Circulating Serum MiRNA-8074 as a Novel Prognostic Biomarker for Multiple Myeloma
Abstract
:1. Introduction
2. Material and Methods
2.1. Patients
2.2. Serum Collection
2.3. MiRNA Analysis in Blood Serum
2.4. Plasma Cell Isolation
2.5. MiRNA and Gene Expression in Bone Marrow Plasma Cells
2.6. Cytogenetic Assessment
2.7. Statistical Analysis
3. Results
3.1. Comparison of the MiR-8074 Expression Depending on Demographic, Clinical and Molecular Factors
3.2. Correlation between Serum MiRNA-8074 and Selected Demographic, Clinical and Molecular Variables
3.3. Correlation between MiRNA-8074 Expression and Chemotherapy Response
3.4. Outcomes: ProgressionFree Survival
3.4.1. Univariate Analysis
3.4.2. Multivariate Analysis
3.5. Overall Survival
3.5.1. Univariate Analysis
3.5.2. Multivariate Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, H.; Novis Durie, S.; Meckl, A.; Hinke, A.; Durie, B. Multiple Myeloma Incidence and Mortality around the Globe; Interrelations between Health Access and Quality, Economic Resources, and Patient Empowerment. Oncologist 2020, 25, e1406–e1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazmi, S.M.; Nusrat, M.; Gunaydin, H.; Cornelison, A.M.; Shah, N.; Kebriaei, P.; Nieto, Y.; Parmar, S.; Popat, U.R.; Oran, B.; et al. Outcomes among High-Risk and Standard-Risk Multiple Myeloma Patients Treated with High-Dose Chemotherapy and Autologous Hematopoietic Stem-Cell Transplantation. Clin. Lymphoma Myeloma Leuk. 2015, 15, 687–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braunlin, M.; Belani, R.; Buchanan, J.; Wheeling, T.; Kim, C. Trends in the multiple myeloma treatment landscape and survival: A U.S. analysis using 2011–2019 oncology clinic electronic health record data. Leuk. Lymphoma 2021, 62, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, M.; Agnelli, L.; Lombardi, L.; Tassone, P.; Neri, A. MicroRNAs in the pathobiology of multiple myeloma. Curr. Cancer Drug Targets 2012, 12, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Amodio, N.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P.; Cho, W.C. MicroRNA and multiple myeloma: From laboratory findings to translational therapeutic approaches. Curr. Pharm. Biotechnol. 2014, 15, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yang, X.; Liu, M.; Zhang, Z.; Xing, E. Roles of miRNA dysregulation in the pathogenesis of multiple myeloma. Cancer Gene Ther. 2021, 28, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.targetscan.org/cgi-bin/targetscan/vert_72/targetscan.cgi?species=Human&gid=&mir_sc=&mir_c=&mir_nc=&mir_vnc=&mirg=miR-8074 (accessed on 1 December 2021).
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Durie, B.G.; Salmon, S.E. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer 1975, 36, 842–854. [Google Scholar] [CrossRef]
- International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: A report of the International Myeloma Working Group. Br. J. Haematol. 2003, 121, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef]
- Common Terminology Criteria for Adverse Events (CTCAE v5.0). Available online: http://evsncinihgov/ftp1/CTCAE/CTCAE_403_2010-06-14_QuickReference_5x7pdf (accessed on 11 August 2021).
