Lifespan Increase of Podospora anserina by Oleic Acid Is Linked to Alterations in Energy Metabolism, Membrane Trafficking and Autophagy
Abstract
1. Introduction
2. Materials and Methods
2.1. P. anserina Strains and Cultivation
2.2. Cloning Procedure and Generation of P. anserina Mutants
2.3. Growth Rate and Lifespan Determination
2.4. Southern Blot Analyses
2.5. Isolation of Total Protein Extracts
2.6. Isolation of Secreted Proteins
2.7. Western Blot Analysis
2.8. Measuring the Rate of Peroxisomal β-Oxidation of Fatty Acids
2.9. In-Gel Peroxidase and Catalase Activity Assay
2.10. Hydrogen Peroxide Emission Measurements
2.11. Fluorescence Microscopy
2.12. Statistical Analysis
3. Results and Discussion
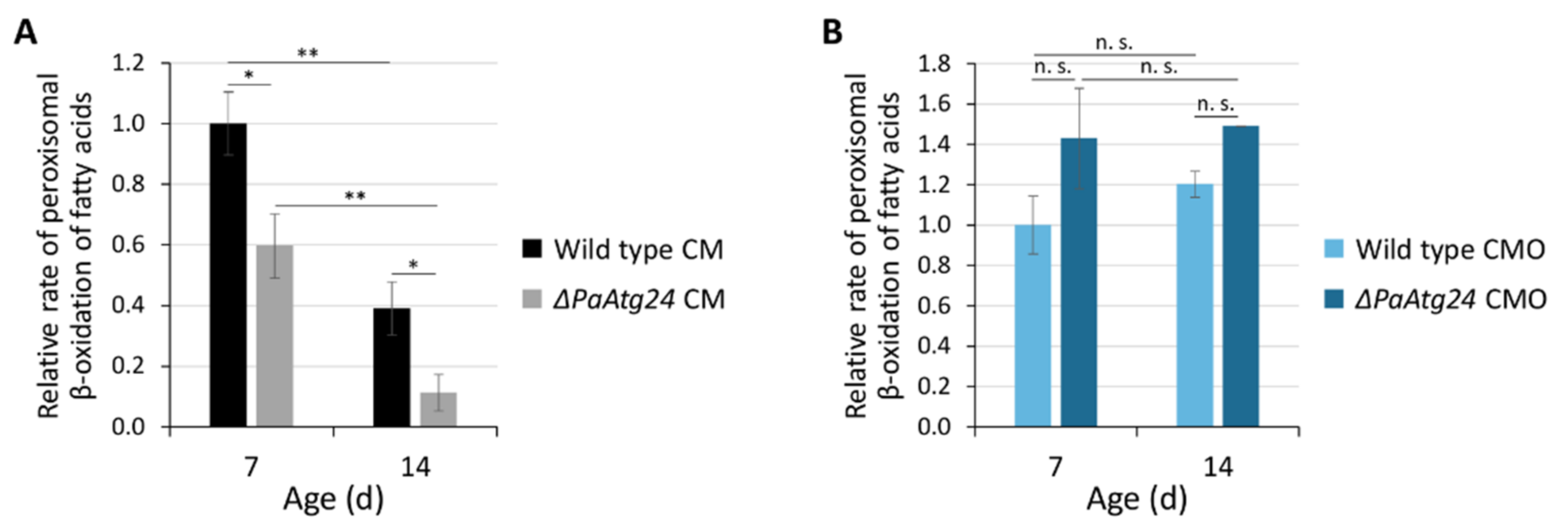
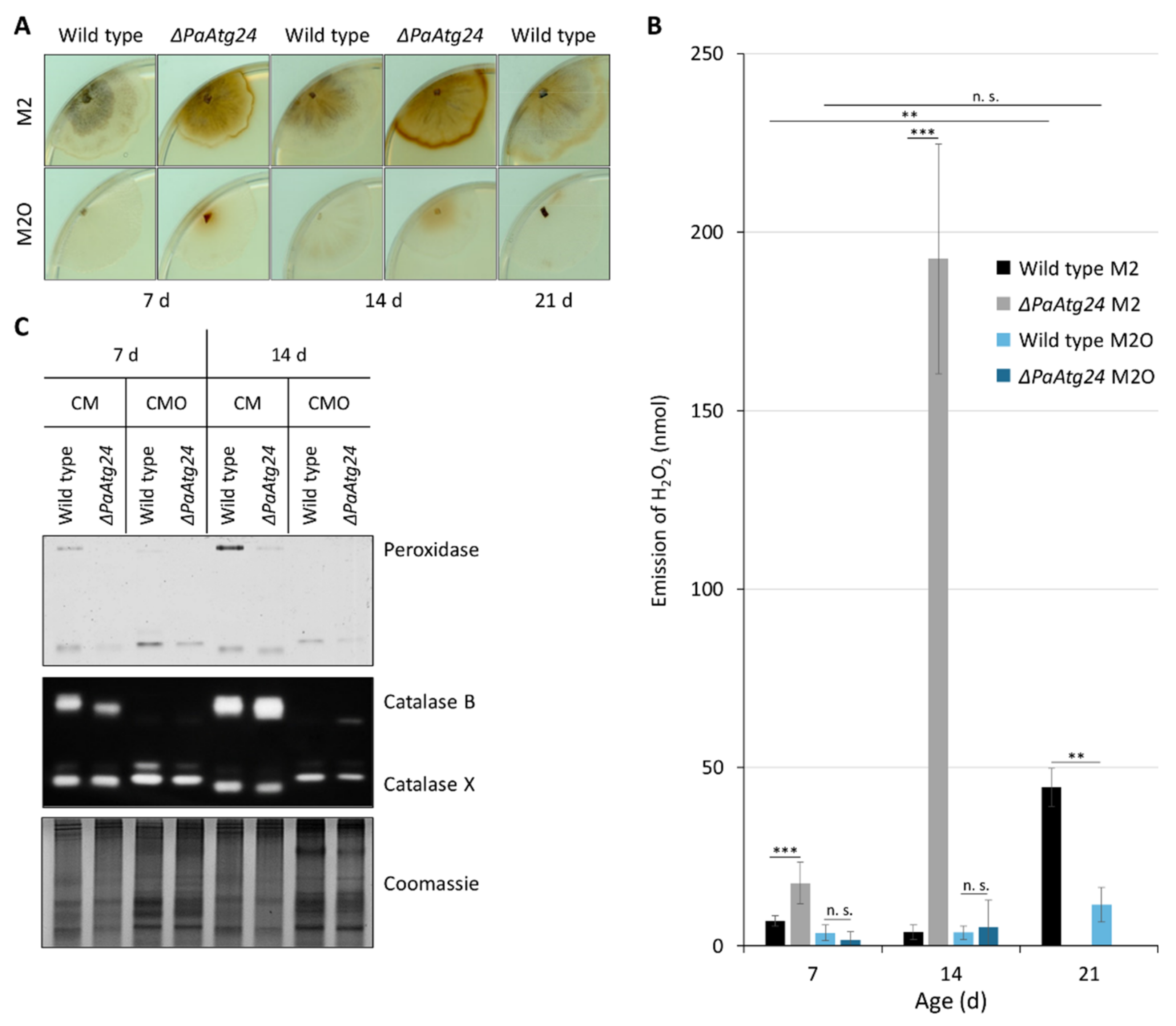
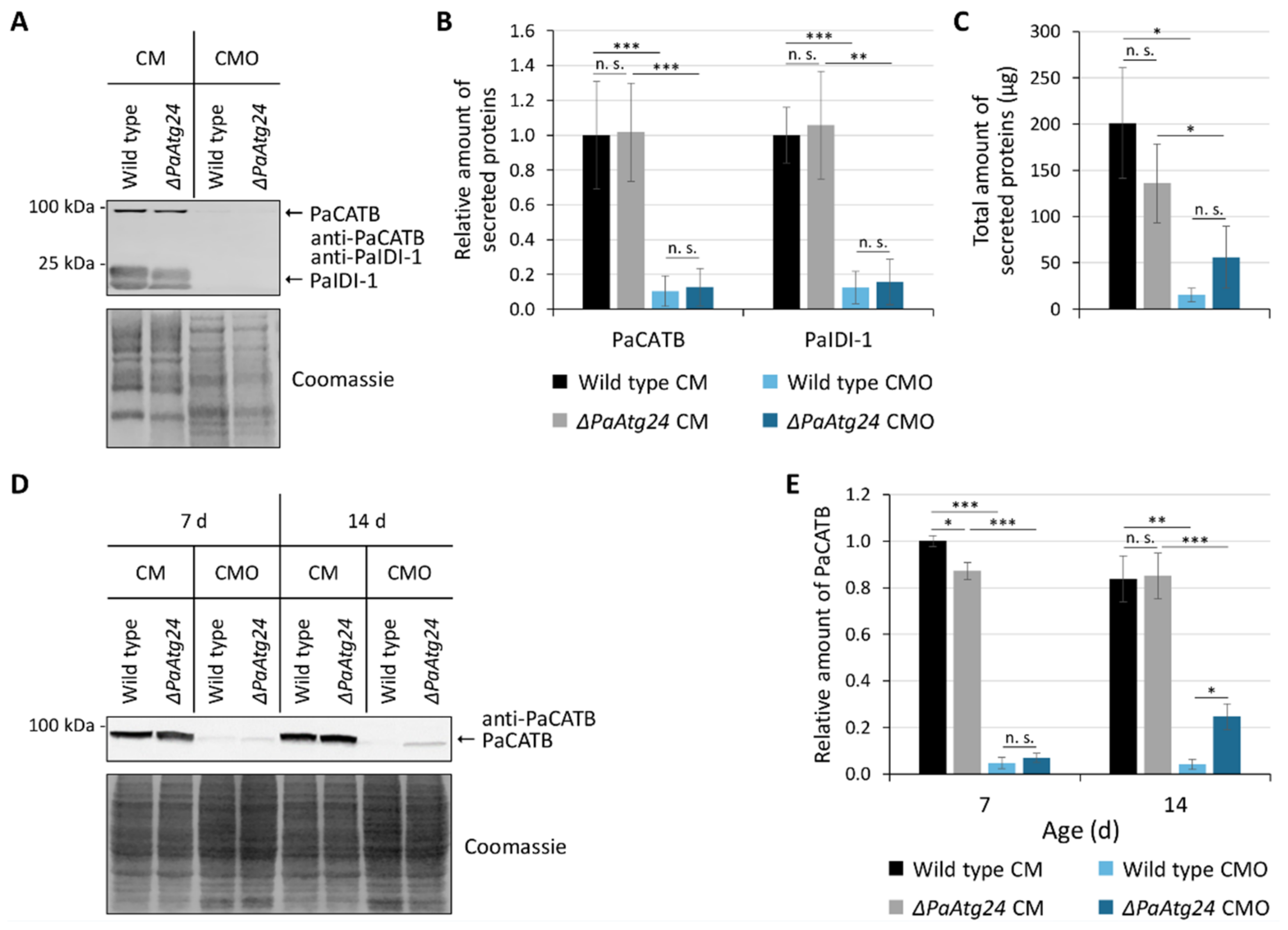
3.1. Oleic Acid Diet Rescues Shortened Lifespan and Impaired Peroxisomal Functionality of ΔPaAtg24
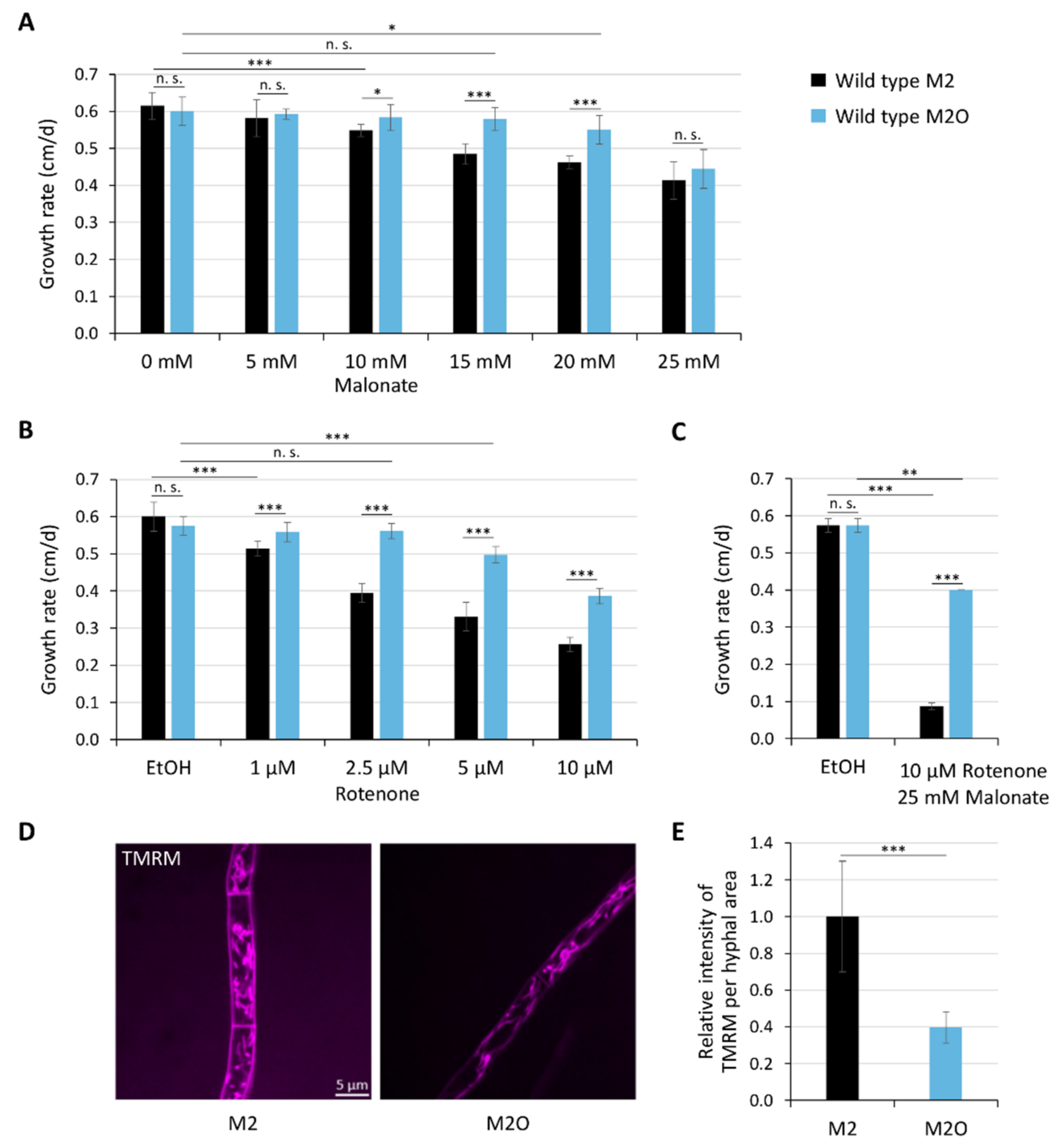
3.2. Oleic Acid Diet Affects Mitochondrial ROS Metabolism
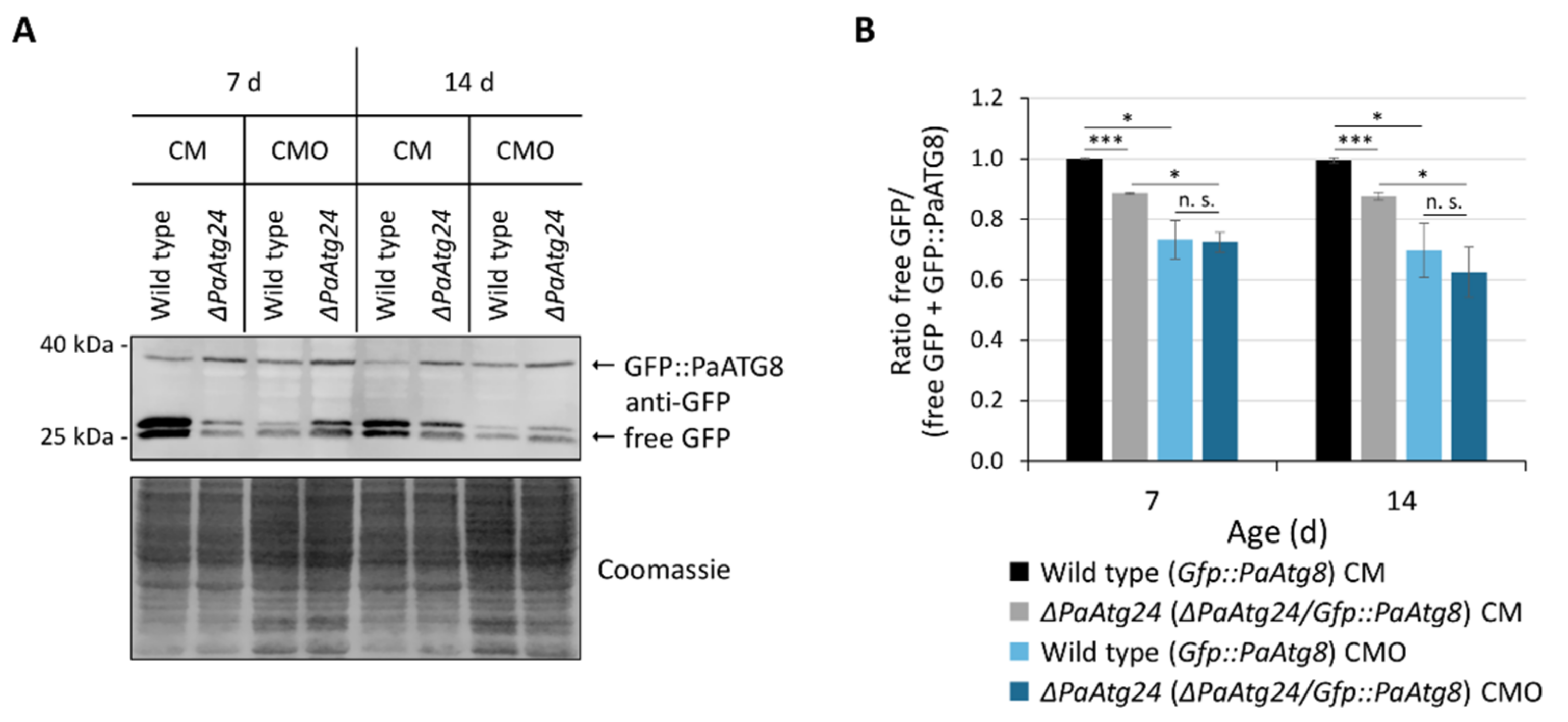
3.3. Oleic Acid Diet Abrogates the Autophagy Defect of ΔPaAtg24
3.4. Oleic Acid Diet Abolishes the Vacuole Formation Defect of ΔPaAtg24 by Normalizing Membrane Trafficking of ΔPaAtg24
3.5. Oleic Acid Diet Alters Membrane Trafficking in P. anserina
3.6. Oleic Acid Diet Abolishes Mislocalization of SNARE PaSNC1 in ΔPaAtg24
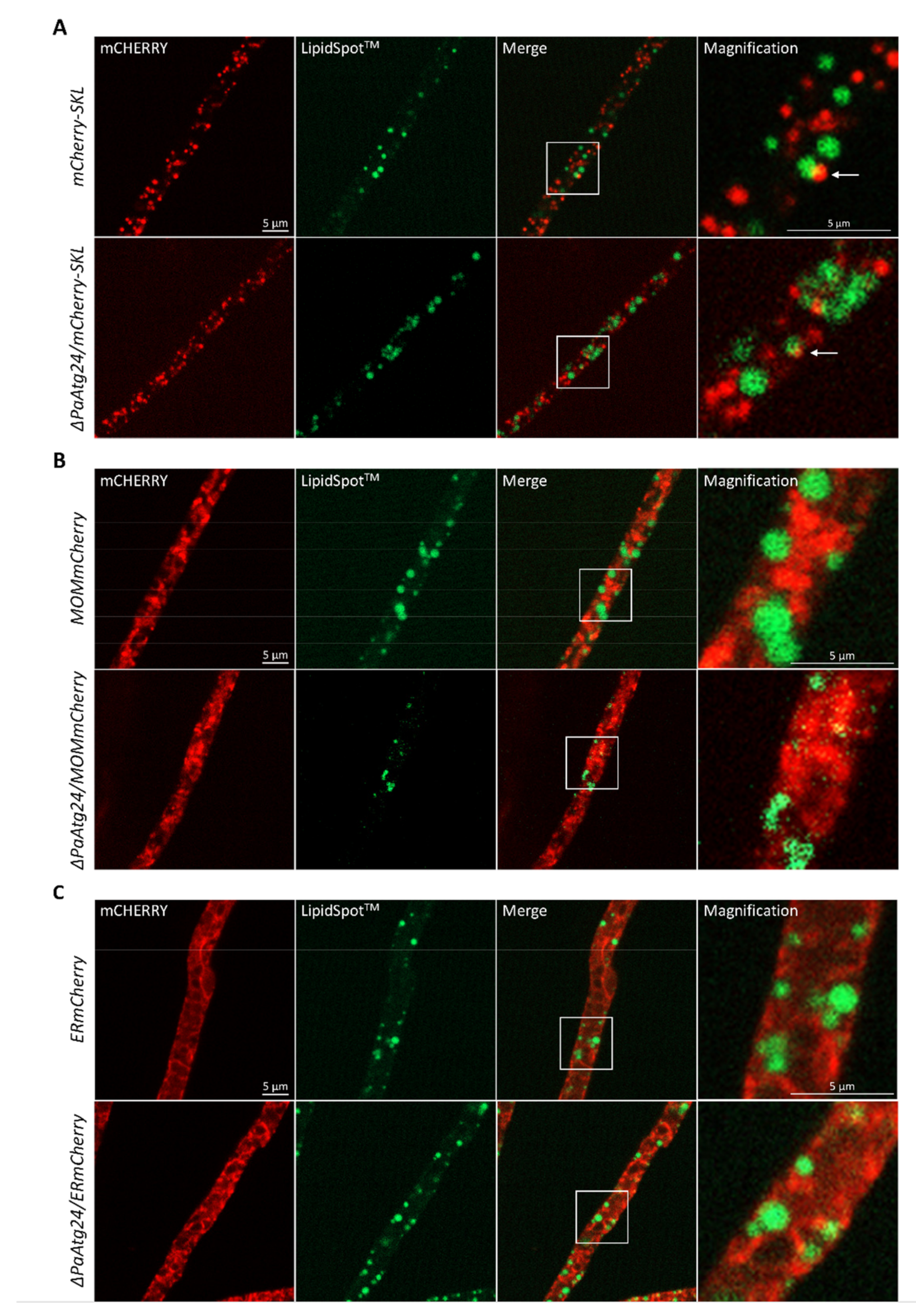
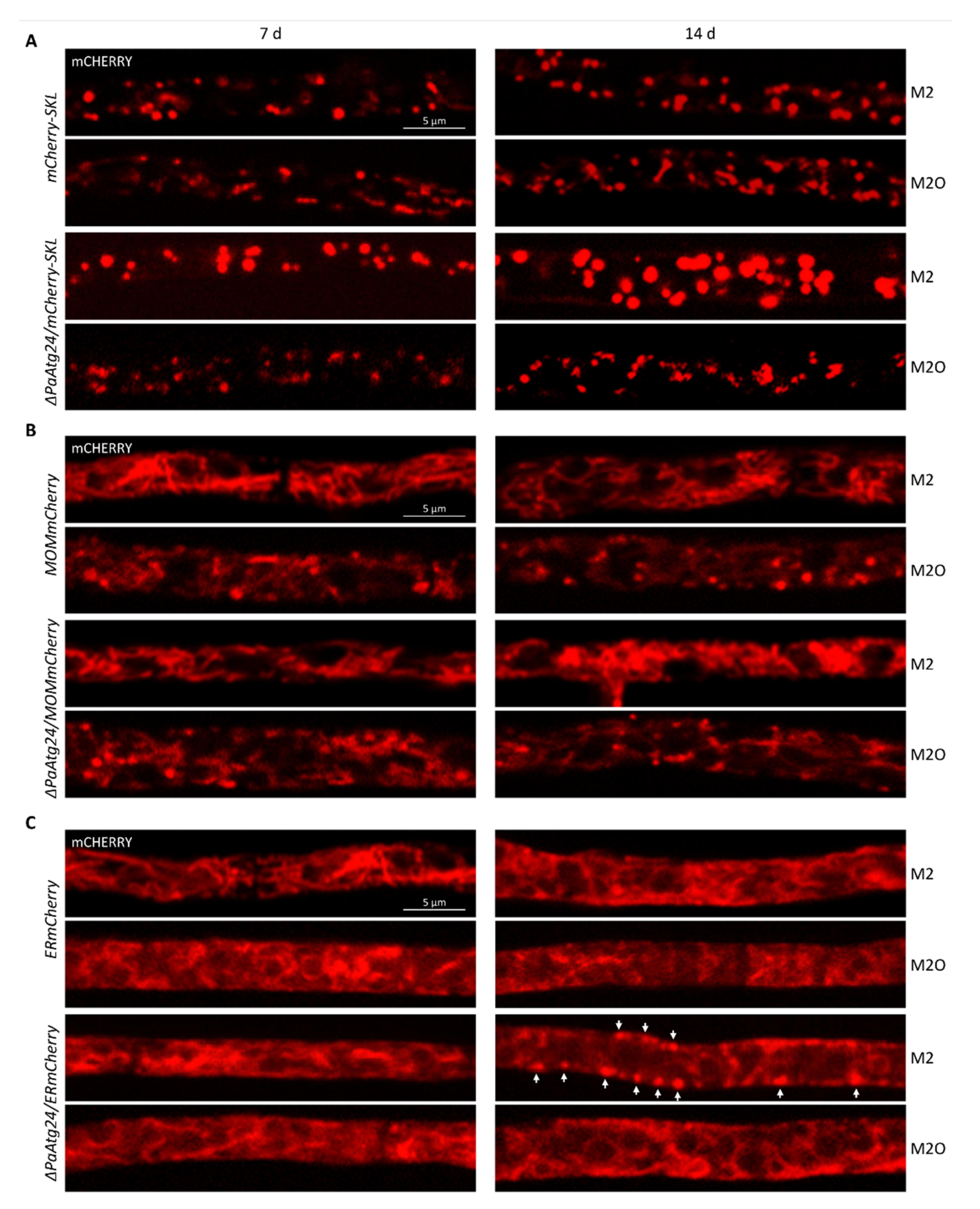
3.7. Impact of Oleic Acid Diet on the Abundance and Morphology of Peroxisomes, Mitochondria and ER
