Regulation of Nucleolar Activity by MYC
Abstract
1. Nucleolar Structure and Organization Allow for Flexibility in the Rate of Ribosome Biogenesis
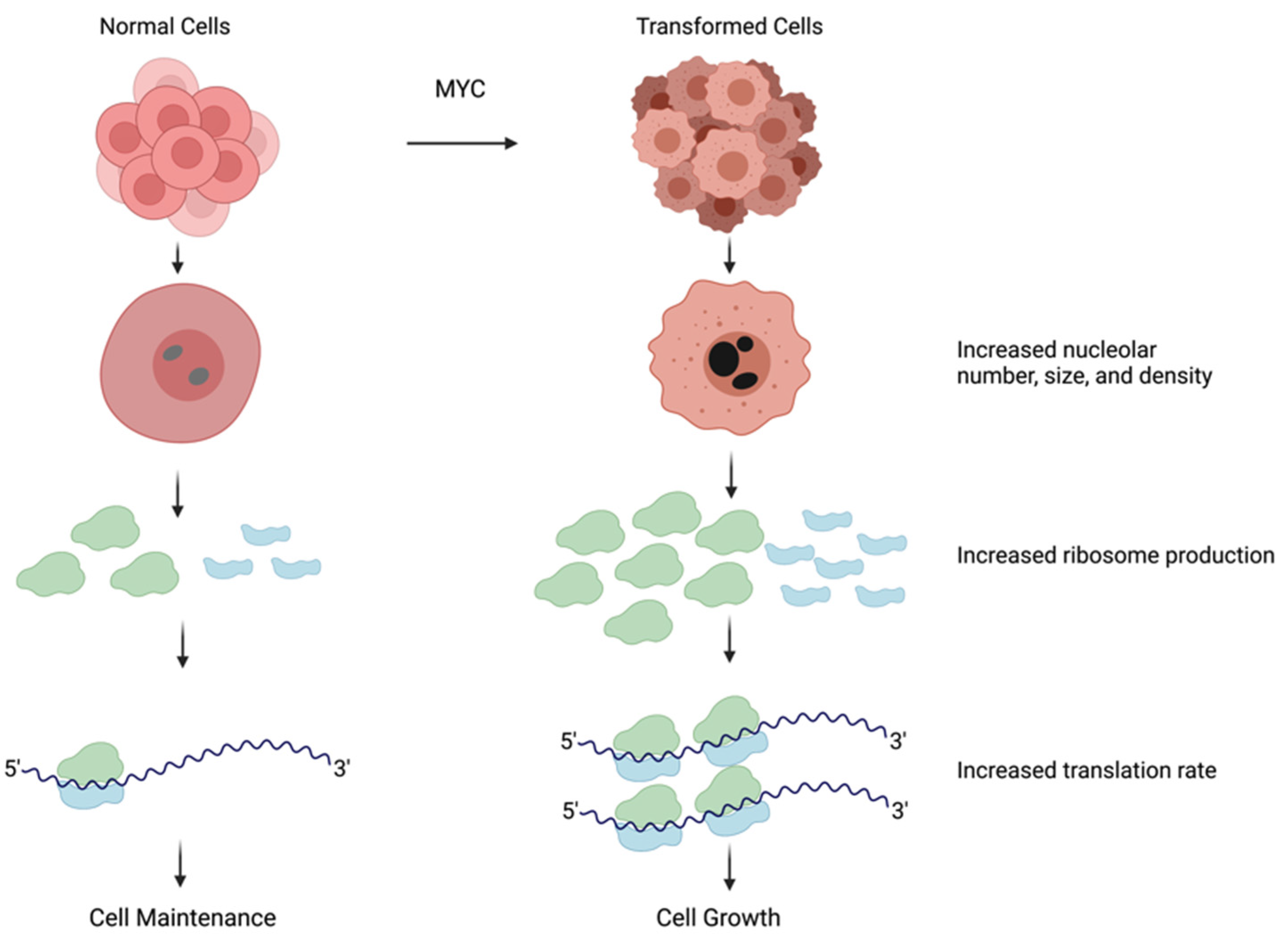
2. Nucleolar Size and Activity Are Increased in Hyperproliferative Cells
3. The MYC Family of Transcription Factors Are Key Regulators of Cell Growth
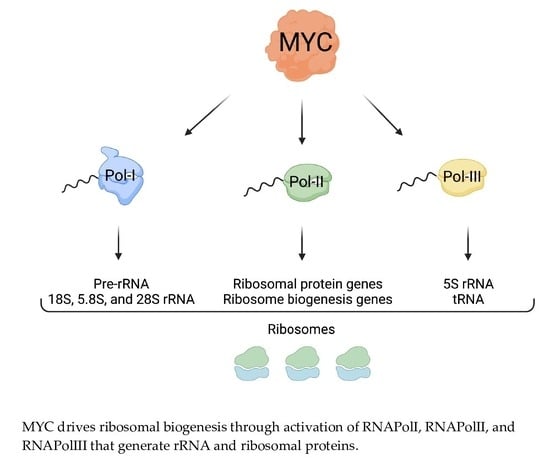
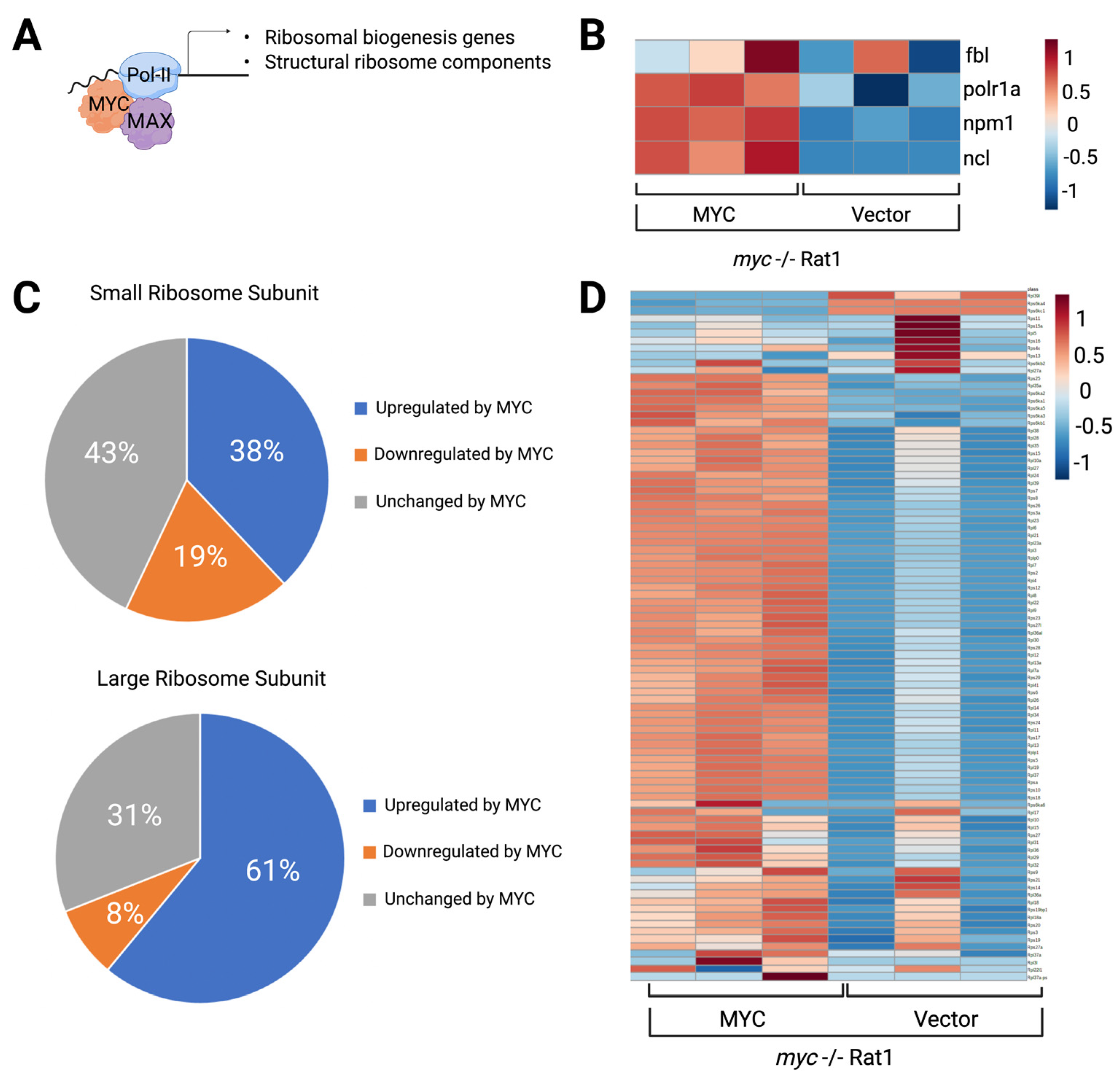
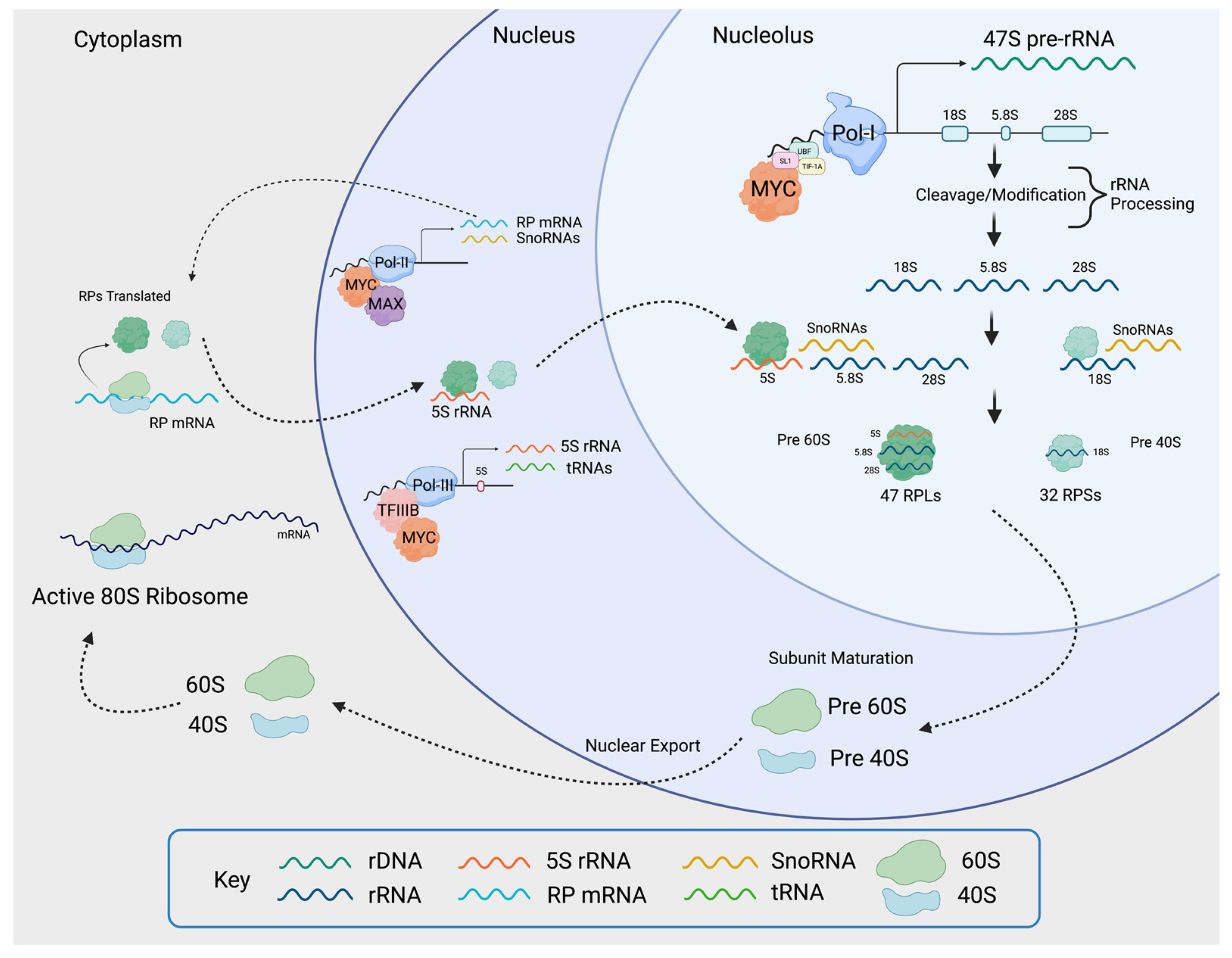
4. MYC Promotes Transcription of rDNA
5. MYC Promotes the Transcription of Genes Encoding RPs and Regulators of Nucleolar Assembly and Activity
6. MYC Promotes the Expression of Genes That Activate Ribosome Biogenesis and Protein Synthesis
7. Nucleolar Assembly and Function Regulate MYC Levels and Activity
8. Targeting Aberrant Nucleolar Activity to Inhibit Cancer Growth
9. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montanaro, L.; Treré, D.; Derenzini, M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef]
- Derenzini, M.; Trerè, D.; Pession, A.; Montanaro, L.; Sirri, V.; Ochs, R.L. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998, 152, 1291–1297. [Google Scholar]
- Thomas, G. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2000, 2, E71–E72. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA 1972, 69, 3394–3398. [Google Scholar] [CrossRef]
- Bersaglieri, C.; Santoro, R. Genome Organization in and around the Nucleolus. Cells 2019, 8, 579. [Google Scholar] [CrossRef] [PubMed]
- Tsekrekou, M.; Stratigi, K.; Chatzinikolaou, G. The Nucleolus: In Genome Maintenance and Repair. Int. J. Mol. Sci. 2017, 18, 1411. [Google Scholar] [CrossRef]
- Anger, A.M.; Armache, J.P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef]
- Martinez-Calvillo, S.; Florencio-Martinez, L.E.; Nepomuceno-Mejia, T. Nucleolar Structure and Function in Trypanosomatid Protozoa. Cells 2019, 8, 421. [Google Scholar] [CrossRef]
- Thiry, M.; Lafontaine, D.L. Birth of a nucleolus: The evolution of nucleolar compartments. Trends Cell Biol. 2005, 15, 194–199. [Google Scholar] [CrossRef]
- Thiry, M.; Lamaye, F.; Lafontaine, D.L. The nucleolus: When 2 became 3. Nucleus 2011, 2, 289–293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davidian, A.; Koshel, E.; Dyomin, A.; Galkina, S.; Saifitdinova, A.; Gaginskaya, E. On some structural and evolutionary aspects of rDNA amplification in oogenesis of Trachemys scripta turtles. Cell Tissue Res. 2021, 383, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Verdun, D.; Roussel, P.; Thiry, M.; Sirri, V.; Lafontaine, D.L. The nucleolus: Structure/function relationship in RNA metabolism. Wiley Interdiscip Rev. RNA 2010, 1, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D. Revisiting the nucleolus: From marker to dynamic integrator of cancer signaling. Sci. Signal. 2012, 5, pe38. [Google Scholar] [CrossRef]
- Destefanis, F.; Manara, V.; Bellosta, P. Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer. Int. J. Mol. Sci. 2020, 21, 4037. [Google Scholar] [CrossRef]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Jantzen, H.M.; Admon, A.; Bell, S.P.; Tjian, R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 1990, 344, 830–836. [Google Scholar] [CrossRef]
- Grummt, I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003, 17, 1691–1702. [Google Scholar] [CrossRef]
- Jin, R.; Zhou, W. TIF-IA: An oncogenic target of pre-ribosomal RNA synthesis. Biochim. Biophys. Acta 2016, 1866, 189–196. [Google Scholar] [CrossRef]
