Salvia miltiorrhiza Extract and Individual Synthesized Component Derivatives Induce Activating-Transcription-Factor-3-Mediated Anti-Obesity Effects and Attenuate Obesity-Induced Metabolic Disorder by Suppressing C/EBPα in High-Fat-Induced Obese Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Studies
2.2. Glucose and Insulin Tolerance Tests
2.3. Measurement of Biochemical Parameters
2.4. Histology, Adipocyte Size Measurement, and Adipocyte Number Estimation
2.5. TF Activation Profiling Analysis
2.6. Cell Culture
2.7. Oil Red O Staining
2.8. Real-Time Polymerase Chain Reaction
2.9. Chromatin Immunoprecipitation Assay
2.10. Statistical Analysis
3. Results
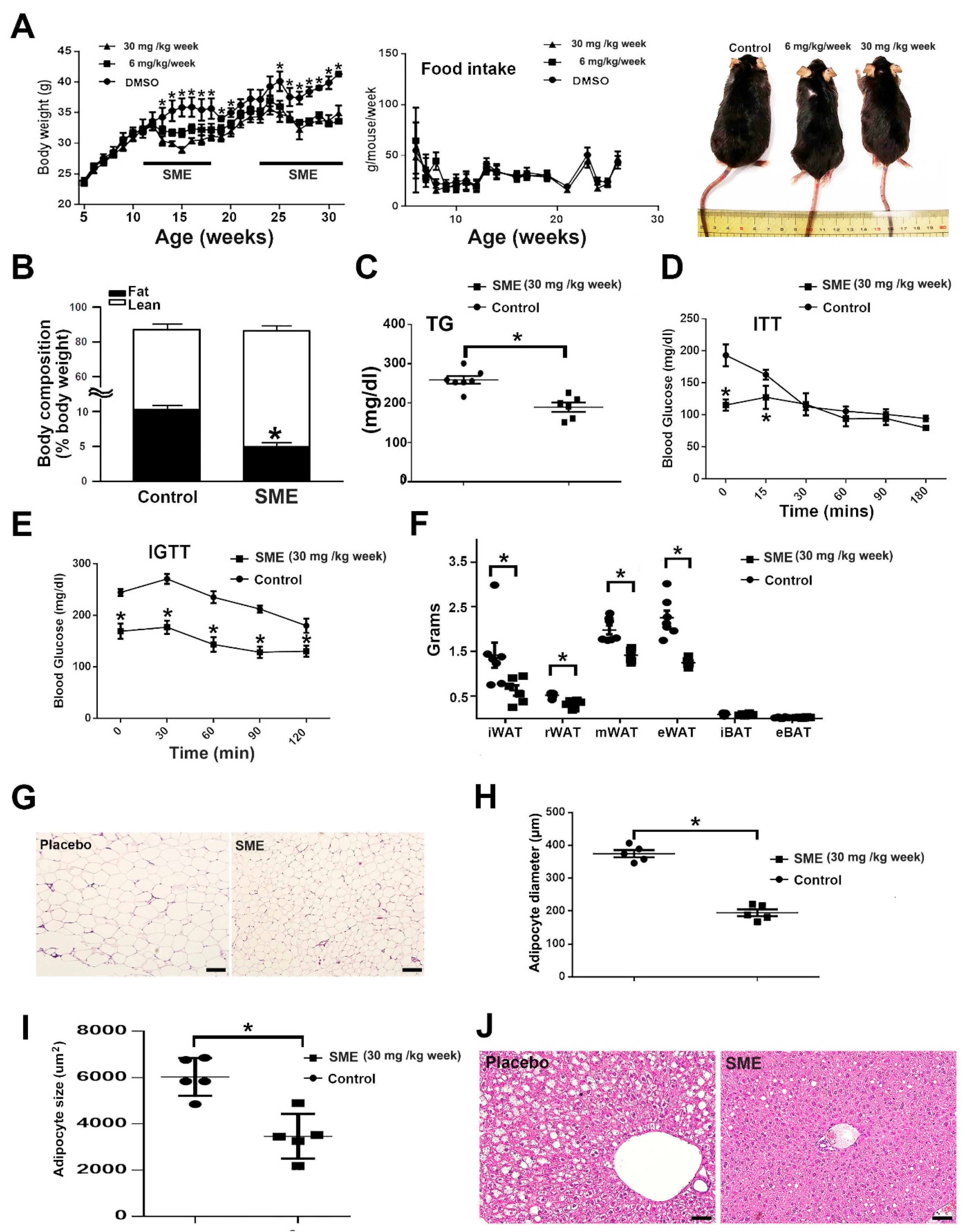
3.1. SME Treatment Ameliorates HFD-Induced Metabolic Dyshomeostasis
3.2. SME Treatment Inhibits Adipogenesis and Ameliorates Lipogenesis/Lipolysis Imbalance in HFD-Fed Mice
3.3. SME Treatment Changes the Expression of Numerous Transcriptional Factors in 3T3-L1 Adipocytes
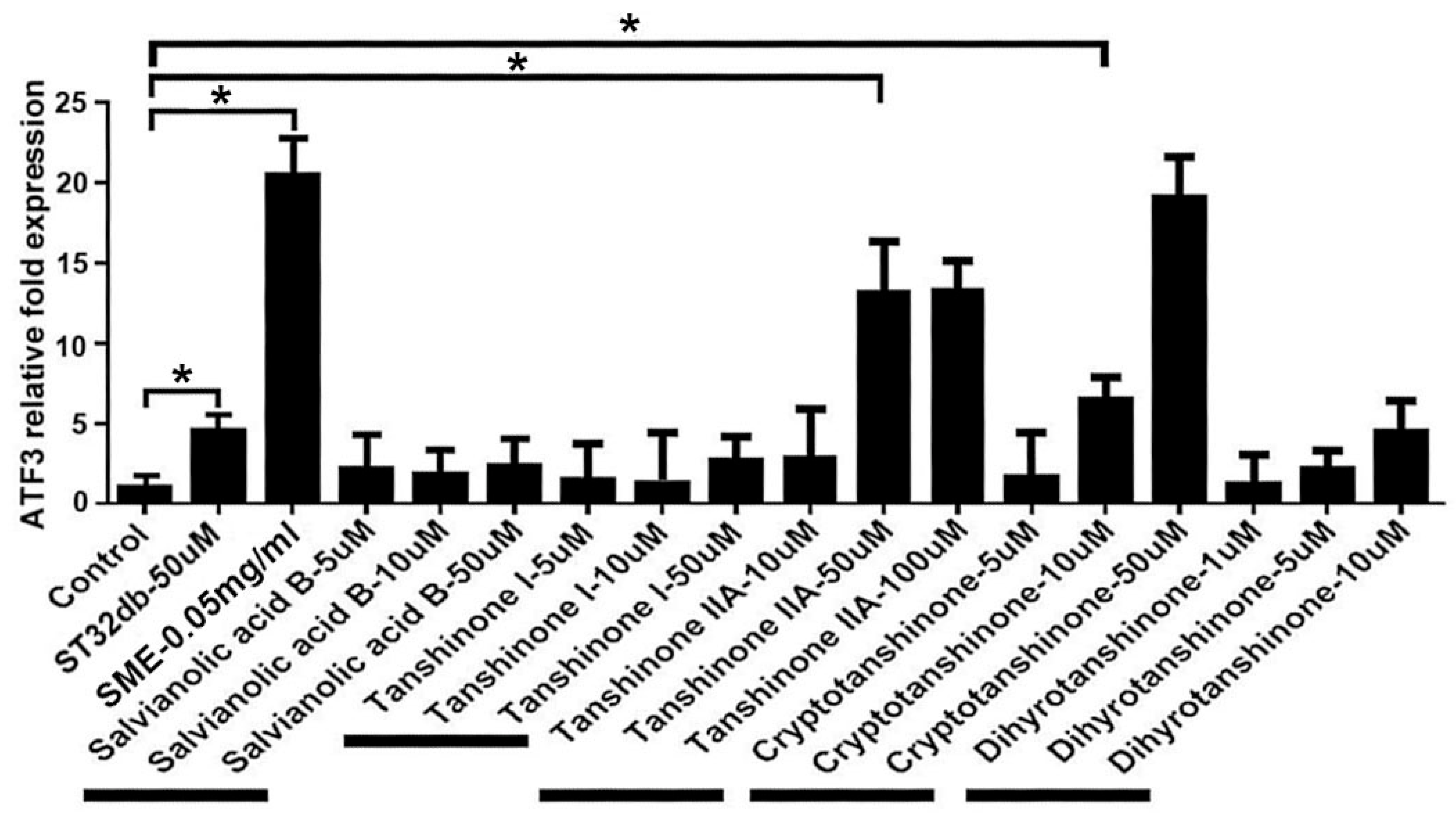
3.4. Synthesizing Individual Compounds as Candidate ATF3 Inducers
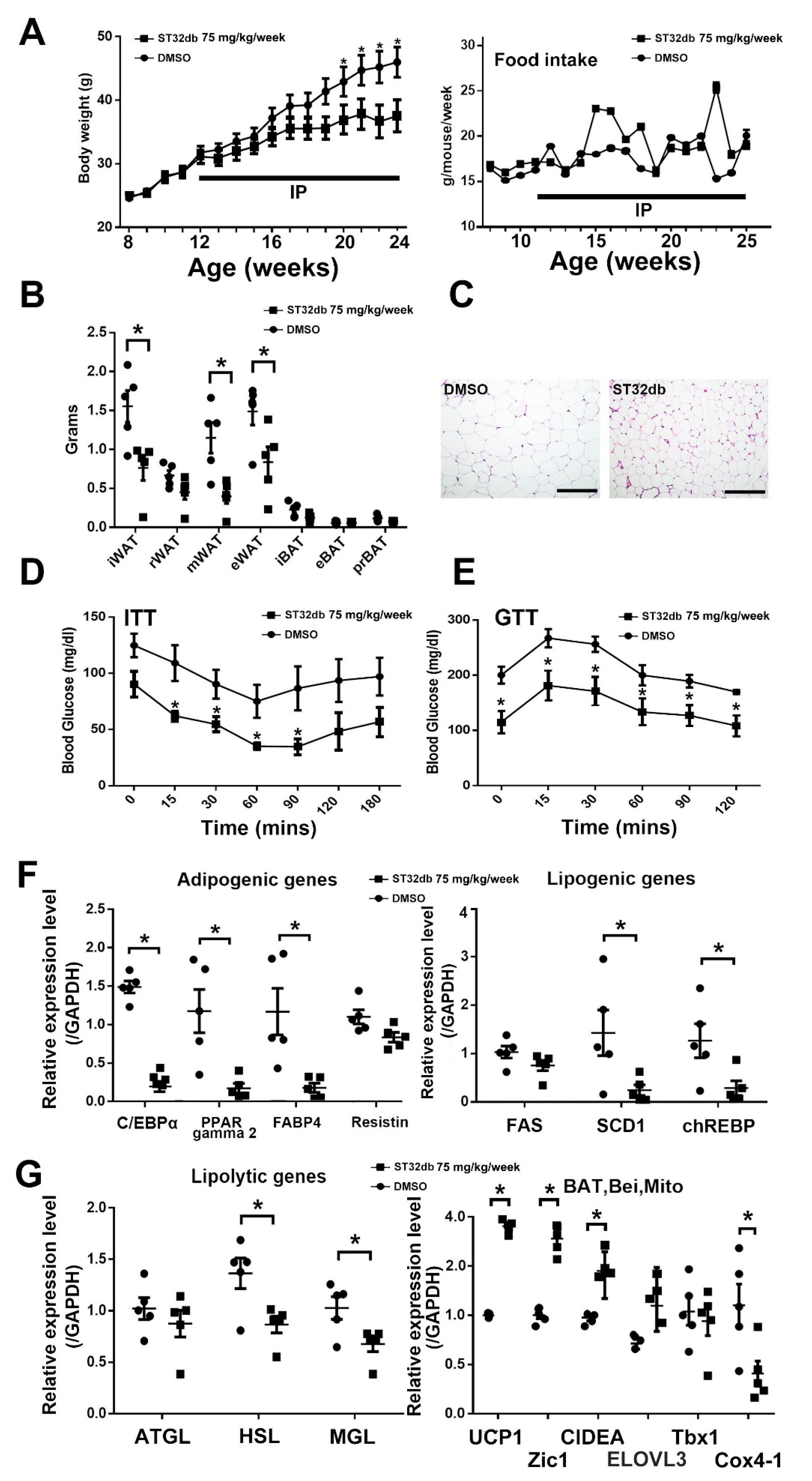
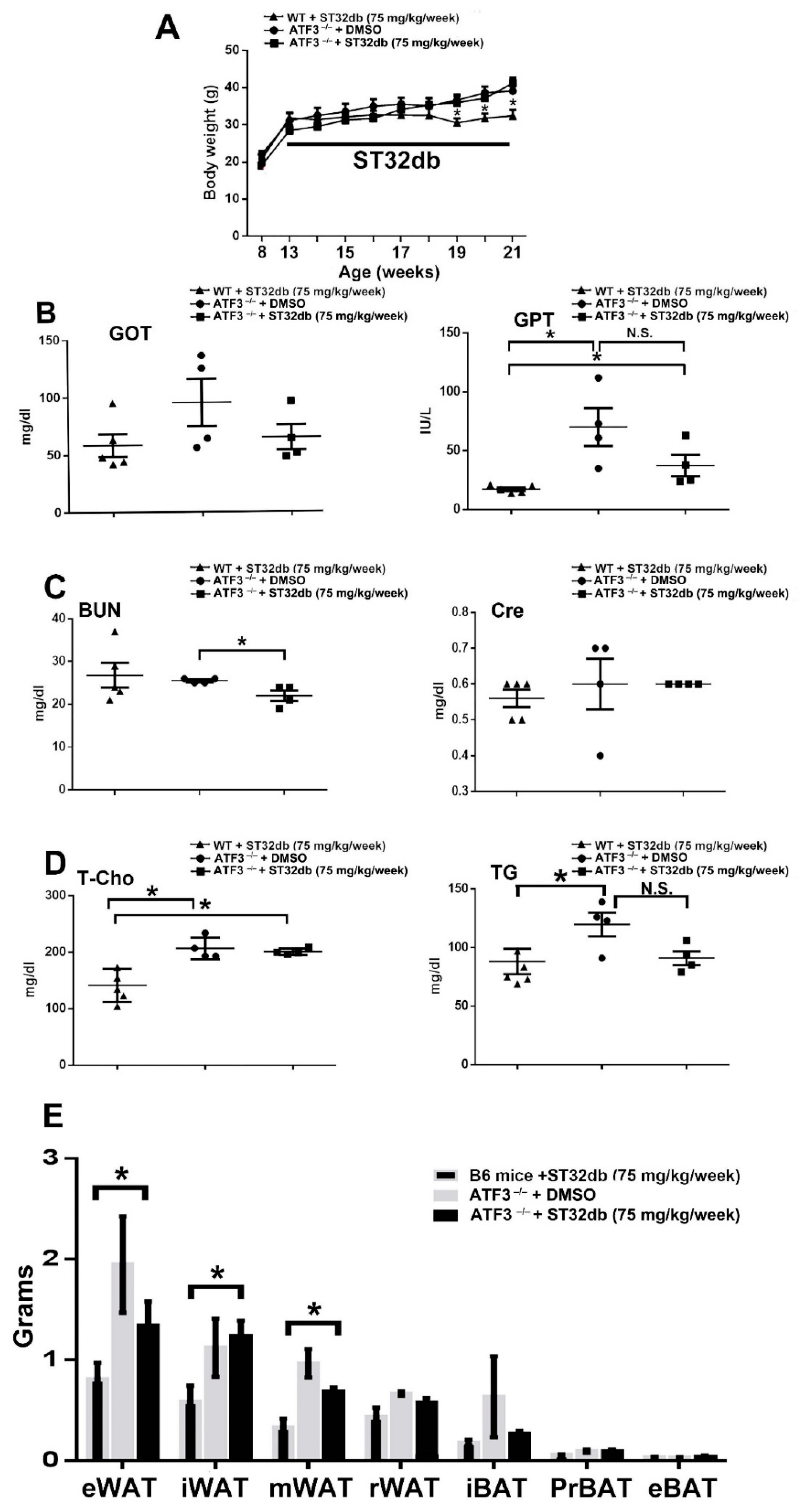
3.5. Individual Compounds as ATF3 Inducers Ameliorate HFD-Induced Metabolic Dyshomeostasis
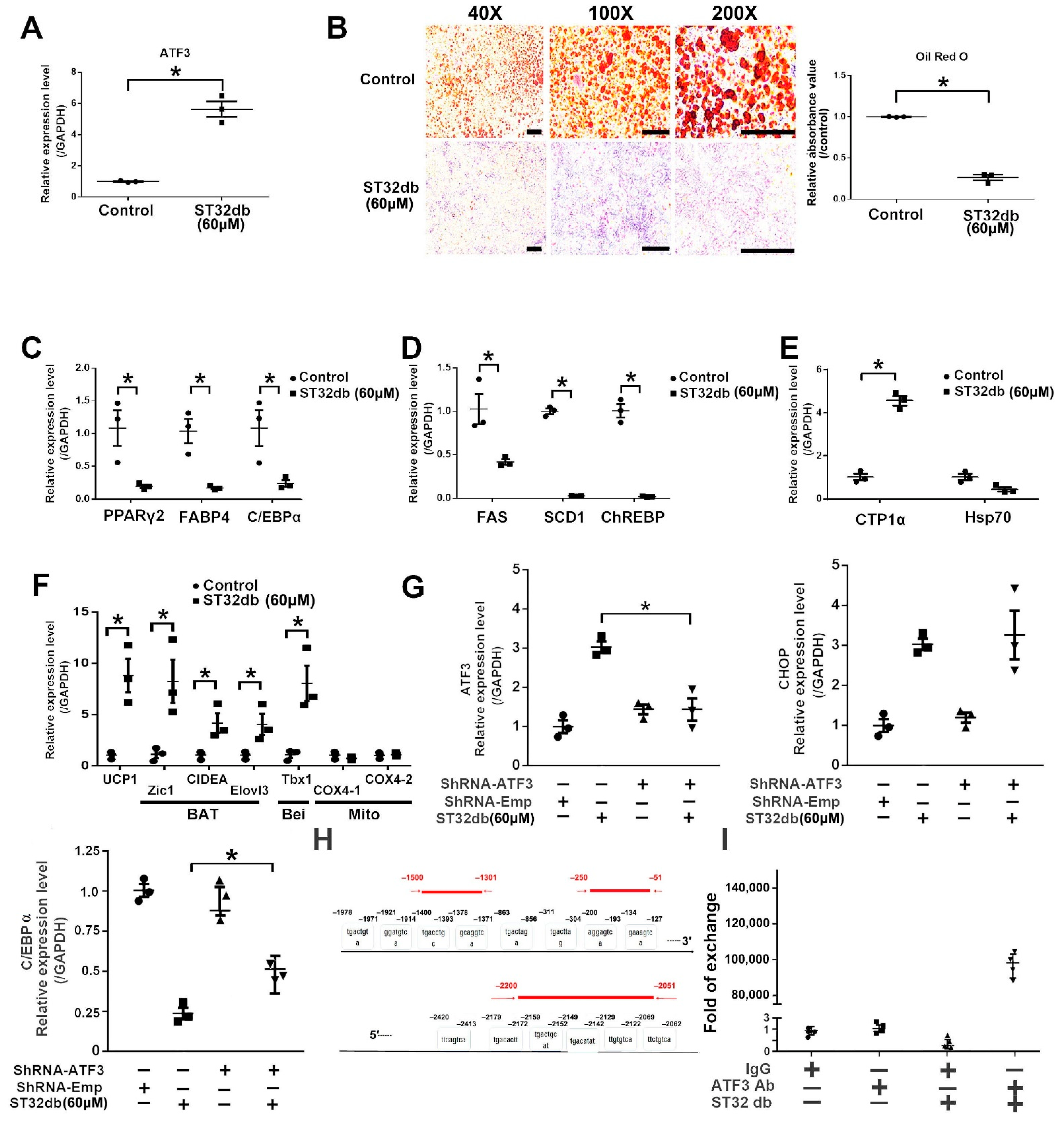
3.6. ATF3 Regulates Adipogenesis through ATF3-Mediated Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chu, D.T.; Tao, Y.; Tasken, K. OPA1 in Lipid Metabolism: Function of OPA1 in Lipolysis and Thermogenesis of Adipocytes. Horm. Metab. Res. 2017, 49, 276–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lidell, M.E.; Betz, M.J.; Enerback, S. Two types of brown adipose tissue in humans. Adipocyte 2014, 3, 63–66. [Google Scholar] [CrossRef] [Green Version]