- Ross, F.; Avet-Loiseau, H.; Ameye, G.; Gutiérrez, N.C.; Liebisch, P.; O’Connor, S.; Dalva, K.; Fabris, S.; Testi, A.M.; Jarosova, M.; et al. Report from the european myeloma network on interphase fish in multiple myeloma and related disorders. Haematologica 2012, 97, 1272–1277. [Google Scholar] [CrossRef] [Green Version]
- Dmoszynska, A.; Chocholska, S. Molecular biology methods in the diagnosis of multiple myeloma. In Molecular Aspects of Hematologic Malignancies: Diagnostic Tools and Clinical Applications; Witt, M., Dawidowska, M., Szczepanski, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 443–449. [Google Scholar]
- Durie, B.G.; Harousseau, J.L.; Miguel, J.S.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.V.; et al. International uniform response criteria for multiple myeloma International uniform response criteria for multiple myeloma. Leukemia 2007, 21, 1134. [Google Scholar] [CrossRef] [Green Version]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised International Staging System for Multiple Myeloma: A Report from International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Bergsagel, P.L.; Drach, J.; Shaughnessy, J.; Gutierrez, N.; Stewart, A.K.; Morgan, G.; Van Ness, B.; Chesi, M.; Minvielle, S.; et al. International Myeloma Working Group molecular classification of multiple myeloma: Spotlight review. Leukemia 2009, 23, 2210–2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, A.K.; Bergsagel, P.L.; Greipp, P.R.; Dispenzieri, A.; Gertz, M.A.; Hayman, S.R.; Kumar, S.; Lacy, M.Q.; Lust, J.A.; Russell, S.J.; et al. A practical guide to defining high-risk myeloma for clinical trials, patient counseling and choice of therapy. Leukemia 2007, 21, 529–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldenhuis, C.N.; Oosting, S.F.; Gietema, J.A.; de Vries, E.G. Prognostic versus predictive value of biomarkers in oncology. Eur. J. Cancer 2008, 44, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M. Biomarkers for personalized oncology: Recent advances and future challenges. Metabolism 2015, 64 (Suppl. 1), S16–S21. [Google Scholar] [CrossRef]
- Zen, K.; Zhang, C.Y. Circulating microRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012, 32, 326–348. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Gilad, S.; Meiri, E.; Yogev, Y.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; Bentwich, Z.; et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef] [Green Version]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Sacco, A.; Belotti, A.; Ribolla, R.; Cancelli, V.; Giacomini, A.; Ronca, R.; Chiarini, M.; Imberti, L.; Marini, M.; et al. Circulating microRNAs and Their Role in Multiple Myeloma. Noncoding RNA 2019, 5, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.S.; Wang, Y.J.; Zhang, G.X.; Zhang, W.T. Potential diagnostic value of circulating miRNA for multiple myeloma: A meta-analysis. J. Bone Oncol. 