3.8. Model Explaining the Impact of Oleic Acid on Membrane Trafficking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Osiewacz, H.D.; Schürmanns, L. A network of pathways controlling cellular homeostasis affects the onset of senescence in Podospora anserina. J. Fungi 2021, 7, 263. [Google Scholar] [CrossRef] [PubMed]
- Knuppertz, L.; Hamann, A.; Pampaloni, F.; Stelzer, E.; Osiewacz, H.D. Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy 2014, 10, 822–834. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haft, C.R.; de la Luz Sierra, M.; Barr, V.A.; Haft, D.H.; Taylor, S.I. Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol. Cell Biol. 1998, 18, 7278–7287. [Google Scholar] [CrossRef] [PubMed]
- Leprince, C.; Le Scolan, E.; Meunier, B.; Fraisier, V.; Brandon, N.; De Gunzburg, J.; Camonis, J. Sorting nexin 4 and amphiphysin 2, a new partnership between endocytosis and intracellular trafficking. J. Cell Sci. 2003, 116, 1937–1948. [Google Scholar] [CrossRef]
- Nice, D.C.; Sato, T.K.; Stromhaug, P.E.; Emr, S.D.; Klionsky, D.J. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002, 277, 30198–30207. [Google Scholar] [CrossRef]
- Gallon, M.; Cullen, P.J. Retromer and sorting nexins in endosomal sorting. Biochem. Soc. Trans. 2015, 43, 33–47. [Google Scholar] [CrossRef]
- Ma, M.; Kumar, S.; Purushothaman, L.; Babst, M.; Ungermann, C.; Chi, R.J.; Burd, C.G. Lipid trafficking by yeast Snx4 family SNX-BAR proteins promotes autophagy and vacuole membrane fusion. Mol. Biol. Cell 2018, 29, 2190–2200. [Google Scholar] [CrossRef]
- Ma, M.; Burd, C.G.; Chi, R.J. Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic 2017, 18, 134–144. [Google Scholar] [CrossRef]
- Henkel, V.; Schürmanns, L.; Brunner, M.; Hamann, A.; Osiewacz, H.D. Role of sorting nexin PaATG24 in autophagy, aging and development of Podospora anserina. Mech. Ageing Dev. 2020, 186, 111211. [Google Scholar] [CrossRef]
