The HIFα-Stabilizing Drug Roxadustat Increases the Number of Renal Epo-Producing Sca-1+ Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Isolation of Mesenchymal Cells and Magnetic Cell Separation (MACS)
2.3. Cell Culture of Sca-1+ Cells
2.4. Human Bone Marrow-Derived Mesenchymal Stem Cells
2.5. Tri-Lineage Differentiation
2.6. Cell Culture of L929 Cells
2.7. ELISA
2.8. Protein Extraction and Western Blot Analysis
2.9. FACS Analysis
2.10. RNA Isolation and Quantitative RT-PCR
2.11. Primer Sequences
2.12. Transcriptome and Bioinformatic Analysis
2.12.1. RNAseq Library Preparation
2.12.2. Mapping and Normalization
2.13. Macrophage Isolation and Differentiation
2.14. Statistical Analysis
3. Results
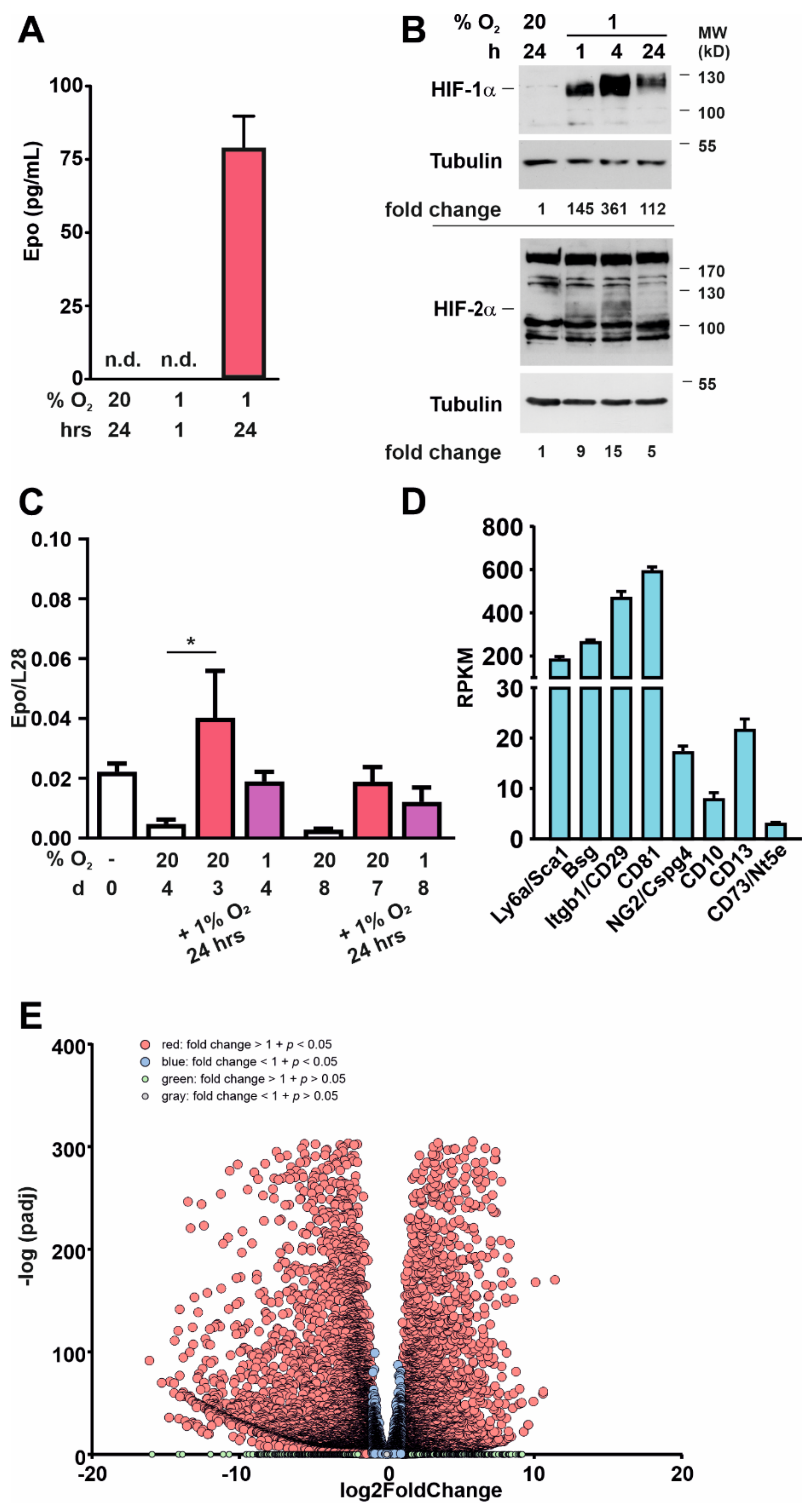
3.1. Roxadustat Treatment Increases the Number of Sca-1-Positive Cells in the Kidneys
3.2. Sca-1+ Cells Display a Mesenchymal Stem-Cell-like Phenotype
3.3. Kidney-Derived Sca-1+ Cells Produce Epo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pugh, C.W.; Ratcliffe, P.J. New horizons in hypoxia signaling pathways. Exp. Cell Res. 2017, 356, 116–121. [Google Scholar] [CrossRef]
- Ivan, M.; Kaelin, W.G., Jr. The EGLN-HIF O2-sensing system: Multiple inputs and feedbacks. Mol. Cell 2017, 66, 772–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenger, R.H.; Hoogewijs, D. Regulated oxygen sensing by protein hydroxylation in renal erythropoietin-producing cells. Am. J. Physiol. Renal. Physiol. 2010, 298, F1287–F1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlando, I.M.C.; Lafleur, V.N.; Storti, F.; Spielmann, P.; Crowther, L.; Santambrogio, S.; Schodel, J.; Hoogewijs, D.; Mole, D.R.; Wenger, R.H. Distal and proximal hypoxia response elements cooperate to regulate organ-specific erythropoietin gene expression. Haematologica 2020, 105, 2774–2784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jelkmann, W. The esa scenario gets complex: From biosimilar epoetins to activin traps. Nephrol. Dial. Transplant. 2015, 30, 553–559. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.C.; Holt-Martyn, J.P.; Schofield, C.J.; Ratcliffe, P.J. Pharmacological targeting of the HIF hydroxylases—A new field in medicine development. Mol. Aspects Med. 2016, 47–48, 54–75. [Google Scholar] [CrossRef]
- Dhillon, S. Roxadustat: First global approval. Drugs 2019, 79, 563–572. [Google Scholar] [CrossRef]
- Eltzschig, H.K.; Bratton, D.L.; Colgan, S.P. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat. Rev. Drug Discov. 2014, 13, 852–869. [Google Scholar] [CrossRef] [Green Version]
- Koury, S.T.; Bondurant, M.C.; Koury, M.J. Localization of erythropoietin synthesizing cells in murine kidneys by in situ hybridization. Blood 1988, 71, 524–527. [Google Scholar] [CrossRef]
- Wenger, R.H.; Kurtz, A. Erythropoietin. Compr. Physiol. 2011, 1, 1759–1794. [Google Scholar] [CrossRef]