2020, 25, 100327. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Xia, T.; Ling, Y.; Chen, B. MiRNAs with prognostic significance in multiple myeloma: A systemic review and meta-analysis. Medicine (Baltimore) 2019, 98, e16711. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, E.; Liu, R.; Chu, Y. miRNA-15a/16: As tumor suppressors and more. Future Oncol. 2015, 11, 2351–2363. [Google Scholar] [CrossRef]
- Kang, W.; Tong, J.H.; Lung, R.W.; Dong, Y.; Zhao, J.; Liang, Q.; Zhang, L.; Pan, Y.; Yang, W.; Pang, J.C.; et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol. Cancer 2015, 14, 52. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.Y.; She, X.M.; Qin, Y.; Chu, Z.B.; Chen, L.; Ai, L.S.; Zhang, L.; Hu, Y. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis 2013, 34, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Tung, Y.T.; Huang, P.W.; Chou, Y.C.; Lai, C.W.; Wang, H.P.; Ho, H.C.; Yen, C.C.; Tu, C.Y.; Tsai, T.C.; Yeh, D.C.; et al. Lung tumorigenesis induced by human vascular endothelial growth factor (hVEGF)-A165 overexpression in transgenic mice and amelioration of tumor formation by miR-16. Oncotarget 2015, 6, 10222–10238. [Google Scholar] [CrossRef] [Green Version]
- Roccaro, A.M.; Sacco, A.; Thompson, B.; Leleu, X.; Azab, A.K.; Azab, F.; Runnels, J.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009, 113, 6669–6680. [Google Scholar] [CrossRef]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhou, L.; Shi, M.; Kuang, Y.; Fang, L. Downregulation of miRNA-15a and miRNA-16 promote tumor proliferation in multiple myeloma by increasing CABIN1 expression. Oncol. Lett. 2018, 15, 1287–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Joosse, S.A.; Muller, V.; Trillsch, F.; Milde-Langosch, K.; Mahner, S.; Geffken, M.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br. J. Cancer 2015, 113, 1358–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birks, D.K.; Barton, V.N.; Donson, A.M.; Handler, M.H.; Vibhakar, R.; Foreman, N.K. Survey of MicroRNA Expression in Pediatric Brain Tumors. Pediatr. Blood Cancer 2011, 56, 211–216. [Google Scholar] [CrossRef]
- Dacic, S.; Kelly, L.; Shuai, Y.; Nikiforova, M. miRNA expression profiling of lung adenocarcinomas: Correlation with mutational status. Mod. Pathol. 2010, 23, 1577–1582. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, C.; Wang, X.; Zhang, J.; Zhang, R.; Liu, R. The expression of miR-25 is increased in colorectal cancer and is associated with patient prognosis. Med. Oncol. 2014, 31, 781. [Google Scholar] [CrossRef]