- Bonnet, C.; Espagne, E.; Zickler, D.; Boisnard, S.; Bourdais, A.; Berteaux-Lecellier, V. The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol. Microbiol. 2006, 62, 157–169. [Google Scholar] [CrossRef]
- Rizet, G. Sur l’impossibilité d’obtenir la multiplication végétative ininterrompue et illimitée de l’ascomycète Podospora anserina. C. R. Hebd. Seances Acad. Sci. 1953, 237, 838–840. [Google Scholar]
- Knuppertz, L.; Warnsmann, V.; Hamann, A.; Grimm, C.; Osiewacz, H.D. Stress-dependent opposing roles for mitophagy in aging of the ascomycete Podospora anserina. Autophagy 2017, 13, 1037–1052. [Google Scholar] [CrossRef]
- Osiewacz, H.D.; Hamann, A.; Zintel, S. Assessing organismal aging in the filamentous fungus Podospora anserina. Methods Mol. Biol. 2013, 965, 439–462. [Google Scholar] [CrossRef]
- Löser, T.; Joppe, A.; Hamann, A.; Osiewacz, H.D. Mitochondrial phospholipid homeostasis is regulated by the i-AAA protease PaIAP and affects organismic aging. Cells 2021, 10, 2775. [Google Scholar] [CrossRef]
- Lacaze, I.; Lalucque, H.; Siegmund, U.; Silar, P.; Brun, S. Identification of NoxD/Pro41 as the homologue of the p22phox NADPH oxidase subunit in fungi. Mol. Microbiol. 2015, 95, 1006–1024. [Google Scholar] [CrossRef]
- Luce, K.; Osiewacz, H.D. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat. Cell Biol. 2009, 11, 852–858. [Google Scholar] [CrossRef]
- Labrousse, A.M.; Zappaterra, M.D.; Rube, D.A.; van der Bliek, A.M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 1999, 4, 815–826. [Google Scholar] [CrossRef]
- Averbeck, N.B.; Borghouts, C.; Hamann, A.; Specke, V.; Osiewacz, H.D. Molecular control of copper homeostasis in filamentous fungi: Increased expression of a metallothionein gene during aging of Podospora anserina. Mol. Gen. Genet. 2001, 264, 604–612. [Google Scholar] [CrossRef]
