Raman Spectroscopy and Its Modifications Applied to Biological and Medical Research
Abstract
1. Introduction
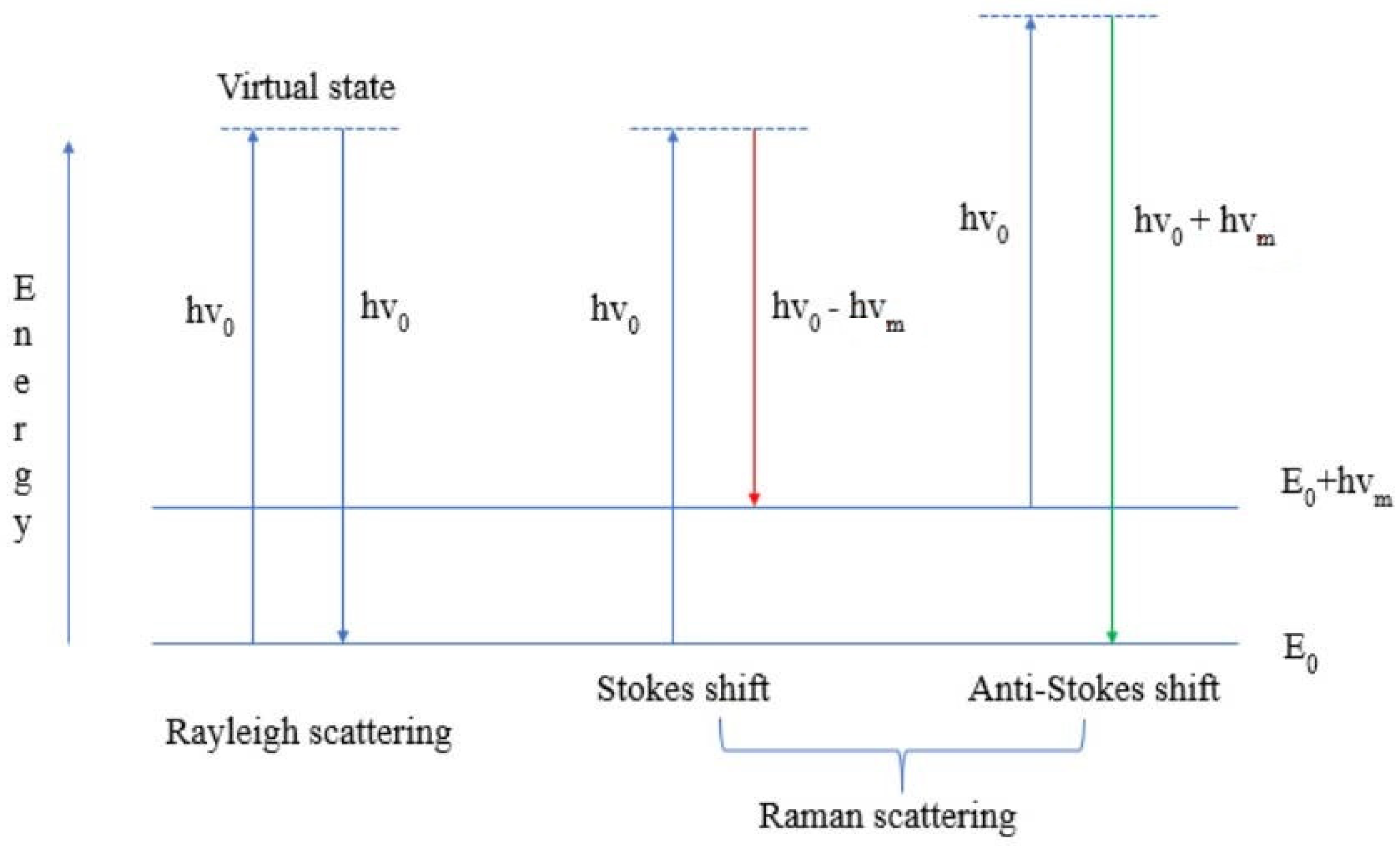
2. The Principles of the Method of Raman Spectroscopy
3. Raman Spectroscopy and Its Modifications: Advantages and Use
3.1. Surface-Enhanced Raman Spectroscopy (SERS)
3.2. Coherent Anti-Stokes Raman Scattering (CARS)
3.3. Resonance Raman Spectroscopy (RRS)
3.4. Spatially Offset Raman Spectroscopy (SORS)
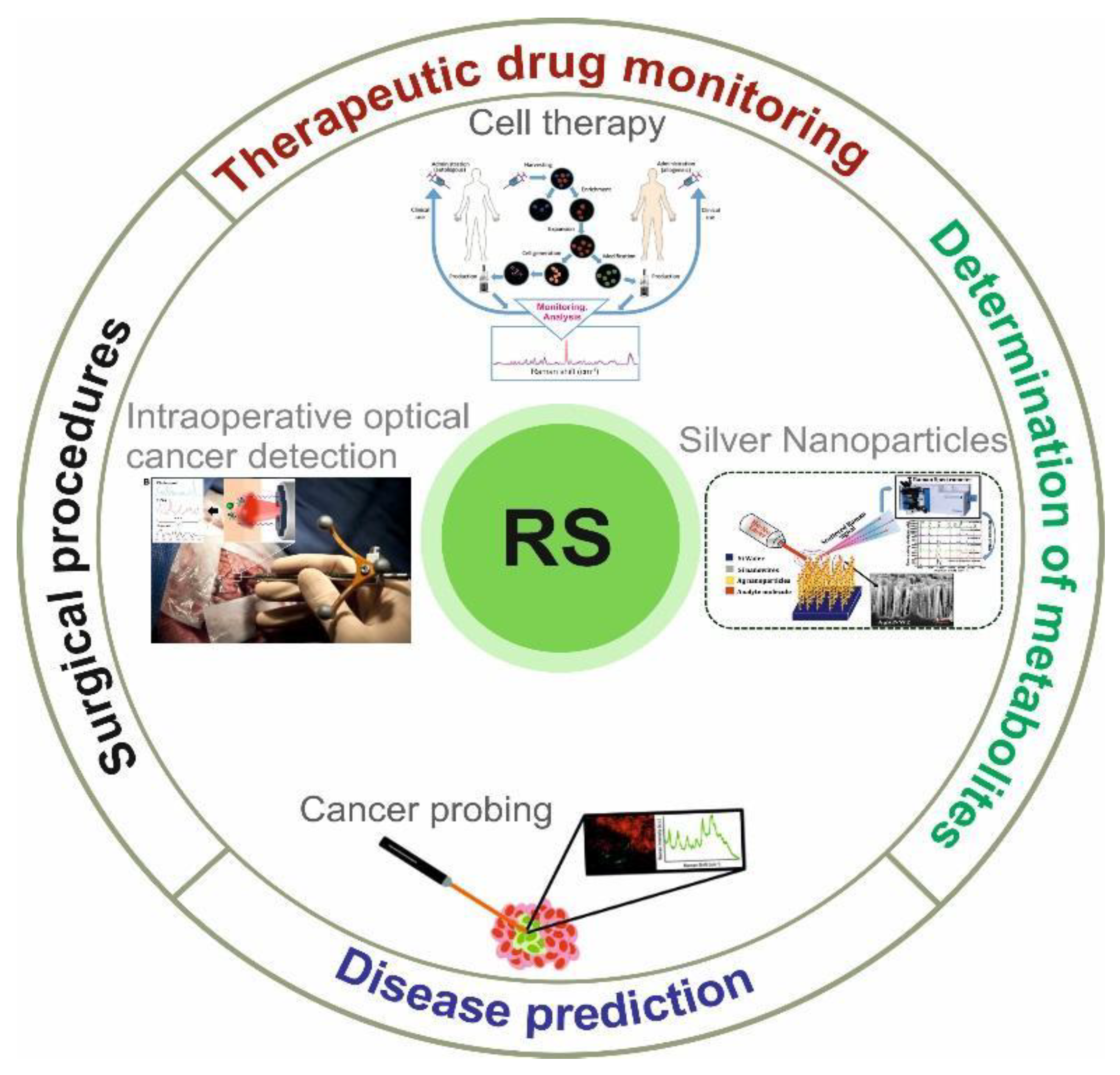
4. Application of Raman Spectroscopy in Biomedical Research
4.1. Disease Prediction
4.2. Surgical Procedures
4.3. Therapeutic Drug Monitoring (TDM)
4.4. Determination of Metabolites
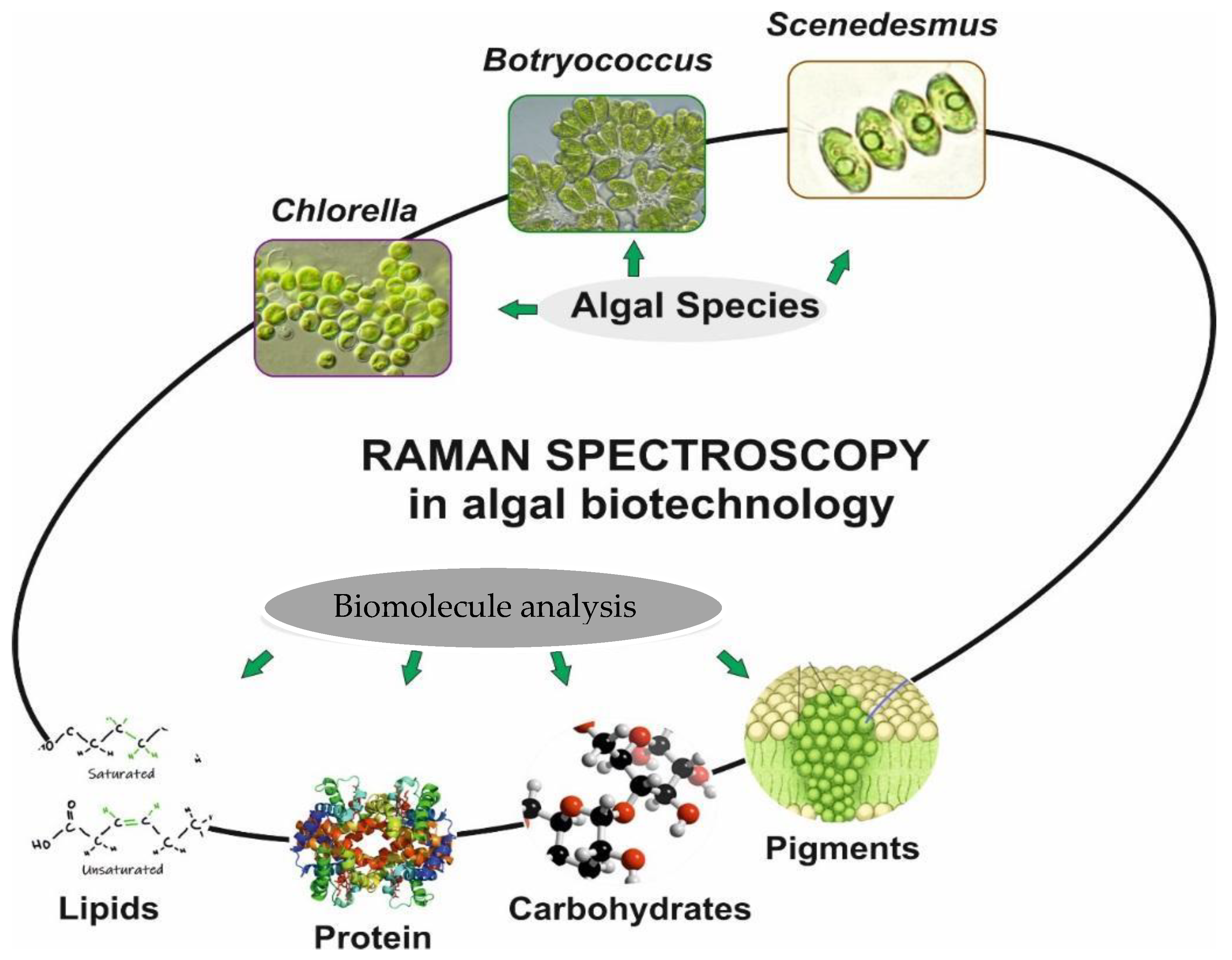
5. Biotechnology Application of Raman Spectroscopy
5.1. Application of Raman Spectroscopy in Algae Studies
5.1.1. Raman Spectroscopy Applied to Lipid
5.1.2. Application of Raman Spectroscopy on Pigment Investigation in Microalgae
6. Raman Spectroscopy for Photosynthetic Studies
7. Raman Spectroscopy for Analytical Studies
8. Future Perspectives
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nikelshparg, E.I.; Grivennikova, V.G.; Baizhumanov, A.A.; Semenova, A.A.; Sosnovtseva, V.; Goodilin, E.A.; Maksimov, G.V.; Brazhe, N.A. Probing Lipids in Biological Membranes Using SERS. Mendeleev Commun. 2019, 29, 635–637. [Google Scholar] [CrossRef]
- Brazhe, N.A.; Evlyukhin, A.B.; Goodilin, E.A.; Semenova, A.A.; Novikov, S.M.; Bozhevolnyi, S.I.; Chichkov, B.N.; Sarycheva, A.S.; Baizhumanov, A.A.; Nikelshparg, E.I.; et al. Probing Cytochrome c in Living Mitochondria with Surface-Enhanced Raman Spectroscopy. Sci. Rep. 2015, 5, 13793. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Thibault, M.-H.; Djaoued, Y.; Pelletier, C.; Touaibia, M.; Tchoukanova, N. Chromatographic, NMR and Vibrational Spectroscopic Investigations of Astaxanthin Esters: Application to “Astaxanthin-Rich Shrimp Oil” Obtained from Processing of Nordic Shrimps. Analyst 2015, 140, 7423–7433. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, D.; Pilger, C.; Hachmeister, H.; Oberländer, E.; Wördenweber, R.; Wichmann, J.; Mussgnug, J.H.; Huser, T.; Kruse, O. Label-Free in vivo Analysis of Intracellular Lipid Droplets in the Oleaginous Microalga Monoraphidium Neglectum by Coherent Raman Scattering Microscopy. Sci. Rep. 2016, 6, 35340. [Google Scholar] [CrossRef]