- Nolan, K.A.; Wenger, R.H. Source and microenvironmental regulation of erythropoietin in the kidney. Curr. Opin. Nephrol. Hypertens. 2018, 27, 277–282. [Google Scholar] [CrossRef]
- Imeri, F.; Nolan, K.A.; Bapst, A.M.; Santambrogio, S.; Abreu-Rodriguez, I.; Spielmann, P.; Pfundstein, S.; Libertini, S.; Crowther, L.; Orlando, I.M.C.; et al. Generation of renal epo-producing cell lines by conditional gene tagging reveals rapid HIF-2 driven epo kinetics, cell autonomous feedback regulation, and a telocyte phenotype. Kidney Int. 2019, 95, 375–387. [Google Scholar] [CrossRef]
- Yutoku, M.; Grossberg, A.L.; Pressman, D. A cell surface antigenic determinant present on mouse plasmacytes and only about half of mouse thymocytes. J. Immunol. 1974, 112, 1774–1781. [Google Scholar]
- Bapst, A.M.; Knöpfel, T.; Nolan, K.A.; Imeri, F.; Schuh, C.D.; Hall, A.M.; Guo, J.; Katschinski, D.M.; Wenger, R.H. Neurogenic and pericytic plasticity of conditionally immortalized cells derived from renal erythropoietin-producing cells. J. Cell Physiol. 2022. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the r/bioconductor package biomart. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [Green Version]
- Guentsch, A.; Beneke, A.; Swain, L.; Farhat, K.; Nagarajan, S.; Wielockx, B.; Raithatha, K.; Dudek, J.; Rehling, P.; Zieseniss, A.; et al. Phd2 is a regulator for glycolytic reprogramming in macrophages. Mol. Cell Biol. 2017, 37, e00236-16. [Google Scholar] [CrossRef] [Green Version]
- Dominici, M.; le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Bernardo, M.E.; Fibbe, W.E. Mesenchymal stromal cells: Sensors and switchers of inflammation. Cell Stem Cell 2013, 13, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Hirano, I.; Sekine, H.; Miyauchi, K.; Nakai, T.; Kato, K.; Ito, S.; Yamamoto, M.; Suzuki, N. An immortalized cell line derived from renal erythropoietin-producing (rep) cells demonstrates their potential to transform into myofibroblasts. Sci. Rep. 2019, 9, 11254. [Google Scholar] [CrossRef]
- Souma, T.; Yamazaki, S.; Moriguchi, T.; Suzuki, N.; Hirano, I.; Pan, X.; Minegishi, N.; Abe, M.; Kiyomoto, H.; Ito, S.; et al. Plasticity of renal erythropoietin-producing cells governs fibrosis. J. Am. Soc. Nephrol. 2013, 24, 1599–1616. [Google Scholar] [CrossRef] [Green Version]
- Hoppe, G.; Yoon, S.; Gopalan, B.; Savage, A.R.; Brown, R.; Case, K.; Vasanji, A.; Chan, E.R.; Silver, R.B.; Sears, J.E. Comparative systems pharmacology of HIF stabilization in the prevention of retinopathy of prematurity. Proc. Natl. Acad. Sci. USA 2016, 113, E2516–E2525. [Google Scholar] [CrossRef] [Green Version]