- El-Khoury, R.; Sellem, C.H.; Coppin, E.; Boivin, A.; Maas, M.F.; Debuchy, R.; Sainsard-Chanet, A. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet 2008, 53, 249–258. [Google Scholar] [CrossRef]
- Osiewacz, H.D. A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr. Genet 1994, 26, 87–90. [Google Scholar] [CrossRef]
- Kunstmann, B.; Osiewacz, H.D. The S-adenosylmethionine dependent O-methyltransferase PaMTH1: A longevity assurance factor protecting Podospora anserina against oxidative stress. Aging 2009, 1, 328–334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lecellier, G.; Silar, P. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 1994, 25, 122–123. [Google Scholar] [CrossRef] [PubMed]
- Zintel, S.; Bernhardt, D.; Rogowska-Wrzesinska, A.; Osiewacz, H.D. PaCATB, a secreted catalase protecting Podospora anserina against exogenous oxidative stress. Aging 2011, 3, 768–781. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Brust, D.; Daum, B.; Breunig, C.; Hamann, A.; Kühlbrandt, W.; Osiewacz, H.D. Cyclophilin D links programmed cell death and organismal aging in Podospora anserina. Aging Cell 2010, 9, 761–775. [Google Scholar] [CrossRef]
- Lazarow, P.B. Assay of peroxisomal beta-oxidation of fatty acids. Methods Enzymol. 1981, 72, 315–319. [Google Scholar] [CrossRef]
- Wayne, L.G.; Diaz, G.A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal. Biochem. 1986, 157, 89–92. [Google Scholar] [CrossRef]
- Munkres, K.D. Histochemical detection of the secretion of superoxide radicals and hydrogen peroxide by age-1 mutants of Neurospora. Fungal Genet. Rep. 1990, 37, 12. [Google Scholar] [CrossRef][Green Version]
