Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities
Abstract
:1. Introduction
1.1. Role of NKT in Virus Infection
1.2. Role of MAIT in Virus Infection
1.3. Role of γδ T Cells in Virus Infection
1.4. Role of ILC in Virus Infection
2. Search Strategy
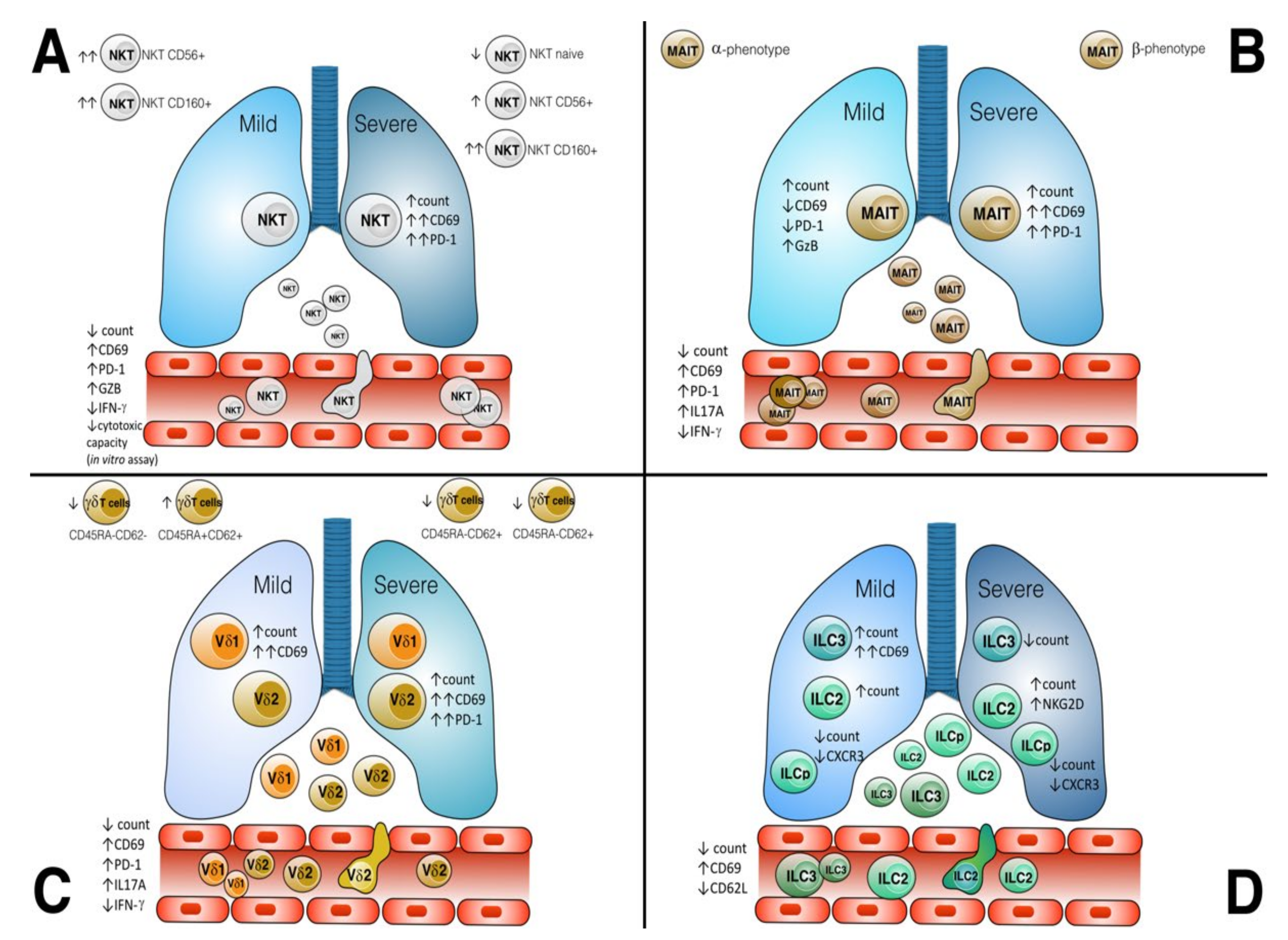
3. Results
3.1. NKT
3.2. MAIT
3.3. γδ T Cells
3.4. ILC
4. Remarkable Considerations and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Fathi, N.; Rezaei, N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol. Int. 2020, 44, 1792–1797. [Google Scholar] [CrossRef]
- Allegra, A.; Di Gioacchino, M.; Tonacci, A.; Musolino, C.; Gangemi, S. Immunopathology of SARS-CoV-2 infection: Immune cells and mediators, prognostic factors, and immune-therapeutic implications. Int. J. Mol. Sci. 2020, 21, 4782. [Google Scholar] [CrossRef]
- Rha, M.-S.; Shin, E.-C. Activation or exhaustion of CD8+ T cells in patients with COVID-19. Cell. Mol. Immunol. 2021, 18, 2325–2333. [Google Scholar] [CrossRef]
- Godfrey, D.; Uldrich, A.; McCluskey, J.; Rossjohn, J.; Moody, D.B. The burgeoning family of unconventional T cells. Nat. Immunol. 2015, 16, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E. The discovery of innate lymphoid cells. Nat. Rev. Immunol. 2021, 21, 616. [Google Scholar] [CrossRef] [PubMed]
- Lantz, O.; Bendelac, A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J. Exp. Med. 1994, 180, 1097–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfrey, D.I.; MacDonald, H.R.; Kronenberg, M.; Smyth, M.J.; Van Kaer, L. NKT cells: What’s in a name? Nat. Rev. Immunol. 2004, 4, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Bendelac, A.; Rivera, M.N.; Park, S.-H.; Roark, J.H. MOUSE CD1-SPECIFIC NK1 T CELLS: Development, Specificity, and Function. Annu. Rev. Immunol. 1997, 15, 535–562. [Google Scholar] [CrossRef]
- Bendelac, A.; Savage, P.B.; Teyton, L. The Biology of NKT Cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef] [Green Version]
- Birkholz, A.; Kronenberg, M. Antigen specificity of invariant natural killer T-cells. Biomed. J. 2015, 38, 470–483. [Google Scholar] [CrossRef] [Green Version]
- Cameron, G.I.; Godfrey, D. Differential surface phenotype and context-dependent reactivity of functionally diverse NKT cells. Immunol. Cell Biol. 2018, 96, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Crosby, C.M.; Kronenberg, M. Tissue-specific functions of invariant natural killer T cells. Nat. Rev. Immunol. 2018, 18, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; McCarthy, C.; Drakesmith, H.; Li, D.; Cerundolo, V.; McMichael, A.J.; Screaton, G.R.; Xu, X.-N. HIV-1 down-regulates the expression of CD1d via Nef. Eur. J. Immunol. 2006, 36, 278–286. [Google Scholar] [CrossRef]
- Van Der Vliet, H.J.J.; Van Vonderen, M.G.A.; Molling, J.W.; Bontkes, H.J.; Reijm, M.; Reiss, P.; Van Agtmael, M.A.; Danner, S.A.; Eertwegh, A.J.M.V.D.; Von Blomberg, B.M.E.; et al. Cutting Edge: Rapid Recovery of NKT Cells upon Institution of Highly Active Antiretroviral Therapy for HIV-1 Infection. J. Immunol. 2006, 177, 5775–5778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessmer, M.S.; Fatima, A.; Paget, C.; Trottein, F.; Brossay, L. NKT cell immune responses to viral infection. Expert Opin. Ther. Targets 2008, 13, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wesley, J.D.; Tessmer, M.S.; Chaukos, D.; Brossay, L. NK Cell–Like Behavior of Vα14i NK T Cells during MCMV Infection. PLOS Pathog. 2008, 4, e1000106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyznik, A.J.; Tupin, E.; Nagarajan, N.A.; HerM, J.; Benedict, C.A.; Kronenberg, M. The mechanism of invariant NKT cell responses to viral danger signals1. J. Immunol. 2008, 181, 4452–4456. [Google Scholar] [CrossRef] [Green Version]
- Ho, L.; Denney, L.; Luhn, K.; Teoh, D.; Clelland, C.; McMichael, A.J. Activation of invariant NKT cells enhances the innate immune response and improves the disease course in influenza A virus infection. Eur. J. Immunol. 2008, 38, 1913–1922. [Google Scholar] [CrossRef]