- Safran, M.; Kim, W.Y.; O’Connell, F.; Flippin, L.; Gunzler, V.; Horner, J.W.; Depinho, R.A.; Kaelin, W.G., Jr. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: Assessment of an oral agent that stimulates erythropoietin production. Proc. Natl. Acad. Sci. USA 2006, 103, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Yeh, T.L.; Leissing, T.M.; Abboud, M.I.; Thinnes, C.C.; Atasoylu, O.; Holt-Martyn, J.P.; Zhang, D.; Tumber, A.; Lippl, K.; Lohans, C.T.; et al. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem. Sci. 2017, 8, 7651–7668. [Google Scholar] [CrossRef] [Green Version]
- Cullen-McEwen, L.A.; Caruana, G.; Bertram, J.F. The where, what and why of the developing renal stroma. Nephron. Exp. Nephrol. 2005, 99, e1–e8. [Google Scholar] [CrossRef]
- Zeisberg, M.; Kalluri, R. Physiology of the renal interstitium. Clin. J. Am. Soc. Nephrol. 2015, 10, 1831–1840. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, A. Endocrine functions of the renal interstitium. Pflugers Arch. 2017, 469, 869–876. [Google Scholar] [CrossRef]
- Bruno, S.; Chiabotto, G.; Camussi, G. Concise review: Different mesenchymal stromal/stem cell populations reside in the adult kidney. Stem Cells Transl. Med. 2014, 3, 1451–1455. [Google Scholar] [CrossRef]
- Plotkin, M.D.; Goligorsky, M.S. Mesenchymal cells from adult kidney support angiogenesis and differentiate into multiple interstitial cell types including erythropoietin-producing fibroblasts. Am. J. Physiol. Renal. Physiol. 2006, 291, F902–F912. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Johnston, P.; Zhang, B.; Zakari, A.; Chowdhry, T.; Smith, R.R.; Marban, E.; Rabb, H.; Womer, K.L. Kidney-derived stromal cells modulate dendritic and T cell responses. J. Am. Soc. Nephrol. 2009, 20, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Dekel, B.; Zangi, L.; Shezen, E.; Reich-Zeliger, S.; Eventov-Friedman, S.; Katchman, H.; Jacob-Hirsch, J.; Amariglio, N.; Rechavi, G.; Margalit, R.; et al. Isolation and characterization of nontubular sca-1+lin- multipotent stem/progenitor cells from adult mouse kidney. J. Am. Soc. Nephrol. 2006, 17, 3300–3314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loughner, C.L.; Bruford, E.A.; McAndrews, M.S.; Delp, E.E.; Swamynathan, S.; Swamynathan, S.K. Organization, evolution and functions of the human and mouse ly6/upar family genes. Hum. Genom. 2016, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vagnozzi, R.J.; Sargent, M.A.; Lin, S.J.; Palpant, N.J.; Murry, C.E.; Molkentin, J.D. Genetic lineage tracing of sca-1+ cells reveals endothelial but not myogenic contribution to the murine heart. Circulation 2018, 138, 2931–2939. [Google Scholar] [CrossRef]
- Zhang, L.; Sultana, N.; Yan, J.; Yang, F.; Chen, F.; Chepurko, E.; Yang, F.C.; Du, Q.; Zangi, L.; Xu, M.; et al. Cardiac sca-1+ cells are not intrinsic stem cells for myocardial development, renewal, and repair. Circulation 2018, 138, 2919–2930. [Google Scholar] [CrossRef]
