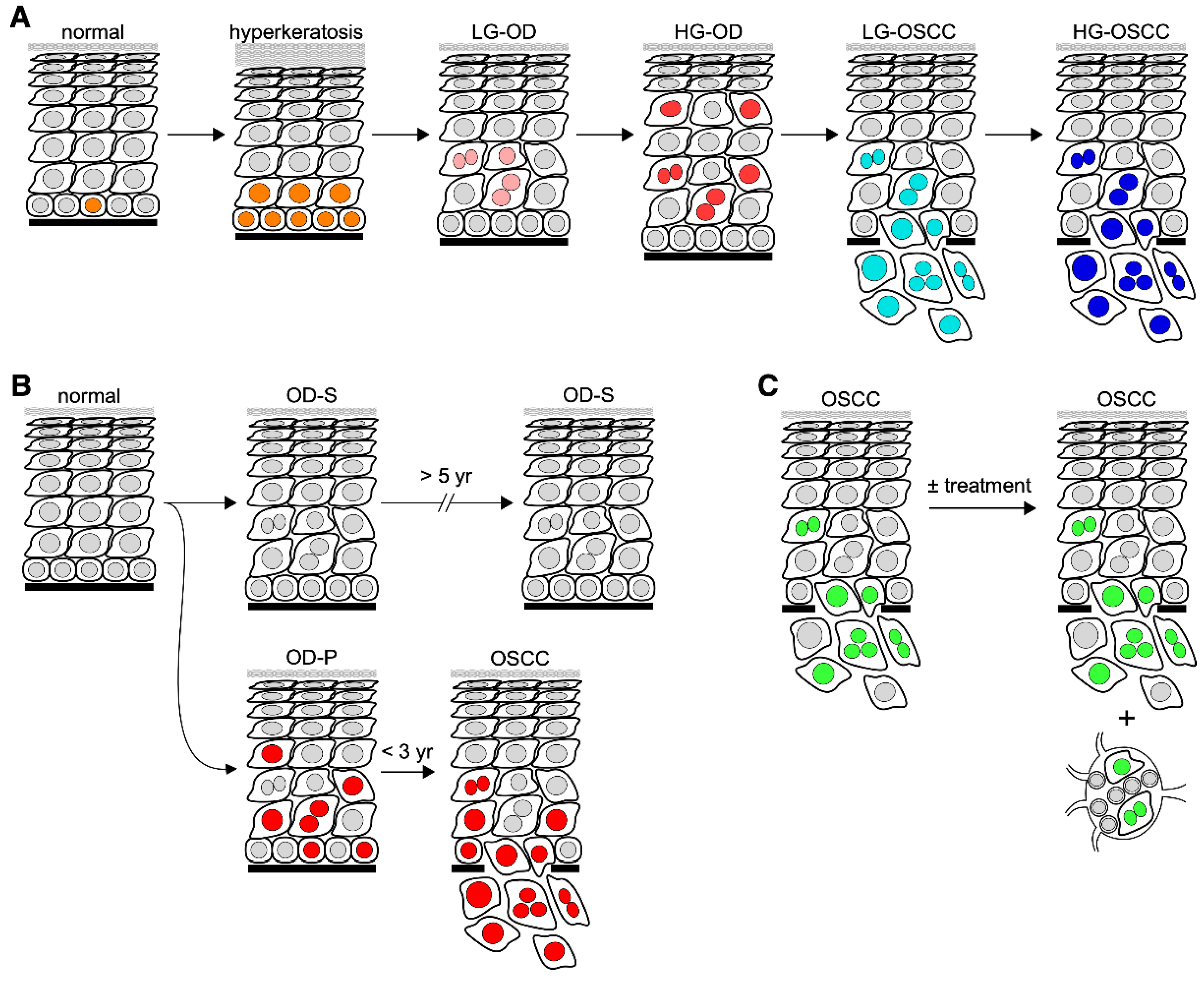
4. Predicting the Cancer Risk of OPLs by Quantitative Pathology
5. Cancer-Risk-Predictive Protein Markers for OPLs
5.1. Stem Cell Self-Renewal Factors
5.2. Tumor Suppressors
5.3. Others
| Reference | Biomarker | Conclusions | Functions | Tissue | F/U (Years) | Strength |
|---|---|---|---|---|---|---|
| Zhang et al. [21] | COX-2, c-Met, β-catenin, CA9, PDPN, Ki-67, p16, p53, IMP3, c-Jun | Expression of all markers potentially risk-predictive. Significant differences in positive expression between groups was observed. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 11.3 (median) | 3.04–29.00 (HR) |
| Zhang et al. (2017) [22] | Axin2, Snail | Elevated expression of Snail and Axin2 significantly correlate to risk of malignant transformation. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 10.8 (median) | 4.41, 7.47 (HR) |
| Sakata et al. [24] | SMAD4 | Low expression combined with elevated lymphocyte infiltration indicative of malignant risk. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | Unknown | 2.63 (HR) |
| Ding et al. [25] | Notch1 | Expression significantly associated with dysplasia severity and OSCC development. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 6.18 (median) | 3.4 (HR) |
| Crawford et al. [36] | Nucleostemin | NS upregulation may be an early event in malignant transformation of low-grade dysplasia. | Stem Cell Self-Renewal | Oral Dysplasia (P/NP) | 2–3 (NP), 7–14 (P) | p = 0.02–0.05 |
| de Vicente et al. [38] | SOX2 | SOX2 is an independent predictor of cancer risk in OL. | Stem Cell Self-Renewal | Oral Leukoplakia (T/N) | 6.25 (median) | 3.0–5.83 (HR) |
| Habiba et al. [39] | ALDH1, PDPN | Both markers can be used for determining risk of malignant transformation in OL. | Stem Cell Self-Renewal | LG & HG Oral Dysplasia (T/N) | 2.08 (median) | 2.91–3.64 (HR) |
| de Vicente et al. [40] | NANOG | Positive NANOG expression associated with progression to oral cancer-positive expression of both markers demonstrated higher risk. | Stem Cell Self-Renewal | LG & HG Oral Dysplasia (T/N) | 5.08 (median) | 2.01 (HR) |
| Cruz et al. (1998) [42] | p53 | p53 expression pattern has prognostic potential for pre-malignant lesions. | Tumor Suppressor | LG & HG Oral Dysplasia (T/N) | 3 (median) | p = 0.002 |
| Cruz et al. (2002) [43] | P53 | Suprabasal p53 expression is indicative of malignant transformation. | Tumor Suppressor | PMOL (T/N) | 5.0 (mean) | 29–33% Sensitivity,83–100% Specificity |
| Wu et al. [44] | p16 | p16 may predict malignant transformation of OL. | Tumor Suppressor | Oral Leukoplakia (T/N) | Unknown | 3.54 (OR) |
| Baran et al. [47] | MAGE-A | MAGE-A expression can be a reliable predictor of malignant transformation in progressing leukoplakia. | Melanoma Associated Antigen | Oral & Laryngeal Leukoplakia (T/N) | 5 | 96.5% Specificity, 58.2% Sensitivity |
| Ries et al. [46] | MAGE-A | Positive expression in oral leukoplakia is significantly correlated to malignant transformation. | Melanoma Associated Antigen | Oral Leukoplakia (T/N) | 5 | p = 0.0001 |
| Wu et al. [48] | TGM3 | Suggests TGM3 takes part in malignant transformation and may predict progression. | Tumor Suppressor | Oral Leukoplakia (T/N) | 4.75 (T), 7.92 (N) (median) | 5.55 (HR) |
| Kaur et al. [49] | S100A7 | Overexpression demonstrates association with risk of transformation, with cytoplasmic overexpression being most significant. | Cell Cycle & Differentiation | Oral Leukoplakia (T/N) | 3.04 (median) | 2.36 (HR) |
| de Vicente et al. [50] | Cortactin, FAK | Pre-malignant oral lesions with co-expression of both markers demonstrate high risk of OSCC development. | Tumor Progression & Metastasis | Oral dysplasia- leukoplakia, erythroplakia (T/N) | 5 (minimum) | 6.30 (HR) |