- Giralt, M.; Villarroya, F. White, brown, beige/brite: Different adipose cells for different functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Despres, J.P. Abdominal obesity as important component of insulin-resistance syndrome. Nutrition 1993, 9, 452–459. [Google Scholar] [PubMed]
- Song, A.; Dai, W.; Jang, M.J.; Medrano, L.; Li, Z.; Zhao, H.; Shao, M.; Tan, J.; Li, A.; Ning, T.; et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J. Clin. Investig. 2020, 130, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Chondronikola, M.; Volpi, E.; Borsheim, E.; Porter, C.; Annamalai, P.; Enerback, S.; Lidell, M.E.; Saraf, M.K.; Labbe, S.M.; Hurren, N.M.; et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 2014, 63, 4089–4099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heaton, J.M. The distribution of brown adipose tissue in the human. J. Anat. 1972, 112, 35–39. [Google Scholar] [PubMed]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef]
- Chu, D.T.; Gawronska-Kozak, B. Brown and brite adipocytes: Same function, but different origin and response. Biochimie 2017, 138, 102–105. [Google Scholar] [CrossRef] [Green Version]
- Hai, T.; Wolfgang, C.D.; Marsee, D.K.; Allen, A.E.; Sivaprasad, U. ATF3 and stress responses. Gene Expr. 1999, 7, 321–335. [Google Scholar] [PubMed]
- Lu, D.; Wolfgang, C.D.; Hai, T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J. Biol. Chem. 2006, 281, 10473–10481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, T.; Hartman, M.G. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene 2001, 273, 1–11. [Google Scholar] [CrossRef]