- Kowald, A.; Hamann, A.; Zintel, S.; Ullrich, S.; Klipp, E.; Osiewacz, H.D. A systems biological analysis links ROS metabolism to mitochondrial protein quality control. Mech. Ageing Dev. 2012, 133, 331–337. [Google Scholar] [CrossRef]
- Swigonová, Z.; Mohsen, A.W.; Vockley, J. Acyl-CoA dehydrogenases: Dynamic history of protein family evolution. J. Mol. Evol. 2009, 69, 176–193. [Google Scholar] [CrossRef]
- Kunau, W.-H.; Kionka, C.; Ledebur, A.; Mateblowski, M.; Moreno de la Garza, M.; Schultz-Borchard, U.; Thieringer, R.; Veenhuis, M. β-oxidation systems in eukaryotic microorganisms. In Peroxisomes in Biology and Medicin; Fahimi, H.D., Sies, H., Eds.; Springer: Berlin/Heidelberg, Germany, 1987; pp. 128–140. [Google Scholar]
- Gredilla, R.; Grief, J.; Osiewacz, H.D. Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp. Gerontol. 2006, 41, 439–447. [Google Scholar] [CrossRef]
- Klein, M.; Swinnen, S.; Thevelein, J.M.; Nevoigt, E. Glycerol metabolism and transport in yeast and fungi: Established knowledge and ambiguities. Environ. Microbiol. 2017, 19, 878–893. [Google Scholar] [CrossRef]
- Mráček, T.; Drahota, Z.; Houštěk, J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta 2013, 1827, 401–410. [Google Scholar] [CrossRef]
- Ehrenberg, B.; Montana, V.; Wei, M.D.; Wuskell, J.P.; Loew, L.M. Membrane potential can be determined in individual cells from the nernstian distribution of cationic dyes. Biophys. J. 1988, 53, 785–794. [Google Scholar] [CrossRef]
- Boutouja, F.; Stiehm, C.M.; Reidick, C.; Mastalski, T.; Brinkmeier, R.; Magraoui, F.E.; Platta, H.W. Vac8 controls vacuolar membrane dynamics during different autophagy pathways in Saccharomyces cerevisiae. Cells 2019, 8, 661. [Google Scholar] [CrossRef]