- Samek, O.; Jonáš, A.; Pilát, Z.; Zemánek, P.; Nedbal, L.; Tříska, J.; Kotas, P.; Trtílek, M. Raman Microspectroscopy of Individual Algal Cells: Sensing Unsaturation of Storage Lipids in vivo. Sensors 2010, 10, 8635–8651. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, X.; Wang, T.; Li, Z.; Han, D.; Yu, C.; Yang, C.; Qu, H.; Chi, H.; Wang, Y.; et al. A Sensitive and Rapid Bacterial Antibiotic Susceptibility Test Method by Surface Enhanced Raman Spectroscopy. Braz. J. Microbiol. 2020, 51, 875–881. [Google Scholar] [CrossRef]
- Jehlička, J.; Edwards, H.G.M.; Osterrothová, K.; Novotná, J.; Nedbalová, L.; Kopecký, J.; Němec, I.; Oren, A. Potential and Limits of Raman Spectroscopy for Carotenoid Detection in Microorganisms: Implications for Astrobiology. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2014, 372, 20140199. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman Spectroscopy to Characterize Biological Materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef]
- Buckley, K.; Ryder, A.G. Applications of Raman Spectroscopy in Biopharmaceutical Manufacturing: A Short Review. Appl. Spectrosc. 2017, 71, 1085–1116. [Google Scholar] [CrossRef]
- Alvarez-Puebla, R.A.; Liz-Marzán, L.M. SERS-Based Diagnosis and Biodetection. Small 2010, 6, 604–610. [Google Scholar] [CrossRef]
- Hudson, S.D.; Chumanov, G. Bioanalytical Applications of SERS (Surface-Enhanced Raman Spectroscopy). Anal. Bioanal. Chem. 2009, 394, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Cialla, D.; März, A.; Böhme, R.; Theil, F.; Weber, K.; Schmitt, M.; Popp, J. Surface-Enhanced Raman Spectroscopy (SERS): Progress and Trends. Anal. Bioanal. Chem. 2012, 403, 27–54. [Google Scholar] [CrossRef] [PubMed]
- Asiala, S.M.; Schultz, Z.D. Surface Enhanced Raman Correlation Spectroscopy of Particles in Solution. Anal. Chem. 2014, 86, 2625–2632. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Beal, C.M.; Cai, W.W.; Ruoff, R.S.; Terentjev, E.M. Micro-Raman Spectroscopy of Algae: Composition Analysis and Fluorescence Background Behavior. Biotechnol. Bioeng. 2010, 105, 889–898. [Google Scholar] [CrossRef]
- Cavonius, L.; Fink, H.; Kiskis, J.; Albers, E.; Undeland, I.; Enejder, A. Imaging of Lipids in Microalgae with Coherent Anti-Stokes Raman Scattering Microscopy. Plant Physiol. 2015, 167, 603–616. [Google Scholar] [CrossRef]
- He, X.N.; Allen, J.; Black, P.N.; Baldacchini, T.; Huang, X.; Huang, H.; Jiang, L.; Lu, Y.F. Coherent Anti-Stokes Raman Scattering and Spontaneous Raman Spectroscopy and Microscopy of Microalgae with Nitrogen Depletion. Biomed. Opt. Express 2012, 3, 2896–2906. [Google Scholar] [CrossRef]