| Saintigny et al. [51] | MET | Overexpression in oral leukoplakia was associated with malignant transformation. | Cell Proliferation | Oral Leukoplakia (T/N) | 6.08 (median) | 3.84 (HR) |
| Weber et al. [52] | CD68, CD163 | Elevated CD68 and CD163 significantly associated with malignant transformation. Suggests the value of macrophages as potential predictive markers. | Macrophage Infiltration | Oral dysplasia- mild, moderate, severe (T/N) | 5 (full) | 55.6–72% Sensitivity, 72.7–73.5% Specificity |
| Ries et al. [53] | PD1, PDL1 | Overexpression of both markers may be indicative of cancer risk. | Cell Proliferation | Oral Leukoplakia (T/N) | 5 (minimum) | 50–76.5% Sensitivity, 72.3–93.6% Specificity |
6. Cancer-Risk-Predictive Genetic Markers for OPLs
| References | Biomarker | Conclusions | Tissue | F/U (Years) | Strength |
|---|---|---|---|---|---|
| Mao et al. [55] | LOH at 3p, 9p | Losses in these regions are frequent early genetic events in OPLs. Cancer developed more quickly in groups with LOH in regions 3p and/or 9p than those without LOH. | Oral Leukoplakia (T/N) | 5.25 (median) | p = 0.039 |
| Rosin et al. [56] | LOH at 3p, 9p, 4q, 8p, 11q, 17p | LOH at 3p and/or 9p exhibit increased risk of cancer development. Risk significantly increased in patients with losses on additional regions. | Hyperplasia, mild and moderate oral dysplasia (P/NP) | 0.5 (minimum) | 3.75, 33.4 (RR) |
| Zhang et al. (2012) [57] | LOH at 3p, 9p, 4q, 17p | LOH at 3p and/or 9p indicates risk for malignant transformation. Risk further increases when combined with LOH at additional sites. | Oral Dysplasia (P/NP) | 3.7 and 3.6 (median) | 22.6 (HR) |
| Graveland et al. [59] | LOH at 9p and p53 mutation | TP53 mutation correlated with losses at 17p and 9p. Losses at 9p significantly associated with risk of transformation. | Oral Leukoplakia (P/NP) | 1.5 (median) | p = 0.014 |
7. Cancer-Risk-Predictive Epigenetic Markers for OPLs
7.1. DNA Methylation
| Reference | Biomarker | Conclusions | Tissue | F/U (Years) | Strength |
|---|---|---|---|---|---|
| Hall et al. [64] | p16 Promoter Methylation | Presence of Promoter methylation of p16 is a potential predictor of malignant transformation. | Oral Leukoplakia, erythroplakia (T/N) | 3 (minimum) | p = 0.002 |
| Cao et al. [67] | p16 Methylation | Higher rate of progression to cancer in patients with positive p16 methylation. | Mild/Moderate Oral Dysplasia (T/N) | 3.82 (median) | 3.7 (OR) |
| Liu et al. (2015) [65] | p16 Hypermethylation | Positive p16 methylation significantly increased in transformed cases. Presents p16 Methylation as a definitive marker for determining malignant risk. | Mild/Moderate Oral Dysplasia (T/N) | 3.42 (median) | p = 0.006 |
| Liu et al. (2018) [66] | p16 Methylation | Progression to malignant transformation was significantly increased for patients with positive p16 methylation. | Oral leukoplakia, lichen planus, or discoid lupus erythematosus (T/N) | 3.42 (median) | 2.67 (OR) |
| Foy et al. [72] | AGTR1, FOXI2, PENK Promoter Methylation; LINE1 Hypomethylation | Patients with a high methylation index experienced worse Oral-Cancer-Free Survival. CpG Island Methylation may be an early event in OSCC. | Oral Premalignant Lesions (Unspecified) (T/N) | 7.64 (N), 2.15 (T) (median) | p = 0.003 |
| Philipone et al. [75] | miRNAs: 208b-3p, 204-5p, 129-2-3p and 3065-5p | Expression of the indicated miRNAs along with age and histology proved to be indicative of at-risk lesions. | Oral Leukoplakia (T/N) | 5 (minimum) | p < 0.05 |
| Cervigne et al. [76] | miR-21, miR-181b, and miR-345, and miR-416a | Overexpression of these miRs in OSCC and progressive tissue suggest their involvement in malignant transformation. | Oral Leukoplakia (T/N) | 5–9 | p < 0.001 |