- Fischer-Parton, S.; Parton, R.M.; Hickey, P.C.; Dijksterhuis, J.; Atkinson, H.A.; Read, N.D. Confocal microscopy of FM4-64 as a tool for analysing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 2000, 198, 246–259. [Google Scholar] [CrossRef]
- Claus, S.; Jezierska, S.; Van Bogaert, I.N.A. Protein-facilitated transport of hydrophobic molecules across the yeast plasma membrane. FEBS Lett. 2019, 593, 1508–1527. [Google Scholar] [CrossRef]
- Kohlwein, S.D.; Veenhuis, M.; van der Klei, I.J. Lipid droplets and peroxisomes: Key players in cellular lipid homeostasis or a matter of fat--store ‘em up or burn ‘em down. Genetics 2013, 193, 1–50. [Google Scholar] [CrossRef]
- Vevea, J.D.; Garcia, E.J.; Chan, R.B.; Zhou, B.; Schultz, M.; Di Paolo, G.; McCaffery, J.M.; Pon, L.A. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 2015, 35, 584–599. [Google Scholar] [CrossRef]
- Geltinger, F.; Tevini, J.; Briza, P.; Geiser, A.; Bischof, J.; Richter, K.; Felder, T.; Rinnerthaler, M. The transfer of specific mitochondrial lipids and proteins to lipid droplets contributes to proteostasis upon stress and aging in the eukaryotic model system Saccharomyces cerevisiae. Geroscience 2020, 42, 19–38. [Google Scholar] [CrossRef]
- Lapierre, L.R.; Gelino, S.; Meléndez, A.; Hansen, M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011, 21, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol. Mol. Biol. Rev. 2018, 82, e00068-17. [Google Scholar] [CrossRef] [PubMed]
- Wessels, J.G.H. Fungal hydrophobins: Proteins that function at an interface. Trends Plant Sci. 1996, 1, 9–15. [Google Scholar] [CrossRef]