- Vogt, S.; Mattner, J. NKT Cells Contribute to the Control of Microbial Infections. Front. Cell. Infect. Microbiol. 2021, 11, 11–877. [Google Scholar] [CrossRef]
- Fergusson, J.; Smith, K.E.; Fleming, V.; Rajoriya, N.; Newell, E.; Simmons, R.; Marchi, E.; Björkander, S.; Kang, Y.-H.; Swadling, L.; et al. CD161 Defines a Transcriptional and Functional Phenotype across Distinct Human T Cell Lineages. Cell Rep. 2014, 9, 1075–1088. [Google Scholar] [CrossRef] [Green Version]
- Chiba, A.; Murayama, G.; Miyake, S. Mucosal-Associated Invariant T Cells in Autoimmune Diseases. Front. Immunol. 2018, 9, 1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahimpour, A.; Koay, H.F.; Enders, A.; Clanchy, R.; Eckle, S.B.; Meehan, B.; Chen, Z.; Whittle, B.; Liu, L.; Fairlie, D.P.; et al. Identification of phenotypically and functionally heterogeneous mouse mucosal- associated invariant T cells using MR1 te-tramers. J. Exp. Med. 2015, 212, 1095–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wilgenburg, B.; Loh, L.; Chen, Z.; Pediongco, T.J.; Wang, H.; Shi, M.; Zhao, Z.; Koutsakos, M.; Nüssing, S.; Sant, S.; et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayday, A.C. γδ T Cell Update: Adaptate Orchestrators of Immune Surveillance. J. Immunol. 2019, 203, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Räikkönen, J.; Crockett, J.C.; Rogers, M.J.; Mönkkönen, H.; Auriola, S.; Mönkkönen, J. Zoledronic acid induces formation of a pro-apoptotic ATP analogue and isopentenyl pyrophosphate in osteoclasts in vivo and in MCF-7 cells in vitro. J. Cereb. Blood Flow Metab. 2009, 157, 427–435. [Google Scholar] [CrossRef] [Green Version]
- Benzaïd, I.; Mönkkönen, H.; Stresing, V.; Bonnelye, E.; Green, J.; Monkkonen, J.; Touraine, J.L.; Clezardin, P. High phos-phoantigen levels in bisphosphonate-treated human breast tumors promote Vgamma9Vdelta2 T-cell chemotaxis and cyto-toxicity in vivo. Cancer Res. 2011, 71, 4562–4572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Presti, E.; Dieli, F.; Fourniè, J.J.; Meraviglia, S. Deciphering human γδ T cell response in cancer: Lessons from tumor infil-trating γδ T cells. Immunol. Rev. 2020, 298, 153–164. [Google Scholar] [CrossRef]
- Davey, M.S.; Willcox, C.R.; Hunter, S.; Kasatskaya, S.A.; Remmerswaal, E.B.M.; Salim, M.; Mohammed, F.; Bemelman, F.J.; Chudakov, D.M.; Oo, Y.H.; et al. The human Vδ2+ T-cell compartment comprises distinct innate- like Vγ9+ and adaptive Vγ9- subsets. Nat. Comm. 2018, 9, 1760. [Google Scholar] [CrossRef]
- Juno, J.A.; Kent, S.J. What can gamma delta t cells contribute to an HIV cure? Front. Cell. Inf. Microb. 2020, 10, 233. [Google Scholar] [CrossRef]
- Poccia, F.; Agrati, C.; Martini, F.; Capobianchi, M.R.; Wallace, M.; Miroslav, M. Antiviral reactivities of t cells. Micr. Inf. 2005, 7, 518–528. [Google Scholar] [CrossRef]
- Xi, X.; Han, X.; Li, L.; Zhao, Z. Identification of a new tuberculosis antigen recognized by γδ T cell receptor. Clin. Vacc. Immunol. 2013, 20, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Déchanet, J.; Merville, P.; Lim, A.; Retière, C.; Pitard, V.; Lafarge, X.; Michelson, S.; Méric, C.; Hallet, M.-M.; Kourilsky, P.; et al. Implication of γδ T cells in the human immune response to cytomegalovirus. J. Clin. Investig. 1999, 103, 1437–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buonocore, S.; Ahern, P.P.; Uhlig, H.H.; Ivanov, I.I.; Littman, D.R.; Maloy, K.J.; Powrie, F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 2010, 464, 1371–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.S.; Artis, D. Group 2 Innate Lymphoid Cells in Health and Disease. Cold Spring Harb. Perspect. Biol. 2015, 7, a016337. [Google Scholar] [CrossRef] [PubMed]