| Hung et al. [77] | miR-31 | A significant difference in miR-31 expression was observed between transformed and non-transformed leukoplakia. | Oral Leukoplakia (T/N) | 2.25 (mean) | 8.34 (HR) |
| Harrandah et al. [78] | miR-375 | Downregulation of miR-375 is associated with malignant transformation in OPLs | Dysplasia, CIS, and verrucous hyperplasia/verrucopapillary hyperkeratosis (T/N) | 0.5 (minimum) | p < 0.0001 |
7.2. miRNA and Histone Modification
8. Challenges Facing the Discovery of Predictive Biomarkers
9. Future Research Trends
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Neville, B.W.; Damm, D.D.; Allen, C.M.; Bouquot, J.E. Squamous cell carcinoma. In Oral and Maxillofacial Pathology; Saunders Elsevier: St. Louis, MO, USA, 2016; pp. 409–421. [Google Scholar]
- Chaturvedi, A.K.; Anderson, W.F.; Lortet-Tieulent, J.; Curado, M.P.; Ferlay, J.; Franceschi, S.; Rosenberg, P.S.; Bray, F.; Gillison, M.L. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J. Clin. Oncol. 2013, 31, 4550–4559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, K.; Barrowman, R.; McCullough, M.; Iseli, T.; Wiesenfeld, D. Non-smoking non-drinking elderly females: A clinically distinct subgroup of oral squamous cell carcinoma patients. Int. J. Oral Maxillofac. Surg. 2013, 42, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Katzel, J.A.; Merchant, M.; Chaturvedi, A.K.; Silverberg, M.J. Contribution of demographic and behavioral factors on the changing incidence rates of oropharyngeal and oral cavity cancers in northern California. Cancer Epidemiol. Biomark. Prev. 2015, 24, 978–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conway, D.I.; Brenner, D.R.; McMahon, A.D.; Macpherson, L.M.; Agudo, A.; Ahrens, W.; Bosetti, C.; Brenner, H.; Castellsague, X.; Chen, C.; et al. Estimating and explaining the effect of education and income on head and neck cancer risk: INHANCE consortium pooled analysis of 31 case-control studies from 27 countries. Int. J. Cancer 2015, 136, 1125–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reibel, J.; Gale, N.; Hille, J.; Hunt, J.L.; Lingen, M.; Muller, S.; Sloan, P.; Tilakarante, W.M.; Westra, W.H.; Williams, M.D.; et al. Oral potentially malignant disorders and oral epithelial dysplasia. WHO/IARC Classif. Head Neck Tumours 2017, 4, 112–115. [Google Scholar]
- Odell, E.; Kujan, O.; Warnakulasuriya, S.; Sloan, P. Oral epithelial dysplasia: Recognition, grading and clinical significance. Oral Dis. 2021, 27, 1947–1976. [Google Scholar] [CrossRef]
- Garcia-Gimenez, J.L.; Seco-Cervera, M.; Tollefsbol, T.O.; Roma-Mateo, C.; Peiro-Chova, L.; Lapunzina, P.; Pallardo, F.V. Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit. Rev. Clin. Lab. Sci. 2017, 54, 529–550. [Google Scholar] [CrossRef] [PubMed]
- Santosh, A.B.; Jones, T.; Harvey, J. A review on oral cancer biomarkers: Understanding the past and learning from the present. J. Cancer Res. Ther. 2016, 12, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Abati, S.; Bramati, C.; Bondi, S.; Lissoni, A.; Trimarchi, M. Oral cancer and precancer: A narrative review on the relevance of early diagnosis. Int. J. Environ. Res. Public Health 2020, 17, 9160. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, R.; Toyoshima, T.; Tanaka, H.; Kawano, S.; Kiyosue, T.; Matsubara, R.; Goto, Y.; Hirano, M.; Oobu, K.; Nakamura, S. Association of cytokeratin 17 expression with differentiation in oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2012, 138, 1299–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, A.C. Leukoplakia (Leukokeratosis; erythroleukoplakia). In Oral and Maxillofacial Pathology, 4th ed.; Neville, B.W., Damm, D.D., Allen, C.M., Chi, A.C., Eds.; Saunders Elsevier: St. Louis, MO, USA, 2016; pp. 355–363. [Google Scholar]