- Legesse, F.B.; Rüger, J.; Meyer, T.; Krafft, C.; Schmitt, M.; Popp, J. Investigation of Microalgal Carotenoid Content Using Coherent Anti-Stokes Raman Scattering (CARS) Microscopy and Spontaneous Raman Spectroscopy. ChemPhysChem 2018, 19, 1048–1055. [Google Scholar] [CrossRef]
- Sharma, S.K.; Nelson, D.R.; Abdrabu, R.; Khraiwesh, B.; Jijakli, K.; Arnoux, M.; O’Connor, M.J.; Bahmani, T.; Cai, H.; Khapli, S.; et al. An Integrative Raman Microscopy-Based Workflow for Rapid in Situ Analysis of Microalgal Lipid Bodies. Biotechnol. Biofuels 2015, 8, 164. [Google Scholar] [CrossRef]
- He, S.; Xie, W.; Zhang, P.; Fang, S.; Li, Z.; Tang, P.; Gao, X.; Guo, J.; Tlili, C.; Wang, D. Preliminary Identification of Unicellular Algal Genus by Using Combined Confocal Resonance Raman Spectroscopy with PCA and DPLS Analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 190, 417–422. [Google Scholar] [CrossRef]
- Osterrothová, K.; Culka, A.; Němečková, K.; Kaftan, D.; Nedbalová, L.; Procházková, L.; Jehlička, J. Analyzing Carotenoids of Snow Algae by Raman Microspectroscopy and High-Performance Liquid Chromatography. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 212, 262–271. [Google Scholar] [CrossRef]
- Kish, E.; Wang, K.; Llansola-Portoles, M.J.; Ilioaia, C.; Pascal, A.A.; Robert, B.; Yang, C. Probing the Pigment Binding Sites in LHCII with Resonance Raman Spectroscopy: The Effect of Mutations at S123. Biochim. Biophys. Acta BBA- Bioenerg. 2016, 1857, 1490–1496. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ji, Y.; Wang, Y.; Jia, J.; Li, J.; Huang, S.; Han, D.; Hu, Q.; Huang, W.E.; Xu, J. Quantitative Dynamics of Triacylglycerol Accumulation in Microalgae Populations at Single-Cell Resolution Revealed by Raman Microspectroscopy. Biotechnol. Biofuels 2014, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Volponi, J.V.; Oliver, A.E.; Parikh, A.N.; Simmons, B.A.; Singh, S. In Vivo Lipidomics Using Single-Cell Raman Spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 3809–3814. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.-L.; Juang, Y.-J. Black Silicon SERS Substrate: Effect of Surface Morphology on SERS Detection and Application of Single Algal Cell Analysis. Biosens. Bioelectron. 2014, 53, 37–42. [Google Scholar] [CrossRef]
- Wang, Y. Construction of Artificial Intelligence-Assisted Prostate Tumor Early Diagnosis System Based on Surface Enhanced Raman Spectroscopy. 2020. Available online: http://www.chictr.org.cn/showproj.aspx?proj=60141 (accessed on 12 December 2021).