- Ardain, A.; Porterfield, J.Z.; Kløverpris, H.; Leslie, A. Type 3 ILCs in Lung Disease. Front. Immunol. 2019, 10, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrega, P.; Loiacono, F.; Di Carlo, E.; Scaramuccia, A.; Mora, M.; Conte, R.; Benelli, R.; Spaggiari, G.M.; Cantoni, C.; Campana, S.; et al. NCR+ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat. Commun. 2015, 6, 8280. [Google Scholar] [CrossRef] [Green Version]
- Cella, M.; Gamini, R.; Sécca, C.; Collins, P.; Zhao, S.; Peng, V.; Robinette, M.L.; Schettini, J.; Zaitsev, K.; Gordon, W.; et al. Subsets of ILC3−ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 2019, 20, 980–991. [Google Scholar] [CrossRef]
- Hoffmann, J.P.; Kolls, J.K.; McCombs, J.E. Regulation and Function of ILC3s in Pulmonary Infections. Front. Immunol. 2021, 12, 672523. [Google Scholar] [CrossRef] [PubMed]
- Jouan, Y.; Guillon, A.; Gonzalez, L.; Perez, Y.; Boisseau, C.; Ehrmann, S.; Ferreira, M.; Daix, T.; Jeannet, R.; François, B.; et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J. Exp. Med. 2020, 217, 217. [Google Scholar] [CrossRef] [PubMed]
- Parrot, T.; Gorin, J.-B.; Ponzetta, A.; Maleki, K.T.; Kammann, T.; Emgård, J.; Perez-Potti, A.; Sekine, T.; Rivera-Ballesteros, O.; The Karolinska COVID-19 Study Group; et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020, 5, 1670. [Google Scholar] [CrossRef] [PubMed]
- Odak, I.; Barros-Martins, J.; Bošnjak, B.; Stahl, K.; David, S.; Wiesner, O.; Busch, M.; Hoeper, M.M.; Pink, I.; Welte, T.; et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine 2020, 57, 102885. [Google Scholar] [CrossRef] [PubMed]
- Tomi, S.; Aoki, J.; Stevanovi, D.; IliA, N.; Gruden-Movsesijan, A.; DiniÄ, M.; RadojeviÄ, D.; BekiÄ, M.; MitroviÄ, N.; TomaÅ¡eviÄ, R. Reduced expression of autophagy markers and expansion of myeloid-derived suppressor cells correlate with poor t cell response in severe COVID-19 patients. Front. Immunol. 2021, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Deschler, S.; Kager, J.; Erber, J.; Fricke, L.; Koyumdzhieva, P.; Georgieva, A.; Lahmer, T.; Wissner, J.; Voit, F.; Schneider, J.; et al. Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients. Viruses 2021, 13, 241. [Google Scholar] [CrossRef]
- Stephenson, E.; Reynolds, G.; Botting, R.A.; Calero-Nieto, F.J.; Morgan, M.D.; Tuong, Z.K.; Bach, K.; Sungnak, W.; Worlock, K.B.; Yoshida, M.; et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021, 27, 904–916. [Google Scholar] [CrossRef]
- Vigón, L.; Fuertes, D.; García-Pérez, J.; Torres, M.; Rodríguez-Mora, S.; Mateos, E.; Corona, M.; Saez-Marín, A.J.; Malo, R.; Navarro, C.; et al. Impaired Cytotoxic Response in PBMCs From Patients With COVID-19 Admitted to the ICU: Biomarkers to Predict Disease Severity. Front. Immunol. 2021, 12, 665329. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Wang, X.-M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.-W.; Fan, X.; Xia, P.; Fu, J.-L.; Wang, S.-Y.; et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, Y.; Zhang, Y.; Ai, J.; Yang, B.; Cui, M.; Liao, Q.; Chen, H.; Bai, H.; Shang, D.; et al. Differential immune responses in pregnant patients recovered from COVID-19. Signal Transduct. Target. Ther. 2021, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hubrack, S.; Al-Nesf, M.A.; Agrebi, N.; Raynaud, C.; Khattab, M.A.; Thomas, M.; Ibrahim, T.; Taha, S.; Dermime, S.; Merhi, M.; et al. In vitro interleukin-7 treatment partially rescues mait cell dysfunction caused by sars-cov-2 infection. Scient. Rep. 2021, 11, 14090. [Google Scholar] [CrossRef] [PubMed]
- Notarbartolo, S.; Ranzani, V.; Bandera, A.; Gruarin, P.; Bevilacqua, V.; Putignano, A.R.; Gobbini, A.; Galeota, E.; Manara, C.; Bombaci, M.; et al. Integrated longitudinal immunophenotypic, transcriptional, and repertoire analyses delineate immune re-sponses in patients with COVID-19. Sci. Immunol. 2021, 6, eabg5021. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhou, J.; Zhang, X.; Hu, W.; Zhao, J.-F.; Wang, S.; Wang, F.-S.; Zhang, J.-Y. Single-Cell Transcriptomic Profiling of MAIT Cells in Patients With COVID-19. Front. Immunol. 2021, 12, 3112. [Google Scholar] [CrossRef]