- Li, Y.; Tan, W.; Neo, T.W.L.; Aung, M.O.; Wasser, S.; Lim, S.G.; Tan, T.M. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009, 100, 1234–1242. [Google Scholar] [CrossRef]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.G.; Negrini, M.; et al. E2F1-regulated microRNAs impair TGF beta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar] [CrossRef] [Green Version]
- Poliseno, L.; Salmena, L.; Riccardi, L.; Fornari, A.; Song, M.S.; Hobbs, R.M.; Sportoletti, P.; Varmeh, S.; Egia, A.; Fedele, G.; et al. Identification of the miR-106b similar to 25 MicroRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Rocci, A.; Hofmeister, C.C.; Geyer, S.; Stiff, A.; Gambella, M.; Cascione, L.; Guan, J.; Benson, D.M.; Efebera, Y.A.; Talabere, T.; et al. Circulating miRNA markers show promise as new prognosticators for multiple myeloma. Leukemia 2014, 28, 1922–1926. [Google Scholar] [CrossRef] [Green Version]
- Zi, Y.; Zhang, Y.; Wu, Y.; Zhang, L.; Yang, R.; Huang, Y. Downregulation of microRNA-25-3p inhibits the proliferation and promotes the apoptosis of multiple myeloma cells via targeting the PTEN/PI3K/AKT signaling pathway. Int. J. Mol. Med. 2021, 47, 8. [Google Scholar] [CrossRef] [PubMed]
- Kubiczkova, L.; Kryukov, F.; Slaby, O.; Dementyeva, E.; Jarkovsky, J.; Nekvindova, J.; Radova, L.; Greslikova, H.; Kuglik, P.; Vetesnikova, E.; et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica 2014, 99, 511–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, B.; Xiao, C.; Liu, Y.; Zhang, N.; Bai, H.; Yang, T.; Xiang, Y.; Nan, Y.; Li, Q.; Zhang, W.; et al. miR-744-5p Inhibits Multiple Myeloma Proliferation, Epithelial Mesenchymal Transformation and Glycolysis by Targeting SOX12/Wnt/β-Catenin Signaling. Onco Targets Ther. 2021, 14, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Pichiorri, F.; Suh, S.S.; Ladetto, M.; Kuehl, M.; Palumbo, T.; Drandi, D.; Taccioli, C.; Zanesi, N.; Alder, H.; Hagan, J.P.; et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 12885–12890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, X.Y.; Zhang, S.S.; Wu, S.; Hong, M.; Li, J.Y.; Chen, L.J.; Xu, J.R. Expression level of microRNA-92a and its clinical significance in multiple myeloma patients. Zhonghua Xue Ye Xue Za Zhi 2013, 34, 332–336. [Google Scholar] [CrossRef]
- Yoshizawa, S.; Ohyashiki, J.H.; Ohyashiki, M.; Umezu, T.; Suzuki, K.; Inagaki, A.; Iida, S.; Ohyashiki, K. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012, 2, e53. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, G.P.; Gertz, M.A.; Dispenzieri, A.; Lacy, M.Q.; Buadi, F.K.; Dingli, D.; Hayman, S.R.; Kapoor, P.; Lust, J.A.; Russell, S.; et al. Impact of cytogenetic classification on outcomes following early high-dose therapy in multiple myeloma. Leukemia 2016, 30, 633–639. [Google Scholar] [CrossRef]