- U.S. National Library of Medicine Non-Invasive Assessment of Mechano-Chemical Properties of Urine Proteins by Hybrid Brillouin-Raman Spectroscopy. Available online: https://clinicaltrials.gov/ct2/show/NCT04311684 (accessed on 12 December 2021).
- U.S. National Library of Medicine. Raman Analysis of Saliva as Biomarker of COPD (BIO-RAnCh). Available online: https://clinicaltrials.gov/ct2/show/NCT04628962 (accessed on 12 December 2021).
- Zavaleta, C.L.; Garai, E.; Liu, J.T.C.; Sensarn, S.; Mandella, M.J.; de Sompel, D.V.; Friedland, S.; Dam, J.V.; Contag, C.H.; Gambhir, S.S. A Raman-Based Endoscopic Strategy for Multiplexed Molecular Imaging. Proc. Natl. Acad. Sci. USA 2013, 110, E2288–E2297. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, Y.; Song, W.; Lu, L. Delineating the Tumor Margin with Intraoperative Surface-Enhanced Raman Spectroscopy. Anal. Bioanal. Chem. 2019, 411, 3993–4006. [Google Scholar] [CrossRef]
- Evans, C.L.; Xie, X.S. Coherent Anti-Stokes Raman Scattering Microscopy: Chemical Imaging for Biology and Medicine. Annu. Rev. Anal. Chem. 2008, 1, 883–909. [Google Scholar] [CrossRef]
- Evans, C.L.; Potma, E.O.; Puoris’haag, M.; Côté, D.; Lin, C.P.; Xie, X.S. Chemical Imaging of Tissue in vivo with Video-Rate Coherent Anti-Stokes Raman Scattering Microscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 16807–16812. [Google Scholar] [CrossRef]
- Cheng, J.-X.; Xie, X.S. Coherent Anti-Stokes Raman Scattering Microscopy: Instrumentation, Theory, and Applications. J. Phys. Chem. B 2004, 108, 827–840. [Google Scholar] [CrossRef]
- Su, L.-M. Robot-Assisted Radical Prostatectomy: Advances since 2005. Curr. Opin. Urol. 2010, 20, 130–135. [Google Scholar] [CrossRef]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-Free Raman Observation of Cytochrome c Dynamics during Apoptosis. Proc. Natl. Acad. Sci. USA 2012, 109, 28–32. [Google Scholar] [CrossRef]
- Vardaki, M.Z.; Atkins, C.G.; Schulze, H.G.; Devine, D.V.; Serrano, K.; Blades, M.W.; Turner, R.F.B. Raman Spectroscopy of Stored Red Blood Cell Concentrate within Sealed Transfusion Blood Bags. Analyst 2018, 143, 6006–6013. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Ochoa, M.; Maher, J.R.; Awad, H.A.; Berger, A.J. Sensitivity of Spatially Offset Raman Spectroscopy (SORS) to Subcortical Bone Tissue. J. Biophotonics 2017, 10, 990–996. [Google Scholar] [CrossRef]
- Desroches, J.; Jermyn, M.; Pinto, M.; Picot, F.; Tremblay, M.-A.; Obaid, S.; Marple, E.; Urmey, K.; Trudel, D.; Soulez, G.; et al. A New Method Using Raman Spectroscopy for in vivo Targeted Brain Cancer Tissue Biopsy. Sci. Rep. 2018, 8, 1792. [Google Scholar] [CrossRef] [PubMed]
- Choo-Smith, L.-P.; Edwards, H.G.M.; Endtz, H.P.; Kros, J.M.; Heule, F.; Barr, H.; Robinson, J.S., Jr.; Bruining, H.A.; Puppels, G.J. Medical Applications of Raman Spectroscopy: From Proof of Principle to Clinical Implementation. Biopolymers 2002, 67, 1–9. [Google Scholar] [CrossRef]
- Pandey, R.; Paidi, S.K.; Valdez, T.A.; Zhang, C.; Spegazzini, N.; Dasari, R.R.; Barman, I. Noninvasive Monitoring of Blood Glucose with Raman Spectroscopy. Acc. Chem. Res. 2017, 50, 264–272. [Google Scholar] [CrossRef]
- Cui, S.; Zhang, S.; Yue, S. Raman Spectroscopy and Imaging for Cancer Diagnosis. J. Healthc. Eng. 2018, 2018, e8619342. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-W.; Huang, C.-C.; Sheu, J.-H.; Lin, C.-W.; Lin, L.-F.; Jin, J.-S.; Chen, W. Differentiating Gastrointestinal Stromal Tumors from Gastric Adenocarcinomas and Normal Mucosae Using Confocal Raman Microspectroscopy. J. Biomed. Opt. 2016, 21, 075006. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Ding, R.; Shen, L.; Gao, P.; Xu, H.; Xiu, C.; Zhang, H.; Song, D.; Han, B. Characterization and Imaging of Surgical Specimens of Invasive Breast Cancer and Normal Breast Tissues with the Application of Raman Spectral Mapping: A Feasibility Study and Comparison with Randomized Single-point Detection Method. Oncol. Lett. 2020, 20, 2969–2976. [Google Scholar] [CrossRef]
- Kopeć, M.; Abramczyk, H. Angiogenesis—A Crucial Step in Breast Cancer Growth, Progression and Dissemination by Raman Imaging. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 198, 338–345. [Google Scholar] [CrossRef]
- Winnard, P.T.; Zhang, C.; Vesuna, F.; Kang, J.W.; Garry, J.; Dasari, R.R.; Barman, I.; Raman, V. Organ-Specific Isogenic Metastatic Breast Cancer Cell Lines Exhibit Distinct Raman Spectral Signatures and Metabolomes. Oncotarget 2017, 8, 20266–20287. [Google Scholar] [CrossRef] [PubMed]
- Paidi, S.K.; Rizwan, A.; Zheng, C.; Cheng, M.; Glunde, K.; Barman, I. Label-Free Raman Spectroscopy Detects Stromal Adaptations in Premetastatic Lungs Primed by Breast Cancer. Cancer Res. 2017, 77, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Ryzhikova, E.; Ralbovsky, N.M.; Sikirzhytski, V.; Kazakov, O.; Halamkova, L.; Quinn, J.; Zimmerman, E.A.; Lednev, I.K. Raman Spectroscopy and Machine Learning for Biomedical Applications: Alzheimer’s Disease Diagnosis Based on the Analysis of Cerebrospinal Fluid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 248, 119188. [Google Scholar] [CrossRef] [PubMed]