- Yu, C.; Littleton, S.; Giroux, N.S.; Mathew, R.; Ding, S.; Kalnitsky, J.; Yang, Y.; Petzold, E.; Chung, H.A.; Rivera, G.O.; et al. Mucosal Associated Invariant T (MAIT) Cell Responses Differ by Sex in COVID-19. Med 2021, 2, 755. [Google Scholar] [CrossRef]
- Yang, Q.; Wen, Y.; Qi, F.; Gao, X.; Chen, W.; Xu, G.; Wei, C.; Wang, H.; Tang, X.; Lin, J.; et al. Suppressive Monocytes Impair MAIT Cells Response via IL-10 in Patients with Severe COVID-19. J. Immunol. 2021, 207, 1848–1856. [Google Scholar] [CrossRef]
- Flament, H.; Rouland, M.; Beaudoin, L.; Toubal, A.; Bertrand, L.; Lebourgeois, S.; Rousseau, C.; Soulard, P.; Gouda, Z.; Cagninacci, L.; et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021, 22, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Laing, A.G.; Lorenc, A.; del Barrio, I.D.M.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Garca, M.; Kokkinou, E.; Carrasco Garca, A.; Parrot, T.; Palma Medina, L.M.; Maleki, K.T.; Christ, W.; VarnaitÄ, R.; Filipovic, I.; Ljunggren, H.G.; et al. Innate lymphoid cell composition associates with COVID-19 disease severity. Clin. Translat. Immunol 2020, 9, e1224. [Google Scholar] [CrossRef]
- Silverstein, N.J.; Wang, Y.; Manickas-Hill, Z.; Carbone, C.; Dauphin, A.; Boribong, B.P.; Loiselle, M.; Davis, J.; Leonard, M.M.; Kuri-Cervantes, L.; et al. Innate Lymphoid Cells and Disease Tolerance in Sars-Cov-2 Infection; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2021. [Google Scholar]
- Gomez-Cadena, A.; Spehner, L.; Kroemer, M.; Ben Khelil, M.; Bouiller, K.; Verdeil, G.; Trabanelli, S.; Borg, C.; Loyon, R.; Jandus, C. Severe COVID-19 patients exhibit an ILC2 NKG2D+ population in their impaired ILC compartment. Cell. Mol. Immunol. 2021, 18, 484–486. [Google Scholar] [CrossRef]
- Gomes, A.M.C.; Farias, G.B.; Dias-Silva, M.; Laia, J.; Trombetta, A.C.; Godinho-Santos, A.; Rosmaninho, P.; Santos, D.F.; Conceição, C.M.; Costa-Reis, R.; et al. SARS-CoV2 pneumonia recovery is linked to expansion of innate lymphoid cells type 2 expressing CCR10. Eur. J. Immunol. 2021, 51, 3194–3201. [Google Scholar] [CrossRef]
- San Segundo, D.; de las Revillas, F.A.; Lamadrid-Perojo, P.; Comins-Boo, A.; González-Rico, C.; Alonso-Peña, M.; Irure-Ventura, J.; Olmos, J.; Fariñas, M.; López-Hoyos, M. Innate and Adaptive Immune Assessment at Admission to Predict Clinical Outcome in COVID-19 Patients. Biomedicines 2021, 9, 917. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Al-terations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Sen, R.; Garbati, M.R.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Musolino, C.; Tonacci, A.; Pioggia, G.; Gangemi, S. Interactions between the MicroRNAs and Microbiota in Cancer Development: Roles and Therapeutic Opportunities. Cancers 2020, 12, 805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arisan, E.D.; Dart, A.; Grant, G.H.; Arisan, S.; Cuhadaroglu, S.; Lange, S.; Uysal-Onganer, P. The prediction of miRNAs in SARS-CoV-2 genomes: Hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses 2020, 12, 614. [Google Scholar] [CrossRef] [PubMed]
- Canatan, D.; De Sanctis, V. The impact of MicroRNAs (miRNAs) on the genotype of coronaviruses. Acta Bio-Med. Atenei Parmensis 2020, 91, 195–198. [Google Scholar]
- Murdaca, G.; Di Gioacchino, M.; Greco, M.; Borro, M.; Paladin, F.; Petrarca, C.; Gangemi, S. Basophils and Mast Cells in COVID-19 Pathogenesis. Cells 2021, 10, 2754. [Google Scholar] [CrossRef] [PubMed]
- Laterre, P.F.; François, B.; Collienne, C.; Hantson, P.; Jeannet, R.; Remy, K.E.; Hotchkiss, R.S. Association of Interleukin 7 Immunotherapy With Lymphocyte Counts Among Patients With Severe Coronavirus Disease 2019 (COVID-19). JAMA Netw. Open 2020, 3, e2016485. [Google Scholar] [CrossRef]