- Szymanski, M.; Barciszewska, M.Z.; Erdmann, V.A.; Barciszewski, J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002, 30, 176–178. [Google Scholar] [CrossRef]
- Ciganda, M.; Williams, N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev. RNA 2011, 2, 523–533. [Google Scholar] [CrossRef]
- Sharma, S.; Lafontaine, D.L.J. ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef]
- Penzo, M.; Guerrieri, A.N.; Zacchini, F.; Treré, D.; Montanaro, L. RNA Pseudouridylation in Physiology and Medicine: For Better and for Worse. Genes 2017, 8, 301. [Google Scholar] [CrossRef] [PubMed]
- Polikanov, Y.S.; Melnikov, S.V.; Söll, D.; Steitz, T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015, 22, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef]
- Russell, J.; Zomerdijk, J.C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005, 30, 87–96. [Google Scholar] [CrossRef]
- Zhang, W.; Cheng, W.; Parlato, R.; Guo, X.; Cui, X.; Dai, C.; Xu, L.; Zhu, J.; Zhu, M.; Luo, K.; et al. Nucleolar stress induces a senescence-like phenotype in smooth muscle cells and promotes development of vascular degeneration. Aging 2020, 12, 22174–22198. [Google Scholar] [CrossRef]
- Lopez-Candales, A.; Holmes, D.R.; Liao, S.; Scott, M.J.; Wickline, S.A.; Thompson, R.W. Decreased vascular smooth muscle cell density in medial degeneration of human abdominal aortic aneurysms. Am. J. Pathol. 1997, 150, 993–1007. [Google Scholar] [PubMed]
- Kang, H.; Shin, J.H. Repression of rRNA transcription by PARIS contributes to Parkinson’s disease. Neurobiol. Dis. 2015, 73, 220–228. [Google Scholar] [CrossRef]
- Sanchez, E.; Laplace-Builhe, B.; Mau-Them, F.T.; Richard, E.; Goldenberg, A.; Toler, T.L.; Guignard, T.; Gatinois, V.; Vincent, M.; Blanchet, C.; et al. POLR1B and neural crest cell anomalies in Treacher Collins syndrome type 4. Genet. Med. 2020, 22, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Dauwerse, J.G.; Dixon, J.; Seland, S.; Ruivenkamp, C.A.; van Haeringen, A.; Hoefsloot, L.H.; Peters, D.J.; Boers, A.C.; Daumer-Haas, C.; Maiwald, R.; et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011, 43, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Gupta, A.; Dutta, A.; Potluri, P.; Batti, B. A Review of Diamond-Blackfan Anemia: Current Evidence on Involved Genes and Treatment Modalities. Cureus 2020, 12, e10019. [Google Scholar] [CrossRef]
- Farrar, J.E.; Quarello, P.; Fisher, R.; O’Brien, K.A.; Aspesi, A.; Parrella, S.; Henson, A.L.; Seidel, N.E.; Atsidaftos, E.; Prakash, S.; et al. Exploiting pre-rRNA processing in Diamond Blackfan anemia gene discovery and diagnosis. Am. J. Hematol. 2014, 89, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Noel, C.B. Diamond-Blackfan anemia RPL35A: A case report. J. Med. Case Rep. 2019, 13, 185. [Google Scholar] [CrossRef]
- Warda, A.S.; Freytag, B.; Haag, S.; Sloan, K.E.; Gorlich, D.; Bohnsack, M.T. Effects of the Bowen-Conradi syndrome mutation in EMG1 on its nuclear import, stability and nucleolar recruitment. Hum. Mol. Genet. 2016, 25, 5353–5364. [Google Scholar] [CrossRef]
- Farooq, M.; Lindbaek, L.; Krogh, N.; Doganli, C.; Keller, C.; Monnich, M.; Goncalves, A.B.; Sakthivel, S.; Mang, Y.; Fatima, A.; et al. RRP7A links primary microcephaly to dysfunction of ribosome biogenesis, resorption of primary cilia, and neurogenesis. Nat. Commun. 2020, 11, 5816. [Google Scholar] [CrossRef]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef]