- Lakshman, A.; Alhaj Moustafa, M.; Rajkumar, S.V.; Dispenzieri, A.; Gertz, M.A.; Buadi, F.K.; Lacy, M.Q.; Dingli, D.; Fonder, A.L.; Hayman, S.R.; et al. Natural history of t(11;14) multiple myeloma. Leukemia 2018, 32, 131–138. [Google Scholar] [CrossRef]
- Gran, C.; Uttervall, K.; Borg Bruchfeld, J.; Wallblom, A.; Alici, E.; Gahrton, G.; Nahi, H. Translocation (11;14) in newly diagnosed multiple myeloma, time to reclassify this standard risk chromosomal aberration? Eur. J. Haematol. 2019, 103, 588–596. [Google Scholar] [CrossRef]
- Francavilla, A.; Gagliardi, A.; Piaggeschi, G.; Tarallo, S.; Cordero, F.; Pensa, R.G.; Impeduglia, A.; Caviglia, G.P.; Ribaldone, D.G.; Gallo, G.; et al. Faecal miRNA profiles associated with age, sex, BMI, and lifestyle habits in healthy individuals. Sci. Rep. 2021, 11, 20645. [Google Scholar] [CrossRef]
- Kolhe, R.; Hunter, M.; Liu, S.; Jadeja, R.N.; Pundkar, C.; Mondal, A.K.; Mendhe, B.; Drewry, M.; Rojiani, M.V.; Liu, Y.; et al. Gender-specific differential expression of exosomal miRNA in synovial fluid of patients with osteoarthritis. Sci. Rep. 2017, 7, 2029. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Meng, Y.; Zhu, X.; Saadiq, I.M.; Jordan, K.L.; Eirin, A.; Lerman, L.O. Metabolic syndrome increases senescence-associated micro-RNAs in extracellular vesicles derived from swine and human mesenchymal stem/stromal cells. Cell Commun. Signal. 2020, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Feng, Y.; Zheng, Y.; Ogundiran, T.O.; Ojengbede, O.; Zheng, W.; Blot, W.; Ambrosone, C.B.; John, E.M.; Bernstein, L.; et al. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. Hum. Genet. 2016, 135, 1145–1159. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Wang, Z.; Sun, H.; Xin, H. Evaluation of Exosomal miRNA in Blood as a Potential Diagnostic Biomarker for Human Non-Small Cell Lung Cancer. Med. Sci. Monit. 2020, 26, e924721. [Google Scholar] [CrossRef] [PubMed]
| Variable | n = 105 (100%) |
|---|---|
| Sex | |
| Men | 52 (49.5%) |
| Women | 53 (50.6%) |
| Age | |
| <65 | 35 (33.3%) |
| ≥65 | 70 (66.7%) |
| Diagnosis | |
| MM with a monoclonal component | 88 (83.8%) |
| Light chain disease | 16 (15.2%) |
| Non-secretory MM | 1 (0.9) |
| Monoclonal protein class | |
| IgA | 29 (27.6%) |
| IgG | 59 (56.2%) |
| Light chain type | |
| Kappa | 64 (61.5%) |
| Lambda | 40 (38.5%) |
| Durie–Salmon stage | |
| I | 9 (8.6%) |