- Lee, Y.S.; Sasaki, T.; Kobayashi, M.; Kikuchi, O.; Kim, H.J.; Yokota-Hashimoto, H.; Shimpuku, M.; Susanti, V.Y.; Ido-Kitamura, Y.; Kimura, K.; et al. Hypothalamic ATF3 is involved in regulating glucose and energy metabolism in mice. Diabetologia 2013, 56, 1383–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.K.; Son, Y.; Jung, M.H. ATF3 plays a role in adipocyte hypoxia-mediated mitochondria dysfunction in obesity. Biochem. Biophys. Res. Commun. 2013, 431, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Kong, M.; Kim, T.M.; Suh, Y.H.; Kim, W.H.; Lim, J.H.; Song, J.H.; Jung, M.H. NFATc4 and ATF3 negatively regulate adiponectin gene expression in 3T3-L1 adipocytes. Diabetes 2006, 55, 1342–1352. [Google Scholar] [CrossRef] [Green Version]
- Rynes, J.; Donohoe, C.D.; Frommolt, P.; Brodesser, S.; Jindra, M.; Uhlirova, M. Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol. Cell Biol. 2012, 32, 3949–3962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zmuda, E.J.; Qi, L.; Zhu, M.X.; Mirmira, R.G.; Montminy, M.R.; Hai, T. The roles of ATF3, an adaptive-response gene, in high-fat-diet-induced diabetes and pancreatic beta-cell dysfunction. Mol. Endocrinol. 2010, 24, 1423–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.F.; Ku, H.C.; Cheng, J.J.; Chao, S.W.; Li, H.F.; Lai, P.F.; Chang, C.C.; Don, M.J.; Chen, H.H.; Lin, H. Adipocyte browning and resistance to obesity in mice is induced by expression of ATF3. Commun. Biol. 2019, 2, 389. [Google Scholar] [CrossRef]
- Kim, S.; Song, N.J.; Bahn, G.; Chang, S.H.; Yun, U.J.; Ku, J.M.; Jo, D.G.; Park, K.W. Atf3 induction is a therapeutic target for obesity and metabolic diseases. Biochem. Biophys. Res. Commun. 2018, 504, 903–908. [Google Scholar] [CrossRef]
- Ku, H.C.; Chan, T.Y.; Chung, J.F.; Kao, Y.H.; Cheng, C.F. The ATF3 inducer protects against diet-induced obesity via suppressing adipocyte adipogenesis and promoting lipolysis and browning. Biomed. Pharmacother. 2022, 145, 112440. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Kim, C.H.; Seong, J.K.; Jung, M.H. ATF3 inhibits adipocyte differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2012, 421, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Song, N.J.; Chang, S.H.; Bahn, G.; Choi, Y.; Rhee, D.K.; Yun, U.J.; Choi, J.; Lee, J.; Yoo, J.H.; et al. Sulfuretin Prevents Obesity and Metabolic Diseases in Diet Induced Obese Mice. Biomol. Ther. 2019, 27, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.; Kim, H.; Moon, S.; Lee, H.; Kim, B. Overview of Salvia miltiorrhiza as a Potential Therapeutic Agent for Various Diseases: An Update on Efficacy and Mechanisms of Action. Antioxidants 2020, 9, 857. [Google Scholar] [CrossRef]
- Li, Z.M.; Xu, S.W.; Liu, P.Q. Salvia miltiorrhizaBurge (Danshen): A golden herbal medicine in cardiovascular therapeutics. Acta Pharmacol. Sin. 2018, 39, 802–824. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.W.; Lau, K.M.; Hon, P.M.; Mak, T.C.; Woo, K.S.; Fung, K.P. Chemistry and biological activities of caffeic acid derivatives from Salvia miltiorrhiza. Curr. Med. Chem. 2005, 12, 237–246. [Google Scholar] [CrossRef]
- Guo, R.; Li, L.; Su, J.; Li, S.; Duncan, S.E.; Liu, Z.; Fan, G. Pharmacological Activity and Mechanism of Tanshinone IIA in Related Diseases. Drug Des. Dev. Ther. 2020, 14, 4735–4748. [Google Scholar] [CrossRef]
- Park, Y.K.; Obiang-Obounou, B.W.; Lee, J.; Lee, T.Y.; Bae, M.A.; Hwang, K.S.; Lee, K.B.; Choi, J.S.; Jang, B.C. Anti-Adipogenic Effects on 3T3-L1 Cells and Zebrafish by Tanshinone IIA. Int. J. Mol. Sci. 2017, 18, 2065. [Google Scholar] [CrossRef] [Green Version]
- Jung, D.Y.; Kim, J.H.; Jung, M.H. Anti-Obesity Effects of Tanshinone I from Salvia miltiorrhiza Bunge in Mice Fed a High-Fat Diet through Inhibition of Early Adipogenesis. Nutrients 2020, 12, 1242. [Google Scholar] [CrossRef]