- White, R.J. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005, 6, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.Y.; Pederson, T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 2014, 28, 3290–3296. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017, 13, e1006994. [Google Scholar] [CrossRef]
- Feng, L.; Du, J.; Yao, C.; Jiang, Z.; Li, T.; Zhang, Q.; Guo, X.; Yu, M.; Xia, H.; Shi, L.; et al. Ribosomal DNA copy number is associated with P53 status and levels of heavy metals in gastrectomy specimens from gastric cancer patients. Environ. Int 2020, 138, 105593. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, M.; Trerè, D.; Pession, A.; Govoni, M.; Sirri, V.; Chieco, P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 2000, 191, 181–186. [Google Scholar] [CrossRef]
- Bywater, M.J.; Poortinga, G.; Sanij, E.; Hein, N.; Peck, A.; Cullinane, C.; Wall, M.; Cluse, L.; Drygin, D.; Anderes, K.; et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 2012, 22, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Arabi, A.; Wu, S.; Ridderstrale, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlen, S.; Hydbring, P.; Soderberg, O.; Grummt, I.; et al. c-myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. c-myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.S.; Li, L.; Orian, A.; Eisenman, R.N.; Edgar, B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005, 7, 295–302. [Google Scholar] [CrossRef]
- Lee, C.M.; Reddy, E.P. The v-myc oncogene. Oncogene 1999, 18, 2997–3003. [Google Scholar] [CrossRef]
- Roussel, M.; Saule, S.; Lagrou, C.; Rommens, C.; Beug, H.; Graf, T.; Stehelin, D. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature 1979, 281, 452–455. [Google Scholar] [CrossRef]
- Sheiness, D.; Bishop, J.M. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus. J. Virol. 1979, 31, 514–521. [Google Scholar] [CrossRef]
- Payne, G.S.; Bishop, J.M.; Varmus, H.E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 1982, 295, 209–214. [Google Scholar] [CrossRef]
- Hayward, W.S.; Neel, B.G.; Astrin, S.M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 1981, 290, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef] [PubMed]
- Pelengaris, S.; Khan, M.; Evan, G. c-myc: More than just a matter of life and death. Nat. Rev. Cancer 2002, 2, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Lafita-Navarro, M.C.; Kim, M.; Borenstein-Auerbach, N.; Venkateswaran, N.; Hao, Y.H.; Ray, R.; Brabletz, T.; Scaglioni, P.P.; Shay, J.W.; Conacci-Sorrell, M. The aryl hydrocarbon receptor regulates nucleolar activity and protein synthesis in Myc-expressing cells. Genes Dev. 2018, 32, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- van Riggelen, J.; Yetil, A.; Felsher, D.W. Myc as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. Myc on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Cascón, A.; Robledo, M. MAX and Myc: A heritable breakup. Cancer Res. 2012, 72, 3119–3124. [Google Scholar] [CrossRef]
- Eilers, M.; Eisenman, R.N. Myc’s broad reach. Genes Dev. 2008, 22, 2755–2766. [Google Scholar] [CrossRef]
- Grandori, C.; Cowley, S.M.; James, L.P.; Eisenman, R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000, 16, 653–699. [Google Scholar] [CrossRef]
- Sammak, S.; Hamdani, N.; Gorrec, F.; Allen, M.D.; Freund, S.M.V.; Bycroft, M.; Zinzalla, G. Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-myc:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA. Biochemistry 2019, 58, 3144–3154. [Google Scholar] [CrossRef]