| II | 14 (13.3%) |
| III | 82 (78.1%) |
| ISS stage | |
| 1 | 29 (27.9%) |
| 2 | 30 (28.8%) |
| 3 | 45 (43.3%) |
| No data: n = 1 | |
| Renal function | |
| A-creatinine < 2 mg/dL | 87 (82.9%) |
| B-creatinine ≥ 2 mg/dL | 18 (17.1%) |
| Performance status | |
| 0 | 8 (7.9%) |
| 1 | 43 (40.9%) |
| 2 | 42 (40.0%) |
| 3 | 10 (9.9%) |
| 4 | 2 (2.0%) |
| Body weight loss | |
| No | 42 (49.4%) |
| Yes | 43 (50.6%) |
| No data: n = 20 | |
| 5% | 14 (32.6%) |
| 10% | 29 (67.4%) |
| Anemia grade (WHO) | |
| Absent or I | 17 (16.2%) |
| II, III or IV | 88 (83.8%) |
| Treatment protocol | |
| CTD | 27 (25.7%) |
| V(C)D | 41 (39.0%) |
| VTD | 37 (35.2%) |
| del(17p13.1) | |
| Absent | 74 (87.1%) |
| Present | 11 (12.9%) |
| No data: n = 20 | |
| t(4;14) | |
| Absent | 75 (88.2%) |
| Present | 10 (11.8%) |
| No data: n = 20 | |
| t(14;16) | |
| Absent | 84 (98.8%) |
| Present | 1 (1.2%) |
| No data: n = 20 | |
| t(11;14) | 76 (89.4%) |
| Absent | 9 (10.6%) |
| Present | |
| No data: n = 20 | |
| Other IgH gene rearrangement | |
| Absent | 72 (84.7%) |
| Present | 13 (15.3%) |
| No data: n = 20 |
| Variable | Progression-Free Survival | Overall Survival | ||||
|---|---|---|---|---|---|---|
| Univariate | Multivariate c | Univariate | Multivariate d | |||
| Me (months) | p | p | Me (months) | p | p | |
| 16 | HR (95% CI) | HR (95% CI) | 24 | HR (95% CI) | HR (95% CI) | |
| Sex | ||||||
| Men | 24 | 0.0603 | 0.1202 | NR | 0.1665 | 0.1213 |
| Women | 37 | 1.65 (0.97–2.80) | 1.83 (0.86–3.92) | 31 | 1.52 (0.84–2.77) | 2.04 (0.83–4.98) |
| Age | ||||||
| <65 | 39 | 0.0929 | 0.0622 | 33 | 0.8347 | 0.3608 |
| ≥65 | 17 | 0.60 (0.35–1.05) | 0.42 (0.17–1.04) | 41 | 1.07 (0.57–2.01) | 1.64 (0.57–4.69) |
| Diagnosis | ||||||
| Light chain disease, Non-secretory MM | 15 | 0.0987 | 0.5534 | 24 | 0.0004 * | 0.0928 |
| MM with a monoclonal component | 26 | 1.72 (0.78–3.80) | 1.47(0.41–5.25) | NR | 3.01 (1.22–7.42) | 2.77 (0.85–9.06) |
| Monoclonal protein class | ||||||
| IgA | 26 | 0.8713 | 0.13 | NR | 0.2565 | 0.3174 |
| IgG | 25 | 1.05 (0.56–1.97) | 2.11 (0.81–5.54) | NR | 1.50 (0.69–3.23) | 1.73 (0.60–5.00) |
| Light chain type | ||||||
| Lambda | 28 | 0.2866 | 0.7216 | 41 | 0.2857 | 0.992 |
| Kappa | 17 | 0.75 (0.43–1.32) | 0.87 (0.41–1.86) | 33 | 0.72 (0.38–1.37) | 0.99 (0.39–2.54) |
| Durie Salmon stage | ||||||
| III | 17 | 0.9544 | 0.0833 | 31 | 0.0101 * | 0.0014 * |
| I, II | 25 | 1.02 (0.53–1.94) | 2.21 (0.90–5.40) | 41 | 3.06 (1.31–7.15) | 4.84 (1.84–12.72) |
| ISS stage | ||||||
| 3 | 17 | 0.0235 * | 0.0116 * | 33 | 0.1567 | 0.6298 |
| 1, 2 | 39 | 1.82 (1.05–3.15) | 2.59 (1.24–5.41) | 41 | 1.53 (0.83–2.83) | 1.25 (0.51–3.06) |
| Renal function | ||||||