- Ralbovsky, N.M.; Halámková, L.; Wall, K.; Anderson-Hanley, C.; Lednev, I.K. Screening for Alzheimer’s Disease Using Saliva: A New Approach Based on Machine Learning and Raman Hyperspectroscopy. J. Alzheimers Dis. 2019, 71, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Ralbovsky, N.M.; Fitzgerald, G.S.; McNay, E.C.; Lednev, I.K. Towards Development of a Novel Screening Method for Identifying Alzheimer’s Disease Risk: Raman Spectroscopy of Blood Serum and Machine Learning. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 254, 119603. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.; Choi, B.H.; Kang, K.-W.; Jeong, H.; Park, Y.; et al. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Barnas, E.; Skret-Magierlo, J.; Skret, A.; Kaznowska, E.; Depciuch, J.; Szmuc, K.; Łach, K.; Krawczyk-Marć, I.; Cebulski, J. Simultaneous FTIR and Raman Spectroscopy in Endometrial Atypical Hyperplasia and Cancer. Int. J. Mol. Sci. 2020, 21, 4828. [Google Scholar] [CrossRef]
- Lin, J.; Chen, R.; Feng, S.; Pan, J.; Li, Y.; Chen, G.; Cheng, M.; Huang, Z.; Yu, Y.; Zeng, H. A Novel Blood Plasma Analysis Technique Combining Membrane Electrophoresis with Silver Nanoparticle-Based SERS Spectroscopy for Potential Applications in Noninvasive Cancer Detection. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 655–663. [Google Scholar] [CrossRef]
- Batool, F.; Nawaz, H.; Majeed, M.I.; Rashid, N.; Bashir, S.; Akbar, S.; Abubakar, M.; Ahmad, S.; Ashraf, M.N.; Ali, S.; et al. SERS-Based Viral Load Quantification of Hepatitis B Virus from PCR Products. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 255, 119722. [Google Scholar] [CrossRef]
- Shen, L.; Du, Y.; Wei, N.; Li, Q.; Li, S.; Sun, T.; Xu, S.; Wang, H.; Man, X.; Han, B. SERS Studies on Normal Epithelial and Cancer Cells Derived from Clinical Breast Cancer Specimens. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 237, 118364. [Google Scholar] [CrossRef]
- Moisoiu, V.; Badarinza, M.; Stefancu, A.; Iancu, S.D.; Serban, O.; Leopold, N.; Fodor, D. Combining Surface-Enhanced Raman Scattering (SERS) of Saliva and Two-Dimensional Shear Wave Elastography (2D-SWE) of the Parotid Glands in the Diagnosis of Sjögren’s Syndrome. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 235, 118267. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, V.; Siddique, R.H.; Park, H.; Choo, H. Bioinspired Disordered Flexible Metasurfaces for Human Tear Analysis Using Broadband Surface-Enhanced Raman Scattering. ACS Omega 2020, 5, 12915–12922. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xu, Y.; Zhao, H.; Li, S.; Chen, L. Design and Preparation of Centrifugal Microfluidic Chip Integrated with SERS Detection for Rapid Diagnostics. Talanta 2019, 194, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Frimpong, R.; Jang, W.; Kim, J.-H.; Driskell, J.D. Rapid Vertical Flow Immunoassay on AuNP Plasmonic Paper for SERS-Based Point of Need Diagnostics. Talanta 2021, 223, 121739. [Google Scholar] [CrossRef] [PubMed]
- Willner, M.R.; McMillan, K.S.; Graham, D.; Vikesland, P.J.; Zagnoni, M. Surface-Enhanced Raman Scattering Based Microfluidics for Single-Cell Analysis. Anal. Chem. 2018, 90, 12004–12010. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Gandra, N.; Fales, A.M.; Taylor, S.M.; Vo-Dinh, T. Sensitive DNA Detection and SNP Discrimination Using Ultrabright SERS Nanorattles and Magnetic Beads for Malaria Diagnostics. Biosens. Bioelectron. 2016, 81, 8–14. [Google Scholar] [CrossRef]
- Lu, F.-K.; Basu, S.; Igras, V.; Hoang, M.P.; Ji, M.; Fu, D.; Holtom, G.R.; Neel, V.A.; Freudiger, C.W.; Fisher, D.E.; et al. Label-Free DNA Imaging in Vivo with Stimulated Raman Scattering Microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 11624–11629. [Google Scholar] [CrossRef]
- Harmsen, S.; Rogalla, S.; Huang, R.; Spaliviero, M.; Neuschmelting, V.; Hayakawa, Y.; Lee, Y.; Tailor, Y.; Toledo-Crow, R.; Kang, J.W.; et al. Detection of Premalignant Gastrointestinal Lesions Using Surface-Enhanced Resonance Raman Scattering–Nanoparticle Endoscopy. ACS Nano 2019, 13, 1354–1364. [Google Scholar] [CrossRef]
- Ryzhikova, E.; Kazakov, O.; Halamkova, L.; Celmins, D.; Malone, P.; Molho, E.; Zimmerman, E.A.; Lednev, I.K. Raman Spectroscopy of Blood Serum for Alzheimer’s Disease Diagnostics: Specificity Relative to Other Types of Dementia. J. Biophotonics 2015, 8, 584–596. [Google Scholar] [CrossRef]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of Tumors during Tissue-Conserving Surgery with Integrated Autofluorescence and Raman Scattering Microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 15189–15194. [Google Scholar] [CrossRef]