- Monneret, G.; De Marignan, D.; Coudereau, R.; Bernet, C.; Ader, F.; Frobert, E.; Gossez, M.; Viel, S.; Venet, F.; Wallet, F. Immune monitoring of interleukin-7 compassionate use in a critically ill COVID-19 patient. Cell. Mol. Immunol. 2020, 17, 1001–1003. [Google Scholar] [CrossRef] [PubMed]
- Brufsky, A.; Marti, J.L.G.; Nasrazadani, A.; Lotze, M.T. Boning up: Amino- bisphophonates as immunostimulants and endo-somal disruptors of dendritic cell in SARS-CoV-2 infection. J. Transl. Med. 2020, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Market, M.; Angka, L.; Martel, A.B.; Bastin, D.; Olanubi, O.; Tennakoon, G.; Boucher, D.M.; Ng, J.; Ardolino, M.; Auer, R.C. Flattening the COVID-19 Curve With Natural Killer Cell Based Immunotherapies. Front. Immunol. 2020, 11, 1512. [Google Scholar] [CrossRef]
- Presti, E.L.; Dieli, F.; Meraviglia, S. Lymphopenia in COVID-19: γδ T Cells-Based Therapeutic Opportunities. Vaccines 2021, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Björkström, N.K.; Ponzetta, A. Natural killer cells and unconventional T cells in COVID-19. Curr. Opin. Virol. 2021, 49, 176–182. [Google Scholar] [CrossRef]
- Von Massow, G.; Oh, S.; Lam, A.; Gustafsson, K. Gamma Delta T Cells and Their Involvement in COVID-19 Virus Infections. Front. Immunol. 2021, 12, 4428. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Cao, W.; Endrias, K.; Kuchipudi, S.V.; Mittal, S.K.; Sambhara, S. Innate lymphoid cells (ILC) in SARS-CoV-2 in-fection. Mol. Aspects Med. 2021, 80, 101008. [Google Scholar] [CrossRef] [PubMed]
| Adult/Child | Severe | Non-Severe | Non-COVID-19 in ICU | Recovered | Healthy Subject | |
|---|---|---|---|---|---|---|
| Jouan 2020 JEM [39] | Adult. N = 30 pts in ICU for severe COVID-19 | N = 30 pts in ICU 66.7% with IMV | ND | N = 17 critically ill pts without pneumonia, requiring IMV. | N = 14 pts | N = 20 subjects |
| ↑males↓females | MA: 64 y | MA: 64 y | age- sex-matched | |||
| MDS: 10 d | 75% males | 55% males | ||||
| Parrot 2021 Science Immunology [40] | Adult. N = 69 pts with AD or C. from Atlas cohort or Biobank. | N = 15 from Atlas blood pts + N = 14 from Biobank. | N = 9 samples from Atlas blood pts | ND | N = 45 pts N = 23 convalescent pts from mild disease. N = 22 convalescent pts from moderate/severe | N = 14 subjects SARS- CoV-2 IgG seronegative |
| ↑males↓females | MA: 57 y for both cohorts | MA: 56 y | ||||
| MDH: 17 d for Atlas and 34 d for Biobank; | 80% males for Atlas samples | 67% males | 48% males in Mild 82% males in Mod/Sev/conv | |||
| Odak 2020 eBioMedicine [41] | Adult. N = 30 hospitalized COVID-19 pts. | N = 15 pts with non-IMV | N = 15 pts with stable parameters with no oxygen flow | ND | N = 7 pts Sampling: weeks after the resolution of infection | N = 60 matched Healthy Controls |
| ↑ males↓females | MA: 60 y | MA: 68 y | MA: 54 y | |||
| Mean of days after onset of symptoms: 11 d | 86% males | 73% males | 80% males | |||
| Tomi 2021 Frontiers of immunology [42] | Adult. N = 41 pts with moderate and severe symptoms. | N = 20 pts hospedalized and 60% needed IMV. | N = 21 pts with mild symptoms without IMV | ND | ND | N = 16 healthy volunteers |
| ↑ males↓females | MA: 62.5 y | MA: 55 y | ||||
| 60% males | 57% males | |||||
| Deschler 2021 Viruses [43] | Adult. N = 43 hospitalized COVID-19 pts. ↑ males↓females | N = 21 pts in ICU 19/21 pts IMV MA: 65 y 76.2% males | N = 22 pts MA: 62 y 54.5% males | ND | ND | N = 25 MA: 28 y 52.0% males |
| Stephenson 2021 Nature medicine [44] | Adult. N = 107 pts divided for symptoms in severe, critical, moderate, mild, asymptomatic, hospedalized Non-COVID-19. | N = 15 severe N = 17 critical pts were intubated | N = 32 Moderate; N = 26 mild; N = 12 asymptomatic | N = 5 subjects | ND | N = 24 subjects; N = 12 Healthy volunteers administered with intravenous lipopolysaccharide (IV-LPS) as a surrogate for an acute systemic inflammatory response |
| males = females | Severe MA: 54 y; Critical MA: 54 y | Moderate MA: 54 y; Mild MA: 53 y. Asymptomatic MA: 50.5 y | MA: 56 y | MA 55.5 y | ||