- Novelli, G.; Ciccacci, C.; Borgiani, P.; Papaluca Amati, M.; Abadie, E. Genetic tests and genomic biomarkers: Regulation, qualification and validation. Clin. Cases Min. Bone Metab. 2008, 5, 149–154. [Google Scholar]
- Dumitrescu, R.G. Early epigenetic markers for precision medicine. Methods Mol. Biol. 2018, 1856, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Reinberg, D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011, 21, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, S.; Feldman, M.D.; Abels, E.; Ashfaq, R.; Beltaifa, S.; Cacciabeve, N.G.; Cathro, H.P.; Cheng, L.; Cooper, K.; Dickey, G.E.; et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: A multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am. J. Surg. Pathol. 2018, 42, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Guillaud, M.; Zhang, L.; Poh, C.; Rosin, M.P.; MacAulay, C. Potential use of quantitative tissue phenotype to predict malignant risk for oral premalignant lesions. Cancer Res. 2008, 68, 3099–3107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos, L.L.; Trufelli, D.C.; de Matos, M.G.; da Silva Pinhal, M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark. Insights 2010, 5, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, R.Y. A molecular view of stem cell and cancer cell self-renewal. Int. J. Biochem. Cell Biol. 2004, 36, 684–694. [Google Scholar] [CrossRef]
- Zhang, X.; Kim, K.Y.; Zheng, Z.; Bazarsad, S.; Kim, J. Nomogram for risk prediction of malignant transformation in oral leukoplakia patients using combined biomarkers. Oral Oncol. 2017, 72, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kim, K.Y.; Zheng, Z.; Kim, H.S.; Cha, I.H.; Yook, J.I. Snail and Axin2 expression predict the malignant transformation of oral leukoplakia. Oral Oncol. 2017, 73, 48–55. [Google Scholar] [CrossRef]
- Du, X.; Li, Q.; Yang, L.; Liu, L.; Cao, Q.; Li, Q. SMAD4 activates Wnt signaling pathway to inhibit granulosa cell apoptosis. Cell Death Dis. 2020, 11, 373. [Google Scholar] [CrossRef] [PubMed]
- Sakata, J.; Yoshida, R.; Matsuoka, Y.; Nagata, M.; Hirosue, A.; Kawahara, K.; Nakamura, T.; Nakamoto, M.; Hirayama, M.; Takahashi, N.; et al. Predictive value of the combination of SMAD4 expression and lymphocyte infiltration in malignant transformation of oral leukoplakia. Cancer Med. 2017, 6, 730–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.; Zheng, Y.; Wang, Z.; Zhang, W.; Dong, Y.; Chen, W.; Li, J.; Chu, W.; Zhang, W.; Zhong, Y.; et al. Expression and oncogenic properties of membranous Notch1 in oral leukoplakia and oral squamous cell carcinoma. Oncol. Rep. 2018, 39, 2584–2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, R.Y. Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too? Cell. Mol. Life Sci. 2016, 73, 1803–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Liu, W.; Shi, L.; Xiao, X.; Wu, W.; Wu, L.; Zhou, Z. Expression of DNA doublestrand repair proteins in oral leukoplakia and the risk of malignant transformation. Oncol. Lett. 2018, 15, 9827–9835. [Google Scholar] [CrossRef]
- Tsai, R.Y.; McKay, R.D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002, 16, 2991–3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, R.Y. Turning a new page on nucleostemin and self-renewal. J. Cell Sci. 2014, 127, 3885–3891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Lin, T.; Peng, G.; Hsu, J.K.; Lee, S.; Lin, S.-Y.; Tsai, R.Y. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 11415–11420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.; Ibrahim, W.; Peng, C.-Y.; Finegold, M.J.; Tsai, R.Y. A novel role of nucleostemin in maintaining the genome integrity of dividing hepatocytes during mouse liver development and regeneration. Hepatology 2013, 58, 2176–2187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.; Meng, L.; Wu, L.J.; Pederson, T.; Tsai, R.Y. Nucleostemin and GNL3L exercise distinct functions in genome protection and ribosome synthesis, respectively. J. Cell Sci. 2014, 127, 2302–2312. [Google Scholar]