- Gong, Z.; Huang, C.; Sheng, X.; Zhang, Y.; Li, Q.; Wang, M.W.; Peng, L.; Zang, Y.Q. The role of tanshinone IIA in the treatment of obesity through peroxisome proliferator-activated receptor gamma antagonism. Endocrinology 2009, 150, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Hartman, M.G.; Lu, D.; Kim, M.L.; Kociba, G.J.; Shukri, T.; Buteau, J.; Wang, X.; Frankel, W.L.; Guttridge, D.; Prentki, M.; et al. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol. Cell Biol. 2004, 24, 5721–5732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.T.; Malinowska, E.; Jura, M.; Kozak, L.P. C57BL/6J mice as a polygenic developmental model of diet-induced obesity. Physiol. Rep. 2017, 5, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Baranova, A.; Schlauch, K.; Gowder, S.; Collantes, R.; Chandhoke, V.; Younossi, Z.M. Microarray technology in the study of obesity and non-alcoholic fatty liver disease. Liver Int. 2005, 25, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Frederiksen, C.M.; Hojlund, K.; Hansen, L.; Oakeley, E.J.; Hemmings, B.; Abdallah, B.M.; Brusgaard, K.; Beck-Nielsen, H.; Gaster, M. Transcriptional profiling of myotubes from patients with type 2 diabetes: No evidence for a primary defect in oxidative phosphorylation genes. Diabetologia 2008, 51, 2068–2077. [Google Scholar] [CrossRef] [Green Version]
- Kurland, L.; Liljedahl, U.; Karlsson, J.; Kahan, T.; Malmqvist, K.; Melhus, H.; Syvanen, A.C.; Lind, L. Angiotensinogen gene polymorphisms: Relationship to blood pressure response to antihypertensive treatment. Results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation vs Atenolol (SILVHIA) trial. Am. J. Hypertens. 2004, 17, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Don, M.J.; Shen, C.C.; Syu, W.J.; Ding, Y.H.; Sun, C.M. Cytotoxic and aromatic constituents from Salvia miltiorrhiza. Phytochemistry 2006, 67, 497–503. [Google Scholar] [CrossRef]