- Kretzner, L.; Blackwood, E.M.; Eisenman, R.N. Myc and Max proteins possess distinct transcriptional activities. Nature 1992, 359, 426–429. [Google Scholar] [CrossRef]
- Amati, B.; Brooks, M.W.; Levy, N.; Littlewood, T.D.; Evan, G.I.; Land, H. Oncogenic activity of the c-myc protein requires dimerization with Max. Cell 1993, 72, 233–245. [Google Scholar] [CrossRef]
- Izumi, H.; Imamura, T.; Nagatani, G.; Ise, T.; Murakami, T.; Uramoto, H.; Torigoe, T.; Ishiguchi, H.; Yoshida, Y.; Nomoto, M.; et al. Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3’-->5’ exonuclease activity. Nucleic Acids Res. 2001, 29, 1200–1207. [Google Scholar] [CrossRef]
- Gomez-Roman, N.; Grandori, C.; Eisenman, R.N.; White, R.J. Direct activation of RNA polymerase III transcription by c-myc. Nature 2003, 421, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Kenneth, N.S.; Ramsbottom, B.A.; Gomez-Roman, N.; Marshall, L.; Cole, P.A.; White, R.J. TRRAP and GCN5 are used by c-myc to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 14917–14922. [Google Scholar] [CrossRef]
- Conacci-Sorrell, M.; Ngouenet, C.; Eisenman, R.N. Myc-nick: A cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 2010, 142, 480–493. [Google Scholar] [CrossRef]
- Conacci-Sorrell, M.; Ngouenet, C.; Anderson, S.; Brabletz, T.; Eisenman, R.N. Stress-induced cleavage of Myc promotes cancer cell survival. Genes Dev. 2014, 28, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Poudel, K.R.; Roh-Johnson, M.; Brabletz, T.; Yu, M.; Borenstein-Auerbach, N.; Grady, W.N.; Bai, J.; Moens, C.B.; Eisenman, R.N.; et al. Myc-nick promotes cell migration by inducing fascin expression and Cdc42 activation. Proc. Natl. Acad. Sci. USA 2016, 113, E5481–E5490. [Google Scholar] [CrossRef] [PubMed]
- Flinn, E.M.; Busch, C.M.; Wright, A.P. Myc boxes, which are conserved in Myc family proteins, are signals for protein degradation via the proteasome. Mol. Cell. Biol. 1998, 18, 5961–5969. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R.; Thompson, C.B.; Eisenman, R.N. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature 1985, 314, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Welcker, M.; Orian, A.; Grim, J.E.; Grim, J.A.; Eisenman, R.N.; Clurman, B.E. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-myc and cell size. Curr. Biol. 2004, 14, 1852–1857. [Google Scholar] [CrossRef]
- Choi, S.H.; Wright, J.B.; Gerber, S.A.; Cole, M.D. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010, 24, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Bahram, F.; von der Lehr, N.; Cetinkaya, C.; Larsson, L.G. c-myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 2000, 95, 2104–2110. [Google Scholar] [CrossRef] [PubMed]
- Ekholm-Reed, S.; Spruck, C.H.; Sangfelt, O.; van Drogen, F.; Mueller-Holzner, E.; Widschwendter, M.; Zetterberg, A.; Reed, S.I.; Reed, S.E. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004, 64, 795–800. [Google Scholar] [CrossRef]
- Spruck, C.H.; Strohmaier, H.; Sangfelt, O.; Müller, H.M.; Hubalek, M.; Müller-Holzner, E.; Marth, C.; Widschwendter, M.; Reed, S.I. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002, 62, 4535–4539. [Google Scholar]
- Rajagopalan, H.; Jallepalli, P.V.; Rago, C.; Velculescu, V.E.; Kinzler, K.W.; Vogelstein, B.; Lengauer, C. Inactivation of hCDC4 can cause chromosomal instability. Nature 2004, 428, 77–81. [Google Scholar] [CrossRef]