- Lin, T.; Lin, T.C.; McGrail, D.J.; Bhupal, P.K.; Ku, Y.H.; Zhang, W.; Meng, L.; Lin, S.Y.; Peng, G.; Tsai, R.Y.L. Nucleostemin reveals a dichotomous nature of genome maintenance in mammary tumor progression. Oncogene 2019, 38, 3919–3931. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; McGrail, D.J.; Bhupal, P.K.; Zhang, W.; Lin, K.Y.; Ku, Y.H.; Lin, T.; Wu, H.; Tsai, K.C.; Li, K.; et al. Nucleostemin modulates outcomes of hepatocellular carcinoma via a tumor adaptive mechanism to genomic stress. Mol. Cancer Res. 2020, 18, 723–734. [Google Scholar] [CrossRef] [Green Version]
- Yasumoto, H.; Meng, L.; Lin, T.; Zhu, Q.; Tsai, R.Y. GNL3L inhibits activity of estrogen-related receptor-gamma by competing for coactivator binding. J. Cell Sci. 2007, 120, 2532–2543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, M.; Liu, X.; Cheng, Y.L.; Tsai, R.Y. Nucleostemin upregulation and STAT3 activation as early events in oral epithelial dysplasia progression to squamous cell carcinoma. Neoplasia 2021, 23, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.C.; Kang, H.J.; Kim, Y.S.; Choi, E.C. All-trans-retinoic acid inhibits growth of head and neck cancer stem cells by suppression of Wnt/beta-catenin pathway. Eur. J. Cancer 2012, 48, 3310–3318. [Google Scholar] [CrossRef]
- de Vicente, J.C.; Donate-Perez Del Molino, P.; Rodrigo, J.P.; Allonca, E.; Hermida-Prado, F.; Granda-Diaz, R.; Rodriguez Santamarta, T.; Garcia-Pedrero, J.M. SOX2 expression is an independent predictor of oral cancer progression. J. Clin. Med. 2019, 8, 1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habiba, U.; Hida, K.; Kitamura, T.; Matsuda, A.Y.; Higashino, F.; Ito, Y.M.; Ohiro, Y.; Totsuka, Y.; Shindoh, M. ALDH1 and podoplanin expression patterns predict the risk of malignant transformation in oral leukoplakia. Oncol. Lett. 2017, 13, 321–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vicente, J.C.; Rodriguez-Santamarta, T.; Rodrigo, J.P.; Allonca, E.; Vallina, A.; Singhania, A.; Donate-Perez Del Molino, P.; Garcia-Pedrero, J.M. The emerging role of NANOG as an early cancer risk biomarker in patients with oral potentially malignant disorders. J. Clin. Med. 2019, 8, 1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murti, P.R.; Warnakulasuriya, K.A.; Johnson, N.W.; Bhonsle, R.B.; Gupta, P.C.; Daftary, D.K.; Mehta, F.S. p53 expression in oral precancer as a marker for malignant potential. J. Oral Pathol. Med. 1998, 27, 191–196. [Google Scholar] [CrossRef]
- Cruz, I.B.; Snijders, P.J.; Meijer, C.J.; Braakhuis, B.J.; Snow, G.B.; Walboomers, J.M.; van der Waal, I. p53 expression above the basal cell layer in oral mucosa is an early event of malignant transformation and has predictive value for developing oral squamous cell carcinoma. J. Pathol. 1998, 184, 360–368. [Google Scholar] [CrossRef]
- Cruz, I.; Napier, S.S.; van der Waal, I.; Snijders, P.J.; Walboomers, J.M.; Lamey, P.J.; Cowan, C.G.; Gregg, T.A.; Maxwell, P.; Meijer, C.J. Suprabasal p53 immunoexpression is strongly associated with high grade dysplasia and risk for malignant transformation in potentially malignant oral lesions from Northern Ireland. J. Clin. Pathol. 2002, 55, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, Z.; Zhou, Z. Role of the human papillomavirus in malignant transformation of oral leukoplakia distinct from oropharyngeal squamous cell carcinoma: A study of 76 patients with internal-control specimens. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Chen, H.S. Biological functions of melanoma-associated antigens. World J. Gastroenterol. 2004, 10, 1849–1853. [Google Scholar] [CrossRef]