- Hettema, E.H.; Lewis, M.J.; Black, M.W.; Pelham, H.R. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003, 22, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Best, J.T.; Xu, P.; McGuire, J.G.; Leahy, S.N.; Graham, T.R. Yeast synaptobrevin, Snc1, engages distinct routes of postendocytic recycling mediated by a sorting nexin, Rcy1-COPI, and retromer. Mol. Biol. Cell 2020, 31, 944–962. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Liu, X.; Feng, Y.; Klionsky, D.J. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 2014, 10, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, S.; de Boer, R.; Veenhuis, M.; van der Klei, I.J. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy 2013, 9, 1044–1056. [Google Scholar] [CrossRef]
- Saheki, Y.; De Camilli, P. Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 2017, 86, 659–684. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Baumann, N.A.; Sullivan, D.P.; Ohvo-Rekilä, H.; Simonot, C.; Pottekat, A.; Klaassen, Z.; Beh, C.T.; Menon, A.K. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 2005, 44, 5816–5826. [Google Scholar] [CrossRef]
- Sandhu, J.; Li, S.; Fairall, L.; Pfisterer, S.G.; Gurnett, J.E.; Xiao, X.; Weston, T.A.; Vashi, D.; Ferrari, A.; Orozco, J.L.; et al. Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell 2018, 175, 514–529.e520. [Google Scholar] [CrossRef]
- Ganesan, S.; Sosa Ponce, M.L.; Tavassoli, M.; Shabits, B.N.; Mahadeo, M.; Prenner, E.J.; Terebiznik, M.R.; Zaremberg, V. Metabolic control of cytosolic-facing pools of diacylglycerol in budding yeast. Traffic 2019, 20, 226–245. [Google Scholar] [CrossRef]
- Ganesan, S.; Tavassoli, M.; Shabits, B.N.; Zaremberg, V. Tubular ER associates with diacylglycerol-rich structures during lipid droplet consumption. Front. Cell Dev. Biol. 2020, 8, 700. [Google Scholar] [CrossRef]