- Koepp, D.M.; Schaefer, L.K.; Ye, X.; Keyomarsi, K.; Chu, C.; Harper, J.W.; Elledge, S.J. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 2001, 294, 173–177. [Google Scholar] [CrossRef]
- Gupta-Rossi, N.; Le Bail, O.; Gonen, H.; Brou, C.; Logeat, F.; Six, E.; Ciechanover, A.; Israël, A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 2001, 276, 34371–34378. [Google Scholar] [CrossRef]
- Wu, G.; Lyapina, S.; Das, I.; Li, J.; Gurney, M.; Pauley, A.; Chui, I.; Deshaies, R.J.; Kitajewski, J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 2001, 21, 7403–7415. [Google Scholar] [CrossRef]
- Nateri, A.S.; Riera-Sans, L.; Da Costa, C.; Behrens, A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 2004, 303, 1374–1378. [Google Scholar] [CrossRef]
- Welcker, M.; Orian, A.; Jin, J.; Grim, J.E.; Grim, J.A.; Harper, J.W.; Eisenman, R.N.; Clurman, B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-myc protein degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 9085–9090. [Google Scholar] [CrossRef]
- Yada, M.; Hatakeyama, S.; Kamura, T.; Nishiyama, M.; Tsunematsu, R.; Imaki, H.; Ishida, N.; Okumura, F.; Nakayama, K.; Nakayama, K.I. Phosphorylation-dependent degradation of c-myc is mediated by the F-box protein Fbw7. EMBO J. 2004, 23, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.; de Lange, T.; Ramsay, G.; Jakobovits, E.; Bishop, J.M.; Varmus, H.; Lee, W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol. Cell. Biol. 1987, 7, 1697–1709. [Google Scholar] [CrossRef]
- Lee, K.K.; Workman, J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007, 8, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.D.; Cowling, V.H. Transcription-independent functions of Myc: Regulation of translation and DNA replication. Nat. Rev. Mol. Cell Biol. 2008, 9, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.R.; Schroeder, M.; Fernandez, P.; Taubert, S.; Amati, B. Binding of c-myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001, 15, 2069–2082. [Google Scholar] [CrossRef]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-myc and E2F oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef]
- Vervoorts, J.; Lüscher-Firzlaff, J.M.; Rottmann, S.; Lilischkis, R.; Walsemann, G.; Dohmann, K.; Austen, M.; Lüscher, B. Stimulation of c-myc transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003, 4, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Herbst, A.; Hemann, M.T.; Tworkowski, K.A.; Salghetti, S.E.; Lowe, S.W.; Tansey, W.P. A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO Rep. 2005, 6, 177–183. [Google Scholar] [CrossRef]
- Cowling, V.H.; Chandriani, S.; Whitfield, M.L.; Cole, M.D. A conserved Myc protein domain, MBIV, regulates DNA binding, apoptosis, transformation, and G2 arrest. Mol. Cell. Biol. 2006, 26, 4226–4239. [Google Scholar] [CrossRef] [PubMed]
- Panov, K.I.; Friedrich, J.K.; Russell, J.; Zomerdijk, J.C. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006, 25, 3310–3322. [Google Scholar] [CrossRef]
- Friedrich, J.K.; Panov, K.I.; Cabart, P.; Russell, J.; Zomerdijk, J.C. TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J. Biol. Chem. 2005, 280, 29551–29558. [Google Scholar] [CrossRef]