- Ries, J.; Agaimy, A.; Vairaktaris, E.; Gorecki, P.; Neukam, F.W.; Strassburg, L.H.; Nkenke, E. Detection of MAGE-A expression predicts malignant transformation of oral leukoplakia. Cancer Investig. 2012, 30, 495–502. [Google Scholar] [CrossRef]
- Baran, C.A.; Agaimy, A.; Wehrhan, F.; Weber, M.; Hille, V.; Brunner, K.; Wickenhauser, C.; Siebolts, U.; Nkenke, E.; Kesting, M.; et al. MAGE-A expression in oral and laryngeal leukoplakia predicts malignant transformation. Mod. Pathol. 2019, 32, 1068–1081. [Google Scholar] [CrossRef]
- Wu, X.; Wang, R.; Jiao, J.; Li, S.; Yu, J.; Yin, Z.; Zhou, L.; Gong, Z. Transglutaminase 3 contributes to malignant transformation of oral leukoplakia to cancer. Int. J. Biochem. Cell Biol. 2018, 104, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Matta, A.; Kak, I.; Srivastava, G.; Assi, J.; Leong, I.; Witterick, I.; Colgan, T.J.; Macmillan, C.; Siu, K.W.; et al. S100A7 overexpression is a predictive marker for high risk of malignant transformation in oral dysplasia. Int. J. Cancer 2014, 134, 1379–1388. [Google Scholar] [CrossRef]
- de Vicente, J.C.; Rodrigo, J.P.; Rodriguez-Santamarta, T.; Lequerica-Fernandez, P.; Allonca, E.; Garcia-Pedrero, J.M. Cortactin and focal adhesion kinase as predictors of cancer risk in patients with premalignant oral epithelial lesions. Oral Oncol. 2012, 48, 641–646. [Google Scholar] [CrossRef]
- Saintigny, P.; William, W.N., Jr.; Foy, J.P.; Papadimitrakopoulou, V.; Lang, W.; Zhang, L.; Fan, Y.H.; Feng, L.; Kim, E.S.; El-Naggar, A.K.; et al. Met receptor tyrosine kinase and chemoprevention of oral cancer. J. Natl. Cancer Inst. 2018, 110, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Wehrhan, F.; Baran, C.; Agaimy, A.; Buttner-Herold, M.; Ozturk, H.; Neubauer, K.; Wickenhauser, C.; Kesting, M.; Ries, J. Malignant transformation of oral leukoplakia is associated with macrophage polarization. J. Transl. Med. 2020, 18, 11. [Google Scholar] [CrossRef] [Green Version]
- Ries, J.; Agaimy, A.; Wehrhan, F.; Baran, C.; Bolze, S.; Danzer, E.; Frey, S.; Jantsch, J.; Most, T.; Buttner-Herold, M.; et al. Importance of the PD-1/PD-L1 axis for malignant transformation and risk assessment of oral leukoplakia. Biomedicines 2021, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Seidal, T.; Balaton, A.J.; Battifora, H. Interpretation and quantification of immunostains. Am. J. Surg. Pathol. 2001, 25, 1204–1207. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Lee, J.S.; Fan, Y.H.; Ro, J.Y.; Batsakis, J.G.; Lippman, S.; Hittelman, W.; Hong, W.K. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat. Med. 1996, 2, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Rosin, M.P.; Cheng, X.; Poh, C.; Lam, W.L.; Huang, Y.; Lovas, J.; Berean, K.; Epstein, J.B.; Priddy, R.; Le, N.D.; et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin. Cancer Res. 2000, 6, 357–362. [Google Scholar] [PubMed]
- Zhang, L.; Poh, C.F.; Williams, M.; Laronde, D.M.; Berean, K.; Gardner, P.J.; Jiang, H.; Wu, L.; Lee, J.J.; Rosin, M.P. Loss of heterozygosity (LOH) profiles—Validated risk predictors for progression to oral cancer. Cancer Prev. Res. 2012, 5, 1081–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.J.; Hong, W.K.; Hittelman, W.N.; Mao, L.; Lotan, R.; Shin, D.M.; Benner, S.E.; Xu, X.C.; Lee, J.S.; Papadimitrakopoulou, V.M.; et al. Predicting cancer development in oral leukoplakia: Ten years of translational research. Clin. Cancer Res. 2000, 6, 1702–1710. [Google Scholar] [PubMed]
- Graveland, A.P.; Bremmer, J.F.; de Maaker, M.; Brink, A.; Cobussen, P.; Zwart, M.; Braakhuis, B.J.; Bloemena, E.; van der Waal, I.; Leemans, C.R.; et al. Molecular screening of oral precancer. Oral Oncol. 2013, 49, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Siwko, S.; Tsai, R.Y.L. Sex and race-related DNA methylation changes in hepatocellular carcinoma. Int. J. Mol. Sci. 2021, 22, 3820. [Google Scholar] [CrossRef]