| MND: for severe 15 d; for critical 12.7 d; for moderate 10.5 d; for mild 10 d. | 3/4 samples were females in critical group. 5/7 samples were female for severe group | 11/17 males in moderate group.6/11 females in mild group | 6/12 males in IV-LPS | |||
| Vigon 2021 Frontiers in Immunology [45] | Adult. N = 109 pts divided in 3 groups based on the symptoms. | N = 19 Severe N = 35 Critical (27/35 IMV); 13% of severe and critical pts developed DIC. | N = 55 pts with mild symptoms and who did not develop DIC | ND | ND | N = 20 subjects with similar age and gender distribution as the pts with COVID-19 |
| ↑males↓females | MA: 72 y for Severe. MA: 63 y for Critical. | MA: 46 y | MA: 55.5 y | |||
| MDH: 23 d severe pts; MDH: 50 d critical pts. | 63% males for Severe pts and 74% for Critical | 67% females | 32.7% females | |||
| Zhang 2020 Nature Immunology [46] | Adult. N = 13 pts classified into moderate, severe and convalescent. | N = 4 pts in ICU | N = 7 pts | ND | N = 6 pts but only 4 were paired with moderate pts | N = 5 |
| ↑ males↓females | MA: 64 years | MA: 37 y | MA: 42 y | MA: 35 y | ||
| Mean of days after onset of symptoms: 6 d | 50% males | 57% males | 66% males | 100% males | ||
| Chen 2021 Signal Transduction and Targeted Therapy [47] | Women. N = 179 pts composed by pregnant and non-pregnant COVID-19 pts. | N = 20 PCov; 60% IMV. 5% Non-IMV N = 4 enrolled in the single cell study and divided in two subgroups: PCovM and PCovS | Only one was asymptomatic and her age was higher than others (43 y) | ND | N = 4 Pcov pts | N = 4 PHC pregnant healthy controls |
| MA: 30.5 y | . | MA of PHC: 32 y | ||||
| Hospital staying: 16.5 d for pregnant and 14 d for Non-pregnant pts. | N = 23 NPCov 14% of NP showed critical symptoms; N = 6 enrolled in the single cell study | N = 136 NPCov | N = 6 Ncov pts | N = 6 NHC. Only N = 3 enrolled in the single cell study | ||
| MA: 33 y | MA of NHC: 36 y |
| Adult/Child | Severe | Non-Severe | Non-COVID-19 in ICU | Recovered | Healthy Subject | |
|---|---|---|---|---|---|---|
| Chen 2021 Signal Transduction and Targeted Therapy [47] | Women. N = 179 pts composed by pregnant and NPCov. Hospital staying: 16.5 d for pregnant and 14 d for NP pts. | N = 20 pts. 60% IMV. 5% Non-IMV. N = 4 enrolled in the single cell study as PCovM and PcovS. | Only one was asymtomatic and her age was higher than others (43 y) N = 136 NPCov | ND | N = 4 Pcov pts | N = 4 PHC MA of PHC: 32 y; |
| MA 30.5 y | ||||||
| N = 23 NPCov 14% of NP showed critical symptoms; | N = 6 Ncov pts | N = 6 NHC. Only N = 3 enrolled in the single cell study MA of NHC: 36 y. | ||||
| MA: 33 y | ||||||
| Deschler 2021 Viruses [43] | Adult. N = 43 hospitalized COVID-19 pts. | N = 21 pts in ICU; 19/21 pts (90%) IMV. | N = 22 pts | ND | Sampling: 4–9 weeks after admission to the hospital. | N = 25 |
| ↑ males↓females | MA: 65 y | MA: 62 y | MA: 28 y | |||
| 76.2% males | 54.5% males | 52.0% males | ||||
| Hubrack 2021 Scientific report [48] | Adult. N = 36 pts classified based on the symptoms. ↑ males↓females | N = 13 pts 92.3% had difficulty of breath. | N = 23 pts 21.7% asymptomatic. | ND | ND | N = 21 |
| MA: 41 years | MA: 38.4 y | MA:38 y | ||||
| 92% males | 91.3% males | 95% males | ||||
| Notarbartolo 2021 Science [49] | Adult. N = 17 pts. For only 4 pts there are paired data of infection and post-infection. | N = 11pts. All required supplementary oxygen support. | N = 6pts | ND | N = 4 | |
| ↑ males↓females | MA: 55 y | MA: 33 y | Sampling: weeks after the resolution of infection. | |||
| 54% females | 83% males | |||||
| Shi 2021 Frontier in Immunology [50] | Adult. N = 13 pts classified in three clinical conditions. Samples data from the G. S. A. of the Beijing Institute of Genomics (BIG) | N = 4 pts MA: ND 50% males | N = 7 pts MA: ND ↑males | ND | N = 6 of whom 4 were paired with moderate cases. | N = 5 |