- Shiue, C.N.; Berkson, R.G.; Wright, A.P. c-myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene 2009, 28, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Shiue, C.N.; Arabi, A.; Wright, A.P. Nucleolar organization, growth control and cancer. Epigenetics 2010, 5, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Shiue, C.N.; Nematollahi-Mahani, A.; Wright, A.P. Myc-induced anchorage of the rDNA IGS region to nucleolar matrix modulates growth-stimulated changes in higher-order rDNA architecture. Nucleic Acids Res. 2014, 42, 5505–5517. [Google Scholar] [CrossRef]
- Patel, J.H.; Loboda, A.P.; Showe, M.K.; Showe, L.C.; McMahon, S.B. Analysis of genomic targets reveals complex functions of Myc. Nat. Rev. Cancer 2004, 4, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Cole, M.D. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 2006, 16, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Perna, D.; Fagà, G.; Verrecchia, A.; Gorski, M.M.; Barozzi, I.; Narang, V.; Khng, J.; Lim, K.C.; Sung, W.K.; Sanges, R.; et al. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene 2012, 31, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.H.; Lafita-Navarro, M.C.; Zacharias, L.; Borenstein-Auerbach, N.; Kim, M.; Barnes, S.; Kim, J.; Shay, J.; DeBerardinis, R.J.; Conacci-Sorrell, M. Induction of LEF1 by Myc activates the WNT pathway and maintains cell proliferation. Cell Commun. Signal. 2019, 17, 129. [Google Scholar] [CrossRef]
- Lafita-Navarro, M.C.; Perez-Castro, L.; Zacharias, L.G.; Barnes, S.; DeBerardinis, R.J.; Conacci-Sorrell, M. The transcription factors aryl hydrocarbon receptor and Myc cooperate in the regulation of cellular metabolism. J. Biol. Chem. 2020, 295, 12398–12407. [Google Scholar] [CrossRef]
- Ruggero, D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009, 69, 8839–8843. [Google Scholar] [CrossRef]
- Ginisty, H.; Amalric, F.; Bouvet, P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998, 17, 1476–1486. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Stanley, C.B.; Nourse, A.; Onuchic, P.L.; Banerjee, P.R.; Phillips, A.H.; Park, C.G.; Deniz, A.A.; Kriwacki, R.W. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 2018, 9, 842. [Google Scholar] [CrossRef]
- Maggi, L.B.; Kuchenruether, M.; Dadey, D.Y.; Schwope, R.M.; Grisendi, S.; Townsend, R.R.; Pandolfi, P.P.; Weber, J.D. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol. Cell. Biol. 2008, 28, 7050–7065. [Google Scholar] [CrossRef]
- Li, Z.; Hann, S.R. The Myc-nucleophosmin-ARF network: A complex web unveiled. Cell Cycle 2009, 8, 2703–2707. [Google Scholar] [CrossRef]
- Lafita-Navarro, M.C.; Venkateswaran, N.; Kilgore, J.A.; Kanji, S.; Han, J.; Barnes, S.; Williams, N.S.; Buszczak, M.; Burma, S.; Conacci-Sorrell, M. Inhibition of the de novo pyrimidine biosynthesis pathway limits ribosomal RNA transcription causing nucleolar stress in glioblastoma cells. PLoS Genet. 2020, 16, e1009117. [Google Scholar] [CrossRef] [PubMed]
- Hubackova, S.; Davidova, E.; Boukalova, S.; Kovarova, J.; Bajzikova, M.; Coelho, A.; Terp, M.G.; Ditzel, H.J.; Rohlena, J.; Neuzil, J. Replication and ribosomal stress induced by targeting pyrimidine synthesis and cellular checkpoints suppress p53-deficient tumors. Cell Death Dis. 2020, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan, T.W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Li, F.; Handler, J.; Huang, C.R.; Xiang, Y.; Neretti, N.; Sedivy, J.M.; Zeller, K.I.; Dang, C.V. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE 2008, 3, e2722. [Google Scholar] [CrossRef]
- Eberhardy, S.R.; Farnham, P.J. c-myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 2001, 276, 48562–48571. [Google Scholar] [CrossRef] [PubMed]