| B | 15 | 0.0044 * | 0.8629 | 25 | 0.0354 * | 0.6035 |
| A | 30 | 2.31 (1.05–5.10) | 1.09 (0.40–3.01) | 41 | 2.03 (0.87–4.76) | 1.39 (0.40–4.86) |
| Stage of chronic kidney disease | ||||||
| G3a/G3b/G4/G5D | 7 | 0.0380 * | 0.6402 | 41 | 0.4027 | 0.8098 |
| G1/G2 | 25 | 2.50 (0.63–9.91) | 1.07 (0.81–1.40) | NR | 1.63 (0.38–6.99) | 1.03 (0.78–1.37) |
| Performance status | ||||||
| 2–4 | 17 | 0.0478 * | 0.2741 | 29 | 0.0023 * | 0.4613 |
| 0, 1 | 42 | 1.73 (1.00–2.98) | 1.51 (0.72–3.17) | NR | 2.64 (1.42–4.88) | 1.40 (0.57–3.42) |
| Treatment protocol (1) | ||||||
| CTD | 24 | 0.2633 | 0.2464 | 33 | 0.9705 | 0.9167 |
| V(C)D, VTD | 25 | 1.35 (0.76–2.40) | 1.83 (0.66–5.07) | 33 | 1.01 (0.53–1.92) | 1.07 (0.31–3.63) |
| Treatment protocol (2) | ||||||
| CTD, V(C)D | 17 | 0.0033 * | 0.0168 * | 33 | 0.3191 | 0.6251 |
| VTD | NR | 2.64 (1.53–4.57) | 2.75 (1.21–6.28) | 41 | 1.41 (0.74–2.66) | 1.28 (0.48–3.43) |
| Body weight loss before treatment | ||||||
| Yes | 18 | 0.0816 | 0.3129 | 30 | 0.0763 | 0.3183 |
| No | 39 | 1.67 (0.93–3.01) | 1.60 (0.64–3.96) | NR | 1.80 (0.93–3.46) | 1.68 (0.61–4.63) |
| Anemia grade before treatment (WHO) | ||||||
| Absent or I | 39 | 0.1645 | 0.5548 | 41 | 0.9537 | 0.3495 |
| II, III or IV | 24 | 0.58 (0.30–1.13) | 0.69 (0.20–2.39) | 41 | 0.98 (0.47–2.03) | 0.37 (0.05–2.93) |
| Hemoglobin | ||||||
| Low | 25 | 0.0459 * | 0.5228 | 41 | 0.8786 | 0.2902 |
| Normal | NR | 2.42 (1.24–4.72) | 1.52 (0.42–5.52) | 33 | 1.06 (0.48–2.32) | 1.89 (0.58–6.12) |
| Platelets | ||||||
| Low | 17 | 0.0233 * | 0.0303 * | 33 | 0.8238 | 0.4022 |
| Normal | 39 | 1.81 (1.04–3.15) | 2.54 (1.10–5.90) | 41 | 1.07 (0.58–1.98) | 1.57 (0.55–4.49) |
| Albumins | ||||||
| Low | 13 | 0.0002 * | 0.0280 * | 25 | 0.0042 * | 0.0899 |
| Normal | 39 | 2.57 (1.45–4.57) | 2.35 (1.10–5.03) | NR | 2.29 (1.21–4.38) | 2.15 (0.89–5.18) |
| CRP | ||||||
| High | 24 | 0.5561 | 0.299 | 31 | 0.1484 | 0.29 |
| Normal | 26 | 1.17 (0.67–2.06) | 1.53 (0.69–3.43) | NR | 1.56 (0.81–3.01) | 1.61 (0.67–3.85) |
| LDH | ||||||
| High | 7 | 0.0041 * | 0.0173 * | 15 | 0.0007 * | 0.0018 * |
| Normal | 25 | 2.58 (0.97–6.87) | 3.28 (1.24–8.66) | NR | 3.01 (1.06–10.30) | 5.94 (1.95–18.11) |
| Calcium | ||||||
| High | 24 | 0.293 | 0.3505 | 28 | 0.0386 * | 0.1036 |
| Normal | 25 | 1.35 (0.73–2.49) | 0.64 (0.26–1.61) | NR | 1.88 (0.93–3.80) | 2.08 (0.86–5.00) |
| B2M | ||||||
| High | 24 | 0.0552 | 0.8337 | 41 | 0.0066 * | 0.5968 |
| Normal | NR | 2.89 (1.35–6.17) | 1.16 (0.28–4.76) | 41 | 3.12 (1.38–7.05) | 0.65 (0.13–3.22) |
| Creatinine | ||||||
| High | 14 | 0.0003 * | 0.0699 | 28 | 0.0622 | 0.3451 |