- Motz, J.T.; Hunter, M.; Galindo, L.H.; Gardecki, J.A.; Kramer, J.R.; Dasari, R.R.; Feld, M.S. Optical Fiber Probe for Biomedical Raman Spectroscopy. Appl. Opt. 2004, 43, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Jermyn, M.; Mercier, J.; Aubertin, K.; Desroches, J.; Urmey, K.; Karamchandiani, J.; Marple, E.; Guiot, M.-C.; Leblond, F.; Petrecca, K. Highly Accurate Detection of Cancer In Situ with Intraoperative, Label-Free, Multimodal Optical Spectroscopy. Cancer Res. 2017, 77, 3942–3950. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.F.; de la Zerda, A.; Jokerst, J.V.; Zavaleta, C.L.; Kempen, P.J.; Mittra, E.; Pitter, K.; Huang, R.; Campos, C.; Habte, F.; et al. A Brain Tumor Molecular Imaging Strategy Using a New Triple-Modality MRI-Photoacoustic-Raman Nanoparticle. Nat. Med. 2012, 18, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Lukic, A.; Dochow, S.; Bae, H.; Matz, G.; Latka, I.; Messerschmidt, B.; Schmitt, M.; Popp, J. Endoscopic Fiber Probe for Nonlinear Spectroscopic Imaging. Optica 2017, 4, 496–501. [Google Scholar] [CrossRef]
- Jaworska, A.; Fornasaro, S.; Sergo, V.; Bonifacio, A. Potential of Surface Enhanced Raman Spectroscopy (SERS) in Therapeutic Drug Monitoring (TDM). A Critical Review. Biosensors 2016, 6, 47. [Google Scholar] [CrossRef]
- Neef, C.; Touw, D.J.; Stolk, L.M. Therapeutic Drug Monitoring in Clinical Research. Pharm. Med. 2008, 22, 235–244. [Google Scholar] [CrossRef]
- Kang, J.-S.; Lee, M.-H. Overview of Therapeutic Drug Monitoring. Korean J. Intern. Med. 2009, 24, 1–10. [Google Scholar] [CrossRef]
- Lennard, L. Therapeutic Drug Monitoring of Antimetabolic Cytotoxic Drugs. Br. J. Clin. Pharmacol. 1999, 47, 131–143. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, H.; Li, R.; Yu, Z.; Lu, F. Raman Spectroscopy and Mapping Technique for the Identification of Expired Drugs. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117407. [Google Scholar] [CrossRef]
- Fei, J.; Wu, L.; Zhang, Y.; Zong, S.; Wang, Z.; Cui, Y. Pharmacokinetics-on-a-Chip Using Label-Free SERS Technique for Programmable Dual-Drug Analysis. ACS Sens. 2017, 2, 773–780. [Google Scholar] [CrossRef]
- Chen, C.; Yang, L.; Li, H.; Chen, F.; Chen, C.; Gao, R.; Lv, X.; Tang, J. Raman Spectroscopy Combined with Multiple Algorithms for Analysis and Rapid Screening of Chronic Renal Failure. Photodiagnosis Photodyn. Ther. 2020, 30, 101792. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Furukawa, H.; Ito, Y.; Nagahama, M.; Hayashi, T.; Ishii-Teshima, M.; Nagai, M. Photocatalytic Nanofabrication and Intracellular Raman Imaging of Living Cells with Functionalized AFM Probes. Micromachines 2020, 11, 495. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, Y.; Cheng, J.; Fujita, A.; Tokumoto, T.; Fujimoto, T. Cytoplasmic Lipid Droplets Are Sites of Convergence of Proteasomal and Autophagic Degradation of Apolipoprotein, B. Mol. Biol. Cell 2006, 17, 2674–2683. [Google Scholar] [CrossRef] [PubMed]
- Samuel, A.Z.; Miyaoka, R.; Ando, M.; Gaebler, A.; Thiele, C.; Takeyama, H. Molecular Profiling of Lipid Droplets inside HuH7 Cells with Raman Micro-Spectroscopy. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Rodionova, N.N.; Allakhverdiev, E.S.; Maksimov, G.V. Study of Myelin Structure Changes during the Nerve Fibers Demyelination. PLoS ONE 2017, 12, e0185170. [Google Scholar] [CrossRef][Green Version]
- Akhalaya, M.Y.; Maksimov, G.V.; Rubin, A.B.; Lademann, J.; Darvin, M.E. Molecular Action Mechanisms of Solar Infrared Radiation and Heat on Human Skin. Ageing Res. Rev. 2014, 16, 1–11. [Google Scholar] [CrossRef]
- Gellermann, W.; Ermakov, I.V.; Ermakova, M.R.; McClane, R.W.; Zhao, D.-Y.; Bernstein, P.S. In Vivo Resonant Raman Measurement of Macular Carotenoid Pigments in the Young and the Aging Human Retina. JOSA A 2002, 19, 1172–1186. [Google Scholar] [CrossRef]
- Ermakov, I.V.; Ermakova, M.R.; Gellermann, W.; Lademann, J. Noninvasive Selective Detection of Lycopene and Beta-Carotene in Human Skin Using Raman Spectroscopy. J. Biomed. Opt. 2004, 9, 332–338. [Google Scholar] [CrossRef]
- Valpapuram, I.; Candeloro, P.; Coluccio, M.L.; Parrotta, E.I.; Giugni, A.; Das, G.; Cuda, G.; Di Fabrizio, E.; Perozziello, G. Waveguiding and SERS Simplified Raman Spectroscopy on Biological Samples. Biosensors 2019, 9, 37. [Google Scholar] [CrossRef]
- Dijkstra, R.J.; Scheenen, W.J.J.M.; Dam, N.; Roubos, E.W.; ter Meulen, J.J. Monitoring Neurotransmitter Release Using Surface-Enhanced Raman Spectroscopy. J. Neurosci. Methods 2007, 159, 43–50. [Google Scholar] [CrossRef]
- Silwal, A.P.; Lu, H.P. Mode-Selective Raman Imaging of Dopamine–Human Dopamine Transporter Interaction in Live Cells. ACS Chem. Neurosci. 2018, 9, 3117–3127. [Google Scholar] [CrossRef] [PubMed]
- Bahadar, H.; Maqbool, F.; Niaz, K.; Abdollahi, M. Toxicity of Nanoparticles and an Overview of Current Experimental Models. Iran. Biomed. J. 2016, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Manciu, F.S.; Lee, K.H.; Durrer, W.G.; Bennet, K.E. Detection and Monitoring of Neurotransmitters—A Spectroscopic Analysis. Neuromodulation Technol. Neural Interface 2013, 16, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Baria, E.; Cicchi, R.; Pavone, F.S. Custom Multiphoton/Raman Microscopy Setup for Imaging and Characterization of Biological Samples. Methods Protoc. 2019, 2, 51. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Hiraishi, N.; Honma, T.; Boschetto, F.; Zanocco, M.; Zhu, W.; Adachi, T.; Kanamura, N.; Yamamoto, T.; Pezzotti, G. Raman Spectroscopy for Early Detection and Monitoring of Dentin Demineralization. Dent. Mater. 2020, 36, 1635–1644. [Google Scholar] [CrossRef]