- Rahl, P.B.; Young, R.A. Myc and transcription elongation. Cold Spring Harb. Perspect. Med. 2014, 4, a020990. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, T.; Price, D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012, 81, 119–143. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.V. The role of c-myc in regulation of translation initiation. Oncogene 2004, 23, 3217–3221. [Google Scholar] [CrossRef]
- Li, Z.; Boone, D.; Hann, S.R. Nucleophosmin interacts directly with c-myc and controls c-myc-induced hyperproliferation and transformation. Proc. Natl. Acad. Sci. USA 2008, 105, 18794–18799. [Google Scholar] [CrossRef]
- Li, Z.; Hann, S.R. Nucleophosmin is essential for c-myc nucleolar localization and c-myc-mediated rDNA transcription. Oncogene 2013, 32, 1988–1994. [Google Scholar] [CrossRef]
- Dai, M.S.; Arnold, H.; Sun, X.X.; Sears, R.; Lu, H. Inhibition of c-myc activity by ribosomal protein L11. EMBO J. 2007, 26, 3332–3345. [Google Scholar] [CrossRef]
- Liao, J.M.; Zhou, X.; Gatignol, A.; Lu, H. Ribosomal proteins L5 and L11 co-operatively inactivate c-myc via RNA-induced silencing complex. Oncogene 2014, 33, 4916–4923. [Google Scholar] [CrossRef]
- Burger, K.; Muhl, B.; Harasim, T.; Rohrmoser, M.; Malamoussi, A.; Orban, M.; Kellner, M.; Gruber-Eber, A.; Kremmer, E.; Holzel, M.; et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 2010, 285, 12416–12425. [Google Scholar] [CrossRef]
- Hannan, K.M.; Sanij, E.; Rothblum, L.I.; Hannan, R.D.; Pearson, R.B. Dysregulation of RNA polymerase I transcription during disease. Biochim. Biophys. Acta 2013, 1829, 342–360. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Rice, W.G.; Grummt, I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharm. Toxicol. 2010, 50, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Hein, N.; Hannan, K.M.; George, A.J.; Sanij, E.; Hannan, R.D. The nucleolus: An emerging target for cancer therapy. Trends Mol. Med. 2013, 19, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Andrews, W.J.; Panova, T.; Normand, C.; Gadal, O.; Tikhonova, I.G.; Panov, K.I. Old drug, new target: Ellipticines selectively inhibit RNA polymerase I transcription. J. Biol. Chem. 2013, 288, 4567–4582. [Google Scholar] [CrossRef]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharm. 2019, 159, 74–81. [Google Scholar] [CrossRef]
- Derenzini, M.; Montanaro, L.; Trere, D. What the nucleolus says to a tumour pathologist. Histopathology 2009, 54, 753–762. [Google Scholar] [CrossRef]
- Woods, S.J.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim. Biophys. Acta 2015, 1849, 821–829. [Google Scholar] [CrossRef]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascues, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Carotenuto, P.; Pecoraro, A.; Palma, G.; Russo, G.; Russo, A. Therapeutic Approaches Targeting Nucleolus in Cancer. Cells 2019, 8, 1090. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Horn, H.F.; Vousden, K.H. Coping with stress: Multiple ways to activate p53. Oncogene 2007, 26, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef] [PubMed]
- Kofuji, S.; Hirayama, A.; Eberhardt, A.O.; Kawaguchi, R.; Sugiura, Y.; Sampetrean, O.; Ikeda, Y.; Warren, M.; Sakamoto, N.; Kitahara, S.; et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat. Cell Biol. 2019, 21, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Ni, M.; Chalishazar, M.D.; Huffman, K.E.; Kim, J.; Cai, L.; Shi, X.; Cai, F.; Zacharias, L.G.; Ireland, A.S.; et al. Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers. Cell Metab. 2018, 28, 369–382 e365. [Google Scholar] [CrossRef]