| Yu 2021 Med [51] | Adult. N = 28 pts. | N = 9 pts. 77.8% requiring intensive care | N = 19 pts N = 11 pts paired with post-infection sampling. | N = 7 exposed subjects with β-phenotype. | . Sampling: weeks after the resolution of infection. | N = 10 Males with β-phenotype, females with α-phenotype. |
| Mean of days after onset to hospitalization:: | 56% males | 58% males | 57% males | 50% males | ||
| 8.5 d; | MA: 59,4 y | MA 36,7 y | MA: 42 y for exposed | MA: 39.7 y | ||
| Yang The Journal of Immunology 2021 [52] | Adult. N = 45 pts among mild and severe; N = 6 asymptomatic. | N = 22 pts. 23% required IMV | N = 23 pts MA: 41.7 y. More females than males. | N = 6 asymptomatic MA: 33 y. | N = 6 convalescent severe; N=8 convalescent mild pts. | N = 44 |
| 54,5% males | 43,7% males | |||||
| MA: 55.7 y | MA: 41.7 y | MA: 33 y | ||||
| Flament 2021 Nature Immunology [53] | Adult. I cohort: N = 182 pts in 3 groups: IDU(moderated) ICU (Severe) and deceased(Death) | N = 51 pts in ICU. 41% Death rate. MA:58 y 82.3% males | N = 51 pts. 11,7% death rate. MA: 61y 66,6% males | ND | ND | N = 80 MA: 27.2 60% males. N = 4 Uninfected MA: 58 y 75% males |
| Adult. II cohort: N = 26 pts in 3 groups: IDU (moderated) ICU (Severe) and deceased (Death). ↑ males↓females for both cohorts | N = 13 ICU. 23% Death MA: 57 y 76,9% males | N = 9 IDU. 11% Death. MA: 79 y 55.5% males | ||||
| Parrot 2020 Science Immunology [40] | Adult. N = 69 pts with acute disease or convalescents from Atlas cohort or Biobank. | N = 15 samples from Atlas blood pts + N = 14 samples from Biobank. | N = 9 samples from Atlas blood pts | ND | N = 45 pts. N = 23 C. pts from mild disease. N = 22 convalescent pts from moderate/severe. | N = 14 Mild convalescent = 23; MA=51 y; 52% females. Moderate/severe convalescent= 22. MA= 56 y; 82% males. |
| ↑males↓females | MA: 57 y for both cohorts | MA: 56 y | Sampling within 1 to 6 weeks from resolution of disease. | |||
| Days in hospital: 17 d for Atlas and 34 d for Biobank; | 80% males for Atlas samples | 67% males. | 48% males in mild convalescent; 82% males in moderate/severe convalescent. | |||
| Zhang 2021 Nature Immunology [46] | Adult. N = 13 pts classified into: moderate, severe and convalescent. | N = 4 pts in ICU | N = 7 pts; | ND | N = 6 pts but only 4 paired with moderate pts; | N = 5 HCs; |
| ↑ males↓females | MA: 6 y | MA: 37 y | MA: 42 y; | MA: 35 y | ||
| Mean of days after onset of symptoms: 6 d | 50% males; | 57% males; | 66% males; | 100% males | ||
| Jouan 2020 JEM [39] | Adult. N = 30 pts in ICU for severe COVID-19; | N = 30 pts in ICU; 66.7% pts IMV; | ND | N = 17 critically ill pts without pneumonia, requiring IMV. | N = 14 discharged pts to ICU ward. | N = 20 age and sex-matched |
| ↑ males↓females | MA: 64 y; | MA: 64 y; | ||||
| Median duration of symptoms before admission in ICU: 10 d | 75% males | 55% males |
| Adult/Child | Severe | Non-Severe | Non-COVID-19 in ICU | Recovered | Healthy Subject | |
|---|---|---|---|---|---|---|
| Odak 2020 eBioMedicine [41] | Adult. N = 30 hospitalized COVID-19 pts | N = 15 pts with non-IMV; | N = 15 pts with stable parameters with no oxygen flow; | ND | N = 7 pts Sampling: weeks after the resolution of infection. | N = 60 Matched HCs |
| ↑ males↓females | MA: 60 y | MA: 68 y | Mean age: 54 y | |||
| Mean of days after onset of symptoms: 11 d | 86% males | 73% males | 80% males | |||
| Zhang 2020 Nature Immunology [46] | Adult. N = 13 pts: moderate, severe and convalescent. | N = 4 pts in ICU | N = 7 pts | ND | N = 6 pts but only 4 were paired with moderate pts; | N = 5 HCs; |
| ↑ males↓females | MA: 64 y | MA: 37 y | MA: 42 y | MA: 35 y | ||
| Mean of days after onset of symptoms: 6 d | 50% males | 57% males | 66% males | 100% males | ||
| Jouan 2020 JEM [39] | Adult. N = 30 pts in ICU for severe COVID-19; ↑ males↓females Median duration symptoms before admission in ICU: 10 d | N = 30 pts in ICU 66.7% pts IMV MA: 64 y 75% males | ND | N = 17 critically ill pts without pneumonia requiring IMV Mean age: 64 years; 55% males | N = 14 discharged pts to ward. Sampling: 15 d | N = 20 subjects age and sex-matched |