| Normal | 39 | 2.54 (1.742–4.54) | 2.31 (0.94–5.66) | NR | 1.74 (0.92–3.29) | 1.74 (0.56–5.42) |
| eGFR | ||||||
| Low | 15 | 0.0051 * | 0.7065 | 28 | 0.0623 | 0.8429 |
| Normal | 39 | 2.08 (1.17–3.70) | 1.21 (0.45–3.26) | NR | 1.75 (0.92–3.31) | 1.13 (0.33–3.95) |
| del(17p) | ||||||
| Present | 15 | 0.0410 * | 0.7902 | 30 | 0.068 | 0.0816 |
| Absent | 28 | 2.08 (0.81–5.37) | 1.14 (0.44–2.96) | NR | 2.21 (0.68–7.23) | 2.85 (0.88–9.22) |
| t(4;14) | ||||||
| Present | NR | 0.343 | 0.6425 | NR | 0.8738 | 0.7077 |
| Absent | 24 | 1.74 (0.68–4.43) | 1.38 (0.35–5.44) | 41 | 1.10 (0.35–3.46) | 1.30(0.34–4.99) |
| t(14;16) | ||||||
| Present | 9 | 0.1089 | 0.1191 | 2 | <0.0001 * | 0.0012 * |
| Absent | 25 | 4.29 (0.07–242.17) | 7.15 (0.61–83.86) | 41 | 21.92 (0.00–18528.31) | 81.76 (5.81–11,150.18) |
| t(11;14) | ||||||
| Present | 8 | 0.0033 * | 0.0003 * | 0.7822 | 0.4937 | |
| Absent | 28 | 2.94 (0.93–9.33) | 5.66 (2.22–14.42) | 31 | 1.16 (0.38–3.51) | 1.55 (0.44–5.45) |
| Other IgH gene rearrangement | ||||||
| Present | 25 | 0.9699 | 0.9648 | NR | 0.3936 | 0.8497 |
| Absent | 25 | 1.01 (0.45–2.27) | 0.98 (0.37–2.61) | 41 | 1.66 (0.62–4.44) | 0.88 (0.24–3.18) |
| miRNA-8074 expression (according to the median value) | ||||||
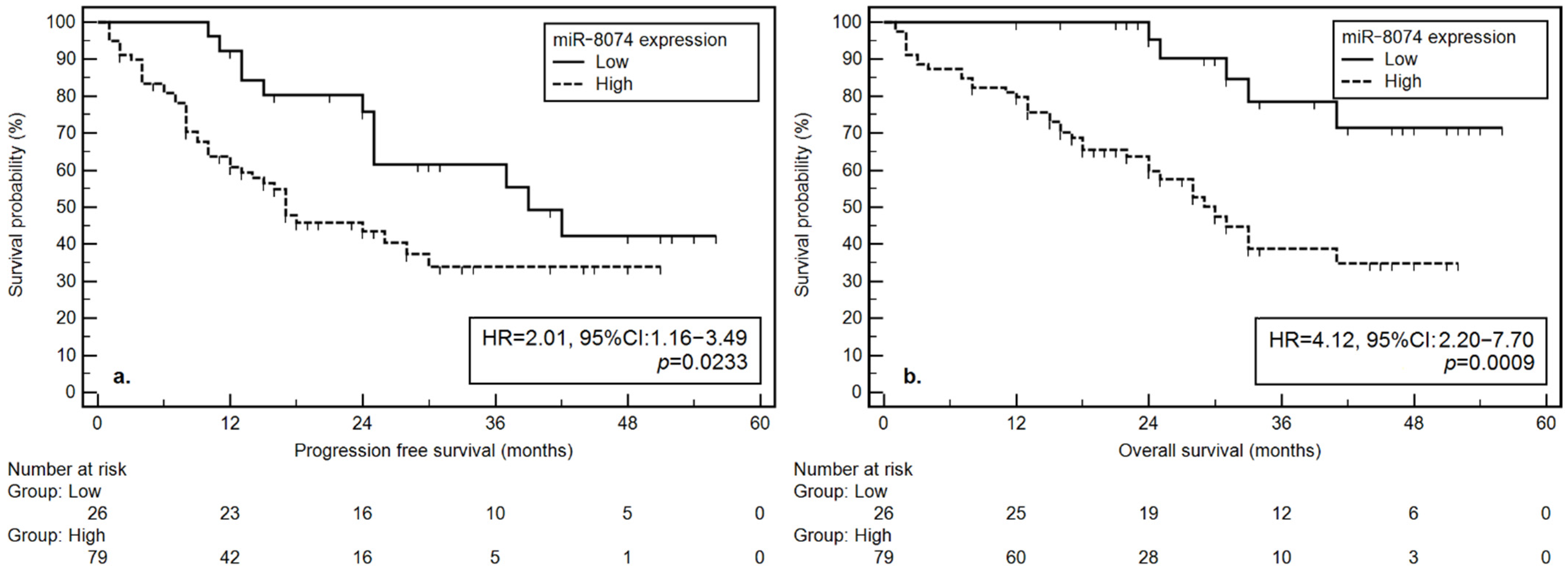
| High | 17 | 0.0233 * | 0.0343 * | 30 | 0.0009 * | 0.0142 * |
| Low | 39 | 2.01 (1.16–3.49) | 2.28 (1.07–4.91) | NR | 4.12 (2.20–7.70) | 3.97 (1.32–11.90) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szudy-Szczyrek, A.; Mlak, R.; Mielnik, M.; Mazurek, M.; Chocholska, S.; Podgajna, M.; Szczyrek, M.; Homa-Mlak, I.; Małecka-Massalska, T.; Hus, M. Circulating Serum MiRNA-8074 as a Novel Prognostic Biomarker for Multiple Myeloma. Cells 2022, 11, 752. https://doi.org/10.3390/cells11040752
Szudy-Szczyrek A, Mlak R, Mielnik M, Mazurek M, Chocholska S, Podgajna M, Szczyrek M, Homa-Mlak I, Małecka-Massalska T, Hus M. Circulating Serum MiRNA-8074 as a Novel Prognostic Biomarker for Multiple Myeloma. Cells. 2022; 11(4):752. https://doi.org/10.3390/cells11040752
Chicago/Turabian StyleSzudy-Szczyrek, Aneta, Radosław Mlak, Michał Mielnik, Marcin Mazurek, Sylwia Chocholska, Martyna Podgajna, Michał Szczyrek, Iwona Homa-Mlak, Teresa Małecka-Massalska, and Marek Hus. 2022. "Circulating Serum MiRNA-8074 as a Novel Prognostic Biomarker for Multiple Myeloma" Cells 11, no. 4: 752. https://doi.org/10.3390/cells11040752
APA StyleSzudy-Szczyrek, A., Mlak, R., Mielnik, M., Mazurek, M., Chocholska, S., Podgajna, M., Szczyrek, M., Homa-Mlak, I., Małecka-Massalska, T., & Hus, M. (2022). Circulating Serum MiRNA-8074 as a Novel Prognostic Biomarker for Multiple Myeloma. Cells, 11(4), 752. https://doi.org/10.3390/cells11040752