- Stanford, W.L.; Haque, S.; Alexander, R.; Liu, X.; Latour, A.M.; Snodgrass, H.R.; Koller, B.H.; Flood, P.M. Altered proliferative response by T lymphocytes of ly-6a (sca-1) null mice. J. Exp. Med. 1997, 186, 705–717. [Google Scholar] [CrossRef] [Green Version]
- Hanson, P.; Mathews, V.; Marrus, S.H.; Graubert, T.A. Enhanced green fluorescent protein targeted to the sca-1 (ly-6a) locus in transgenic mice results in efficient marking of hematopoietic stem cells in vivo. Exp. Hematol. 2003, 31, 159–167. [Google Scholar] [CrossRef]
- Ito, C.Y.; Li, C.Y.; Bernstein, A.; Dick, J.E.; Stanford, W.L. Hematopoietic stem cell and progenitor defects in sca-1/ly-6a-null mice. Blood 2003, 101, 517–523. [Google Scholar] [CrossRef]
- Suzuki, N.; Hirano, I.; Pan, X.; Minegishi, N.; Yamamoto, M. Erythropoietin production in neuroepithelial and neural crest cells during primitive erythropoiesis. Nat. Commun. 2013, 4, 2902. [Google Scholar] [CrossRef] [Green Version]
- Broeker, K.A.E.; Fuchs, M.A.A.; Schrankl, J.; Kurt, B.; Nolan, K.A.; Wenger, R.H.; Kramann, R.; Wagner, C.; Kurtz, A. Different subpopulations of kidney interstitial cells produce erythropoietin and factors supporting tissue oxygenation in response to hypoxia in vivo. Kidney Int. 2020, 98, 918–931. [Google Scholar] [CrossRef]
- Asada, N.; Takase, M.; Nakamura, J.; Oguchi, A.; Asada, M.; Suzuki, N.; Yamamura, K.; Nagoshi, N.; Shibata, S.; Rao, T.N.; et al. Dysfunction of fibroblasts of extrarenal origin underlies renal fibrosis and renal anemia in mice. J. Clin. Invest. 2011, 121, 3981–3990. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jatho, A.; Zieseniss, A.; Brechtel-Curth, K.; Guo, J.; Böker, K.O.; Salinas, G.; Wenger, R.H.; Katschinski, D.M. The HIFα-Stabilizing Drug Roxadustat Increases the Number of Renal Epo-Producing Sca-1+ Cells. Cells 2022, 11, 753. https://doi.org/10.3390/cells11040753
Jatho A, Zieseniss A, Brechtel-Curth K, Guo J, Böker KO, Salinas G, Wenger RH, Katschinski DM. The HIFα-Stabilizing Drug Roxadustat Increases the Number of Renal Epo-Producing Sca-1+ Cells. Cells. 2022; 11(4):753. https://doi.org/10.3390/cells11040753
Chicago/Turabian StyleJatho, Aline, Anke Zieseniss, Katja Brechtel-Curth, Jia Guo, Kai Oliver Böker, Gabriela Salinas, Roland H. Wenger, and Dörthe M. Katschinski. 2022. "The HIFα-Stabilizing Drug Roxadustat Increases the Number of Renal Epo-Producing Sca-1+ Cells" Cells 11, no. 4: 753. https://doi.org/10.3390/cells11040753
APA StyleJatho, A., Zieseniss, A., Brechtel-Curth, K., Guo, J., Böker, K. O., Salinas, G., Wenger, R. H., & Katschinski, D. M. (2022). The HIFα-Stabilizing Drug Roxadustat Increases the Number of Renal Epo-Producing Sca-1+ Cells. Cells, 11(4), 753. https://doi.org/10.3390/cells11040753