- Ghosh, A.; Ghosh, S.; Maiti, G.P.; Sabbir, M.G.; Alam, N.; Sikdar, N.; Roy, B.; Roychoudhury, S.; Panda, C.K. SH3GL2 and CDKN2A/2B loci are independently altered in early dysplastic lesions of head and neck: Correlation with HPV infection and tobacco habit. J. Pathol. 2009, 217, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Ghosh, S.; Maiti, G.P.; Sabbir, M.G.; Zabarovsky, E.R.; Roy, A.; Roychoudhury, S.; Panda, C.K. Frequent alterations of the candidate genes hMLH1, ITGA9 and RBSP3 in early dysplastic lesions of head and neck: Clinical and prognostic significance. Cancer Sci. 2010, 101, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Kresty, L.A.; Mallery, S.R.; Knobloch, T.J.; Song, H.; Lloyd, M.; Casto, B.C.; Weghorst, C.M. Alterations of p16(INK4a) and p14(ARF) in patients with severe oral epithelial dysplasia. Cancer Res. 2002, 62, 5295–5300. [Google Scholar] [PubMed]
- Hall, G.L.; Shaw, R.J.; Field, E.A.; Rogers, S.N.; Sutton, D.N.; Woolgar, J.A.; Lowe, D.; Liloglou, T.; Field, J.K.; Risk, J.M. p16 Promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2174–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Liu, X.W.; Dong, G.; Zhou, J.; Liu, Y.; Gao, Y.; Liu, X.Y.; Gu, L.; Sun, Z.; Deng, D. P16 methylation as an early predictor for cancer development from oral epithelial dysplasia: A double-blind multicentre prospective study. EBioMedicine 2015, 2, 432–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Liu, Z.; Liu, X.W.; Xu, S.; Wang, L.; Liu, Y.; Zhou, J.; Gu, L.; Gao, Y.; Liu, X.Y.; et al. A similar effect of P16 hydroxymethylation and true-methylation on the prediction of malignant transformation of oral epithelial dysplasia: Observation from a prospective study. BMC Cancer 2018, 18, 918. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhou, J.; Gao, Y.; Gu, L.; Meng, H.; Liu, H.; Deng, D. Methylation of p16 CpG island associated with malignant progression of oral epithelial dysplasia: A prospective cohort study. Clin. Cancer Res. 2009, 15, 5178–5183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towle, R.; Truong, D.; Hogg, K.; Robinson, W.P.; Poh, C.F.; Garnis, C. Global analysis of DNA methylation changes during progression of oral cancer. Oral Oncol. 2013, 49, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Khongsti, S.; Lamare, F.A.; Shunyu, N.B.; Ghosh, S.; Maitra, A.; Ghosh, S. Whole genome DNA methylation profiling of oral cancer in ethnic population of Meghalaya, North East India reveals novel genes. Genomics 2018, 110, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Ghosh, S.; Maitra, A.; Biswas, N.K.; Panda, C.K.; Roy, B.; Sarin, R.; Majumder, P.P. Epigenomic dysregulation-mediated alterations of key biological pathways and tumor immune evasion are hallmarks of gingivo-buccal oral cancer. Clin. Epigenet. 2019, 11, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planello, A.C.; Singhania, R.; Kron, K.J.; Bailey, S.D.; Roulois, D.; Lupien, M.; Line, S.R.; de Souza, A.P.; De Carvalho, D.D. Pre-neoplastic epigenetic disruption of transcriptional enhancers in chronic inflammation. Oncotarget 2016, 7, 15772–15786. [Google Scholar] [CrossRef]