- Bailly, A.; Perrin, A.; Bou Malhab, L.J.; Pion, E.; Larance, M.; Nagala, M.; Smith, P.; O’Donohue, M.F.; Gleizes, P.E.; Zomerdijk, J.; et al. The NEDD8 inhibitor MLN4924 increases the size of the nucleolus and activates p53 through the ribosomal-Mdm2 pathway. Oncogene 2016, 35, 415–426. [Google Scholar] [CrossRef]
- Swords, R.T.; Erba, H.P.; DeAngelo, D.J.; Bixby, D.L.; Altman, J.K.; Maris, M.; Hua, Z.; Blakemore, S.J.; Faessel, H.; Sedarati, F.; et al. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: A phase 1 study. Br. J. Haematol. 2015, 169, 534–543. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- CRICK, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar]
- BRENNER, S.; JACOB, F.; MESELSON, M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 1961, 190, 576–581. [Google Scholar] [CrossRef]
- Dinman, J.D. Pathways to Specialized Ribosomes: The Brussels Lecture. J. Mol. Biol. 2016, 428, 2186–2194. [Google Scholar] [CrossRef]
- Shi, Z.; Barna, M. Translating the genome in time and space: Specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 2015, 31, 31–54. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 2017, 67, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, J.C.; Zavolan, M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016, 17, 236. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Warner, J.R. Ribosome-omics of the human ribosome. RNA 2014, 20, 1004–1013. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- Wool, I.G. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 1996, 21, 164–165. [Google Scholar] [CrossRef]
- Lai, M.D.; Xu, J. Ribosomal proteins and colorectal cancer. Curr. Genom. 2007, 8, 43–49. [Google Scholar] [CrossRef]
- Golomb, L.; Volarevic, S.; Oren, M. p53 and ribosome biogenesis stress: The essentials. FEBS Lett. 2014, 588, 2571–2579. [Google Scholar] [CrossRef]
- Russo, A.; Russo, G. Ribosomal Proteins Control or Bypass p53 during Nucleolar Stress. Int. J. Mol. Sci. 2017, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Lee, S.Y.; Gutierrez, A.; Perrigoue, J.; Thapa, R.J.; Tu, Z.; Jeffers, J.R.; Rhodes, M.; Anderson, S.; Oravecz, T.; et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 2012, 120, 3764–3773. [Google Scholar] [CrossRef] [PubMed]
- Pogue-Geile, K.; Geiser, J.R.; Shu, M.; Miller, C.; Wool, I.G.; Meisler, A.I.; Pipas, J.M. Ribosomal protein genes are overexpressed in colorectal cancer: Isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol. Cell. Biol. 1991, 11, 3842–3849. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, I.N.; Lafita-Navarro, M.C.; Conacci-Sorrell, M. Regulation of Nucleolar Activity by MYC. Cells 2022, 11, 574. https://doi.org/10.3390/cells11030574
Brown IN, Lafita-Navarro MC, Conacci-Sorrell M. Regulation of Nucleolar Activity by MYC. Cells. 2022; 11(3):574. https://doi.org/10.3390/cells11030574
Chicago/Turabian StyleBrown, Isabella N., M. Carmen Lafita-Navarro, and Maralice Conacci-Sorrell. 2022. "Regulation of Nucleolar Activity by MYC" Cells 11, no. 3: 574. https://doi.org/10.3390/cells11030574
APA StyleBrown, I. N., Lafita-Navarro, M. C., & Conacci-Sorrell, M. (2022). Regulation of Nucleolar Activity by MYC. Cells, 11(3), 574. https://doi.org/10.3390/cells11030574