- Trichopoulou, A.; Vasilopoulou, E. Mediterranean diet and longevity. Br. J. Nutr. 2000, 84 (Suppl. 2), S205–S209. [Google Scholar] [CrossRef]
- Sales-Campos, H.; Souza, P.R.; Peghini, B.C.; da Silva, J.S.; Cardoso, C.R. An overview of the modulatory effects of oleic acid in health and disease. Mini Rev. Med. Chem. 2013, 13, 201–210. [Google Scholar]
- Trichopoulou, A.; Dilis, V. Olive oil and longevity. Mol. Nutr. Food Res. 2007, 51, 1275–1278. [Google Scholar] [CrossRef]
- Han, S.; Schroeder, E.A.; Silva-García, C.G.; Hebestreit, K.; Mair, W.B.; Brunet, A. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 2017, 544, 185–190. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schürmanns, L.; Hamann, A.; Osiewacz, H.D. Lifespan Increase of Podospora anserina by Oleic Acid Is Linked to Alterations in Energy Metabolism, Membrane Trafficking and Autophagy. Cells 2022, 11, 519. https://doi.org/10.3390/cells11030519
Schürmanns L, Hamann A, Osiewacz HD. Lifespan Increase of Podospora anserina by Oleic Acid Is Linked to Alterations in Energy Metabolism, Membrane Trafficking and Autophagy. Cells. 2022; 11(3):519. https://doi.org/10.3390/cells11030519
Chicago/Turabian StyleSchürmanns, Lea, Andrea Hamann, and Heinz D. Osiewacz. 2022. "Lifespan Increase of Podospora anserina by Oleic Acid Is Linked to Alterations in Energy Metabolism, Membrane Trafficking and Autophagy" Cells 11, no. 3: 519. https://doi.org/10.3390/cells11030519
APA StyleSchürmanns, L., Hamann, A., & Osiewacz, H. D. (2022). Lifespan Increase of Podospora anserina by Oleic Acid Is Linked to Alterations in Energy Metabolism, Membrane Trafficking and Autophagy. Cells, 11(3), 519. https://doi.org/10.3390/cells11030519