- Foy, J.P.; Pickering, C.R.; Papadimitrakopoulou, V.A.; Jelinek, J.; Lin, S.H.; William, W.N., Jr.; Frederick, M.J.; Wang, J.; Lang, W.; Feng, L.; et al. New DNA methylation markers and global DNA hypomethylation are associated with oral cancer development. Cancer Prev. Res. 2015, 8, 1027–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.; Tian, G.; Gao, J. Construction of prognostic risk prediction model of oral squamous cell carcinoma based on co-methylated genes. Int. J. Mol. Med. 2019, 44, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Preston, R.; Soudry, E.; Acero, J.; Orera, M.; Moreno-Lopez, L.; Macia-Colon, G.; Jaffe, A.; Berdasco, M.; Ili-Gangas, C.; Brebi-Mieville, P.; et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev. Res. 2011, 4, 1061–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philipone, E.; Yoon, A.J.; Wang, S.; Shen, J.; Ko, Y.C.; Sink, J.M.; Rockafellow, A.; Shammay, N.A.; Santella, R.M. MicroRNAs-208b-3p, 204–205p, 129-2-3p and 3065-5p as predictive markers of oral leukoplakia that progress to cancer. Am. J. Cancer Res. 2016, 6, 1537–1546. [Google Scholar] [PubMed]
- Cervigne, N.K.; Reis, P.P.; Machado, J.; Sadikovic, B.; Bradley, G.; Galloni, N.N.; Pintilie, M.; Jurisica, I.; Perez-Ordonez, B.; Gilbert, R.; et al. Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum. Mol. Genet. 2009, 18, 4818–4829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, K.F.; Liu, C.J.; Chiu, P.C.; Lin, J.S.; Chang, K.W.; Shih, W.Y.; Kao, S.Y.; Tu, H.F. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016, 53, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Harrandah, A.M.; Fitzpatrick, S.G.; Smith, M.H.; Wang, D.; Cohen, D.M.; Chan, E.K. MicroRNA-375 as a biomarker for malignant transformation in oral lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016, 122, 743–752.e1. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; St John, M.A.; Zhou, X.; Kim, Y.; Sinha, U.; Jordan, R.C.; Eisele, D.; Abemayor, E.; Elashoff, D.; Park, N.H.; et al. Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, H.; Onsongo, G.; Popko, J.; de Jong, E.P.; Cao, J.; Carlis, J.V.; Griffin, R.J.; Rhodus, N.L.; Griffin, T.J. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol. Cell. Proteom. 2008, 7, 486–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagler, R.; Bahar, G.; Shpitzer, T.; Feinmesser, R. Concomitant analysis of salivary tumor markers--a new diagnostic tool for oral cancer. Clin. Cancer Res. 2006, 12, 3979–3984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macaulay, C.; Poh, C.F.; Guillaud, M.; Michele Williams, P.; Laronde, D.M.; Zhang, L.; Rosin, M.P. High throughput image cytometry for detection of suspicious lesions in the oral cavity. J. Biomed. Opt. 2012, 17, 086004. [Google Scholar] [CrossRef] [Green Version]
- Parfenova, E.; Liu, K.Y.P.; Harrison, A.; MacAulay, C.; Guillaud, M.; Poh, C.F. An improved algorithm using a Health Canada-approved DNA-image cytometry system for non-invasive screening of high-grade oral lesions. J. Oral Pathol. Med. 2021, 50, 502–509. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crawford, M.; Johnson, E.H.; Liu, K.Y.P.; Poh, C.; Tsai, R.Y.L. On the Cutting Edge of Oral Cancer Prevention: Finding Risk-Predictive Markers in Precancerous Lesions by Longitudinal Studies. Cells 2022, 11, 1033. https://doi.org/10.3390/cells11061033
Crawford M, Johnson EH, Liu KYP, Poh C, Tsai RYL. On the Cutting Edge of Oral Cancer Prevention: Finding Risk-Predictive Markers in Precancerous Lesions by Longitudinal Studies. Cells. 2022; 11(6):1033. https://doi.org/10.3390/cells11061033
Chicago/Turabian StyleCrawford, Madeleine, Eliza H. Johnson, Kelly Y. P. Liu, Catherine Poh, and Robert Y. L. Tsai. 2022. "On the Cutting Edge of Oral Cancer Prevention: Finding Risk-Predictive Markers in Precancerous Lesions by Longitudinal Studies" Cells 11, no. 6: 1033. https://doi.org/10.3390/cells11061033
APA StyleCrawford, M., Johnson, E. H., Liu, K. Y. P., Poh, C., & Tsai, R. Y. L. (2022). On the Cutting Edge of Oral Cancer Prevention: Finding Risk-Predictive Markers in Precancerous Lesions by Longitudinal Studies. Cells, 11(6), 1033. https://doi.org/10.3390/cells11061033