- Lee, J.E.; Schmidt, H.; Lai, B.; Ge, K. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol. Cell Biol. 2019, 39, e00601-18. [Google Scholar] [CrossRef] [Green Version]
- An, T.; Zhang, J.; Lv, B.; Liu, Y.; Huang, J.; Lian, J.; Wu, Y.; Gao, S.; Jiang, G. Salvianolic acid B plays an anti-obesity role in high fat diet-induced obese mice by regulating the expression of mRNA, circRNA, and lncRNA. PeerJ 2019, 7, e6506. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.W.; Lee, B.; Kim, D.H.; Jeong, H.O.; Moon, K.M.; Kim, M.J.; Yokozawa, T.; Chung, H.Y. Mechanism of Action of Magnesium Lithospermate B against Aging and Obesity-Induced ER Stress, Insulin Resistance, and Inflammsome Formation in the Liver. Molecules 2018, 23, 2098. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Hong, F.; Wang, J.; Wang, J.; Zhao, X.; Wang, S.; Xue, T.; Xu, J.; Zheng, X.; Zhai, Y. DBZ is a putative PPARgamma agonist that prevents high fat diet-induced obesity, insulin resistance and gut dysbiosis. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2690–2701. [Google Scholar] [CrossRef]
- Cao, W.; Medvedev, A.V.; Daniel, K.W.; Collins, S. beta-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 2001, 276, 27077–27082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Chen, J.; Hai, T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem. J. 2007, 401, 559–567. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Linhart, H.G.; Ishimura-Oka, K.; DeMayo, F.; Kibe, T.; Repka, D.; Poindexter, B.; Bick, R.J.; Darlington, G.J. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 2001, 98, 12532–12537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Z.; Umek, R.M.; McKnight, S.L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991, 5, 1538–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pant, R.; Firmal, P.; Shah, V.K.; Alam, A.; Chattopadhyay, S. Epigenetic Regulation of Adipogenesis in Development of Metabolic Syndrome. Front. Cell Dev. Biol. 2020, 8, 619888. [Google Scholar] [CrossRef]
- Kuzmochka, C.; Abdou, H.S.; Hache, R.J.; Atlas, E. Inactivation of histone deacetylase 1 (HDAC1) but not HDAC2 is required for the glucocorticoid-dependent CCAAT/enhancer-binding protein alpha (C/EBPalpha) expression and preadipocyte differentiation. Endocrinology 2014, 155, 4762–4773. [Google Scholar] [CrossRef] [Green Version]
- Li, H.F.; Cheng, C.F.; Liao, W.J.; Lin, H.; Yang, R.B. ATF3-mediated epigenetic regulation protects against acute kidney injury. J. Am. Soc. Nephrol. 2010, 21, 1003–1013. [Google Scholar] [CrossRef] [Green Version]
- Modi, S.; Yaluri, N.; Kokkola, T. Strigolactone GR24 and pinosylvin attenuate adipogenesis and inflammation of white adipocytes. Biochem. Biophys. Res. Commun. 2018, 499, 164–169. [Google Scholar] [CrossRef]
- Grancieri, M.; Martino, H.S.D.; Gonzalez de Mejia, E. Protein Digests and Pure Peptides from Chia Seed Prevented Adipogenesis and Inflammation by Inhibiting PPARgamma and NF-kappaB Pathways in 3T3L-1 Adipocytes. Nutrients 2021, 13, 176. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, M.; Thorsson, V.; Li, B.; Rust, A.G.; Korb, M.; Roach, J.C.; Kennedy, K.; Hai, T.; Bolouri, H.; Aderem, A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 2006, 441, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Batchvarova, N.; Wang, X.Z.; Ron, D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153). EMBO J. 1995, 14, 4654–4661. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.P.; Wolfgang, C.D.; Hai, T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell Biol. 1996, 16, 1157–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, T.A.; Woods, S.C. Overview of animal models of obesity. Curr. Protoc. Pharmacol. 2012, 58, 5–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.-L.; Lin, H.; Li, H.-F.; Don, M.-J.; King, P.-C.; Chen, H.-H. Salvia miltiorrhiza Extract and Individual Synthesized Component Derivatives Induce Activating-Transcription-Factor-3-Mediated Anti-Obesity Effects and Attenuate Obesity-Induced Metabolic Disorder by Suppressing C/EBPα in High-Fat-Induced Obese Mice. Cells 2022, 11, 1022. https://doi.org/10.3390/cells11061022
Wu Y-L, Lin H, Li H-F, Don M-J, King P-C, Chen H-H. Salvia miltiorrhiza Extract and Individual Synthesized Component Derivatives Induce Activating-Transcription-Factor-3-Mediated Anti-Obesity Effects and Attenuate Obesity-Induced Metabolic Disorder by Suppressing C/EBPα in High-Fat-Induced Obese Mice. Cells. 2022; 11(6):1022. https://doi.org/10.3390/cells11061022
Chicago/Turabian StyleWu, Yueh-Lin, Heng Lin, Hsiao-Fen Li, Ming-Jaw Don, Pei-Chih King, and Hsi-Hsien Chen. 2022. "Salvia miltiorrhiza Extract and Individual Synthesized Component Derivatives Induce Activating-Transcription-Factor-3-Mediated Anti-Obesity Effects and Attenuate Obesity-Induced Metabolic Disorder by Suppressing C/EBPα in High-Fat-Induced Obese Mice" Cells 11, no. 6: 1022. https://doi.org/10.3390/cells11061022
APA StyleWu, Y.-L., Lin, H., Li, H.-F., Don, M.-J., King, P.-C., & Chen, H.-H. (2022). Salvia miltiorrhiza Extract and Individual Synthesized Component Derivatives Induce Activating-Transcription-Factor-3-Mediated Anti-Obesity Effects and Attenuate Obesity-Induced Metabolic Disorder by Suppressing C/EBPα in High-Fat-Induced Obese Mice. Cells, 11(6), 1022. https://doi.org/10.3390/cells11061022