- Arabi, M.; Ostovan, A.; Zhang, Z.; Wang, Y.; Mei, R.; Fu, L.; Wang, X.; Ma, J.; Chen, L. Label-Free SERS Detection of Raman-Inactive Protein Biomarkers by Raman Reporter Indicator: Toward Ultrasensitivity and Universality. Biosens. Bioelectron. 2021, 174, 112825. [Google Scholar] [CrossRef]
- Arend, N.; Pittner, A.; Ramoji, A.; Mondol, A.S.; Dahms, M.; Rüger, J.; Kurzai, O.; Schie, I.W.; Bauer, M.; Popp, J.; et al. Detection and Differentiation of Bacterial and Fungal Infection of Neutrophils from Peripheral Blood Using Raman Spectroscopy. Anal. Chem. 2020, 92, 10560–10568. [Google Scholar] [CrossRef]
- Collard, L.; Sinjab, F.; Notingher, I. Raman Spectroscopy Study of Curvature-Mediated Lipid Packing and Sorting in Single Lipid Vesicles. Biophys. J. 2019, 117, 1589–1598. [Google Scholar] [CrossRef]
- Luneva, O.G.; Brazhe, N.A.; Maksimova, N.V.; Rodnenkov, O.V.; Parshina, E.Y.; Bryzgalova, N.Y.; Maksimov, G.V.; Rubin, A.B.; Orlov, S.N.; Chazov, E.I. Ion Transport, Membrane Fluidity and Haemoglobin Conformation in Erythrocyte from Patients with Cardiovascular Diseases: Role of Augmented Plasma Cholesterol. Pathophysiology 2007, 14, 41–46. [Google Scholar] [CrossRef]
- Maksimov, G.V.; Maksimova, N.V.; Churin, A.A.; Orlov, S.N.; Rubin, A.B. Study on Conformational Changes in Hemoglobin Protoporphyrin in Essential Hypertension. Biochem. Mosc. 2001, 66, 295–299. [Google Scholar] [CrossRef]
- Maksimov, G.V.; Luneva, O.G.; Maksimova, N.V.; Matettuchi, E.; Medvedev, E.A.; Pashchenko, V.Z.; Rubin, A.B. Role of Viscosity and Permeability of the Erythrocyte Plasma Membrane in Changes in Oxygen-Binding Properties of Hemoglobin during Diabetes Mellitus. Bull. Exp. Biol. Med. 2005, 140, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Brazhe, N.A.; Baizhumanov, A.A.; Parshina, E.Y.; Yusipovich, A.I.; Akhalaya, M.Y.; Yarlykova, Y.V.; Labetskaya, O.I.; Ivanova, S.M.; Morukov, B.V.; Maksimov, G.V. Studies of the Blood Antioxidant System and Oxygen-Transporting Properties of Human Erythrocytes during 105-Day Isolation. Hum. Physiol. 2014, 40, 804–809. [Google Scholar] [CrossRef]
- Allakhverdiev, E.S.; Slatinskaya, O.V.; Rodnenkov, O.V.; Maksimov, G.V.; Martynyuk, T.V. Evaluation of Hemoglobin Conformation State in Patients with Pulmonary Hypertension. Russ. J. Cardiol. 2021, 25, 23. [Google Scholar]
- Skjånes, K.; Rebours, C.; Lindblad, P. Potential for Green Microalgae to Produce Hydrogen, Pharmaceuticals and Other High Value Products in a Combined Process. Crit. Rev. Biotechnol. 2013, 33, 172–215. [Google Scholar] [CrossRef] [PubMed]
- Schenk, P.M.; Thomas-Hall, S.R.; Stephens, E.; Marx, U.C.; Mussgnug, J.H.; Posten, C.; Kruse, O.; Hankamer, B. Second Generation Biofuels: High-Efficiency Microalgae for Biodiesel Production. BioEnergy Res. 2008, 1, 20–43. [Google Scholar] [CrossRef]
- Sadvakasova, A.K.; Kossalbayev, B.D.; Zayadan, B.K.; Kirbayeva, D.K.; Alwasel, S.; Allakhverdiev, S.I. Potential of Cyanobacteria in the Conversion of Wastewater to Biofuels. World J. Microbiol. Biotechnol. 2021, 37, 140. [Google Scholar] [CrossRef]
- Bolatkhan, K.; Sadvakasova, A.K.; Zayadan, B.K.; Kakimova, A.B.; Sarsekeyeva, F.K.; Kossalbayev, B.D.; Bozieva, A.M.; Alwasel, S.; Allakhverdiev, S.I. Prospects for the Creation of a Waste-Free Technology for Wastewater Treatment and Utilization of Carbon Dioxide Based on Cyanobacteria for Biodiesel Production. J. Biotechnol. 2020, 324, 162–170. [Google Scholar] [CrossRef]
- Zayadan, B.; Usserbayeva, A.; Bolatkhan, K.; Akmukhanova, N.; Kossalbayev, B.; Baizhigitova, A.; Los, D. Screening of Isolated and Collection Strains of Cyanobacteria on Productivity for Determining Their Biotechnological Potential. Eur. J. Entomol. 2018, 55. [Google Scholar] [CrossRef]
- Breuer, G.; de Jaeger, L.; Artus, V.G.; Martens, D.E.; Springer, J.; Draaisma, R.B.; Eggink, G.; Wijffels, R.H.; Lamers, P.P. Superior Triacylglycerol (TAG) Accumulation in Starchless Mutants of Scenedesmus Obliquus: (II) Evaluation of TAG Yield and Productivity in Controlled Photobioreactors. Biotechnol. Biofuels 2014, 7, 70. [Google Scholar] [CrossRef]
- Kaczor, A.; Turnau, K.; Baranska, M. In Situ Raman Imaging of Astaxanthin in a Single Microalgal Cell. Analyst 2011, 136, 1109–1112. [Google Scholar] [CrossRef]
- Hosokawa, M.; Ando, M.; Mukai, S.; Osada, K.; Yoshino, T.; Hamaguchi, H.; Tanaka, T. In Vivo Live Cell Imaging for the Quantitative Monitoring of Lipids by Using Raman Microspectroscopy. Anal. Chem. 2014, 86, 8224–8230. [Google Scholar] [CrossRef] [PubMed]