| Stephenson 2021 Nature medicine [44] | Adult. N = 107pts: severe, critical, moderate, mild, asymptomatic, hospitalized Non-COVID-19. | N = 15 severe and N = 17 critical pts with IMV | N = 32 moderate; N=26 mild; N = 12 asymptomatic. | N =5 subjects MA: 5 y | ND | N = 24 N = 12 Healthy volunteers treated with intravenous lipopolysaccharide (IV-LPS) |
| Males = females | Severe MA: 54 y; Critical MA: 54 y. | Moderate MA: 54 y; Mild MA: 53 y. Asymptomatic MA: 50.5 y. | MA: 55.5 y | |||
| Mean of days from onset of symptoms: for severe 15d; for critical 12.7 d; for moderate 10.5d; for mild 10 d. | 3/4 pts females in critical group. 5/7 pts female for severe group. | 11/17 pts males in moderate group. 6/11 pts female for mild group. | 6/12 pts males in IV-LPS |
| Adult/Child | Severe | Non-Severe | Non-COVID-19 in ICU | Recovered | Healthy Subject | |
|---|---|---|---|---|---|---|
| Garcia 2021 Clinical and translational Immunology [55] | Adult. N = 23 pts; N = 16 HCs. | N = 12 pts in ICU with IMV | N = 11 pts; Non-intubated and Non-oxygen need. Some hospedalized ICU. | ND | ND | N = 16 |
| ↑males↓females | MA: 59.5 y | MA: 56 y | ||||
| Median days hospitalized: 11 d for moderate pts; 22 d for severe pts. | 83% males | 63.6% males. | ||||
| Segundo 2020 Biomedicines [58] | Adult. N = 150 pts divided based on oxygen therapy requirements. | N = 82 mod/sev (ICU with IMV or only hospitalized or deceased) | N = 73 Mild pts | ND | ND | ND |
| MA: 72 y | MA: 59 y | |||||
| ↑ male gender. | ↑ female gender. | |||||
| Silverstein 2021 (preprint) [56] | Adult cohort of samples | N = 40 pts. Among whom N=33 (82.5%) at ICU; N=32 (80%) with IMV; N=7 (17.5%) died. | N = 51 outpatients infected with SARS-CoV-2 who were treated for COVID-19. | ND | ND | N = 86 who donated blood prior to the SARS-CoV-2 outbreak. |
| ↓males ↑ females | MA: 57.6 y | MA: 36.8 y | MA: 50.9 y | |||
| Mean days hospitalized: 34.2 d | 60% males gender | 25.5% males gender | 55.8% males | |||
| Pediatric. N = 30 pts; N = 17 HCs. | N = 11 pts. Among whom N = 1 (5.3%) at ICU with IMV | N = 8 outpatients infected with SARS-CoV-2 | N = 11 MIS-C pts | N = 14 COVID-19 follow-up; N = 7 MIS-C follow-up; N = 10 pts (5 COVID-19 and 5 MIS-C) | N = 17 SARS-CoV-2-uninfected pediatric blood donors | |
| ↑males ↓ females | MA: 13 y | MA: 13 y | ||||
| Gomez-Cadena 2021 Cellular & Molecular Immunology [57] | Adult. N = 60 pts hospitalized for COVID-19. males = females | N = 30 pts MA: 68.3 with 41% of the severe pts were older than 75 y | N = 30 with mild symptoms. MA: 39.8 y | ND | ND | N = 21 |
| Pts were enrolled at least 21 d after the first symptoms of COVID-19 | 76.7% males | 23.3% males | ||||
| Gomez 2021 Eur. J. Immunology [58] | Adult. N = 20 pts hospitalized for COVID-19. Days of symptoms to admission: 12 d. | N = 20 pts 11/20 pts in ICU 80% males MA: 56 y | ND | ND | N = 14 pts in follow-up. Days of symptoms to recovery: 19 d | N = 9MA: 58 y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo Presti, E.; De Gaetano, A.; Pioggia, G.; Gangemi, S. Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities. Cells 2022, 11, 542. https://doi.org/10.3390/cells11030542
Lo Presti E, De Gaetano A, Pioggia G, Gangemi S. Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities. Cells. 2022; 11(3):542. https://doi.org/10.3390/cells11030542
Chicago/Turabian StyleLo Presti, Elena, Andrea De Gaetano, Giovanni Pioggia, and Sebastiano Gangemi. 2022. "Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities" Cells 11, no. 3: 542. https://doi.org/10.3390/cells11030542
APA StyleLo Presti, E., De Gaetano, A., Pioggia, G., & Gangemi, S. (2022). Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities. Cells, 11(3), 542. https://doi.org/10.3390/cells11030542