- Pořízka, P.; Prochazková, P.; Prochazka, D.; Sládková, L.; Novotný, J.; Petrilak, M.; Brada, M.; Samek, O.; Pilát, Z.; Zemánek, P.; et al. Algal Biomass Analysis by Laser-Based Analytical Techniques—A Review. Sensors 2014, 14, 17725–17752. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Jie, D.; Cuello, J.J.; Johnson, D.J.; Qiu, Z.; He, Y. Microalgal Detection by Raman Microspectroscopy. TrAC Trends Anal. Chem. 2014, 53, 33–40. [Google Scholar] [CrossRef]
- Gouda, M.; Chen, K.; Li, X.; Liu, Y.; He, Y. Detection of Microalgae Single-Cell Antioxidant and Electrochemical Potentials by Gold Microelectrode and Raman Micro-Spectroscopy Combined with Chemometrics. Sens. Actuators B Chem. 2021, 329, 129229. [Google Scholar] [CrossRef]
- Brahma, S.K.; Hargraves, P.E.; Howard, W.F.; Nelson, W.H. A Resonance Raman Method for the Rapid Detection and Identification of Algae in Water. Appl. Spectrosc. 1983, 37, 55–58. [Google Scholar] [CrossRef]
- Vítek, P.; Jehlička, J.; Edwards, H.G.M.; Hutchinson, I.; Ascaso, C.; Wierzchos, J. The Miniaturized Raman System and Detection of Traces of Life in Halite from the Atacama Desert: Some Considerations for the Search for Life Signatures on Mars. Astrobiology 2012, 12, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Jehlička, J.; Edwards, H.G.M.; Oren, A. Raman Spectroscopy of Microbial Pigments. Appl. Environ. Microbiol. 2014, 80, 3286–3295. [Google Scholar] [CrossRef]
- Weiss, T.L.; Chun, H.J.; Okada, S.; Vitha, S.; Holzenburg, A.; Laane, J.; Devarenne, T.P. Raman Spectroscopy Analysis of Botryococcene Hydrocarbons from the Green Microalga Botryococcus Braunii*. J. Biol. Chem. 2010, 285, 32458–32466. [Google Scholar] [CrossRef]
- Ramya, A.N.; Ambily, P.S.; Sujitha, B.S.; Arumugam, M.; Maiti, K.K. Single Cell Lipid Profiling of Scenedesmus Quadricauda CASA-CC202 under Nitrogen Starved Condition by Surface Enhanced Raman Scattering (SERS) Fingerprinting. Algal Res. 2017, 25, 200–206. [Google Scholar] [CrossRef]
- Byler, D.M.; Susi, H. Examination of the Secondary Structure of Proteins by Deconvolved FTIR Spectra. Biopolymers 1986, 25, 469–487. [Google Scholar] [CrossRef]
- Barka, A.; Amira, A.B.; Francis, F.; Blecker, C. Physicochemical Characterization of Colored Soluble Protein Fractions Extracted from Spirulina (Spirulina Platensis). Food Sci. Technol. Int. 2018, 24, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, S.; Pugazhendy, K.; Sangeetha, D.; Vasantharaja, C.; Prabakaran, S.; Meenambal, M. Fourier Transform Infrared (FT-IR) Spectoroscopic Analysis of Spirulina. Int. J. Pharm. Biol. Arch. 2012, 3, 969–972. [Google Scholar]
- Moudříková, Š.; Sadowsky, A.; Metzger, S.; Nedbal, L.; Mettler-Altmann, T.; Mojzeš, P. Quantification of Polyphosphate in Microalgae by Raman Microscopy and by a Reference Enzymatic Assay. Anal. Chem. 2017, 89, 12006–12013. [Google Scholar] [CrossRef]
- Jehlička, J.; Culka, A.; Nedbalová, L. Colonization of Snow by Microorganisms as Revealed Using Miniature Raman Spectrometers—Possibilities for Detecting Carotenoids of Psychrophiles on Mars? Astrobiology 2016, 16, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Javee, A.; Sulochana, S.B.; Pallissery, S.J.; Arumugam, M. Major Lipid Body Protein: A Conserved Structural Component of Lipid Body Accumulated during Abiotic Stress in S. quadricauda CASA-CC202. Front. Energy Res. 2016, 4. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, P. The New Face of the Lipid Droplet: Lipid Droplet Proteins. Proteomics 2019, 19, 1700223. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kannegulla, A.; Kong, X.; Sun, R.; Liu, Y.; Wang, R.; Yu, Q.; Wang, A.X. Simultaneous Colorimetric and Surface-Enhanced Raman Scattering Detection of Melamine from Milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 231, 118130. [Google Scholar] [CrossRef] [PubMed]
- Solovchenko, A.; Neverov, K. Carotenogenic Response in Photosynthetic Organisms: A Colorful Story. Photosynth. Res. 2017, 133, 31–47. [Google Scholar] [CrossRef]
- Chekanov, K.; Lukyanov, A.; Boussiba, S.; Aflalo, C.; Solovchenko, A. Modulation of Photosynthetic Activity and Photoprotection in Haematococcus Pluvialis Cells during Their Conversion into Haematocysts and Back. Photosynth. Res. 2016, 128, 313–323. [Google Scholar] [CrossRef]
- Maksimov, E.G.; Mironov, K.S.; Trofimova, M.S.; Nechaeva, N.L.; Todorenko, D.A.; Klementiev, K.E.; Tsoraev, G.V.; Tyutyaev, E.V.; Zorina, A.A.; Feduraev, P.V.; et al. Membrane Fluidity Controls Redox-Regulated Cold Stress Responses in Cyanobacteria. Photosynth. Res. 2017, 133, 215–223. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Structure and Expression of Fatty Acid Desaturases. Biochim. Biophys. Acta BBA- Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Kubo, Y.; Ikeda, T.; Yang, S.-Y.; Tsuboi, M. Orientation of Carotenoid Molecules in the Eyespot of Alga: In Situ Polarized Resonance Raman Spectroscopy. Appl. Spectrosc. 2000, 54, 1114–1119. [Google Scholar] [CrossRef]
- Heraud, P.; Wood, B.R.; Beardall, J.; McNaughton, D. Effects of Pre-Processing of Raman Spectra on in Vivo Classification of Nutrient Status of Microalgal Cells. J. Chemom. 2006, 20, 193–197. [Google Scholar] [CrossRef]
- Heraud, P.; Beardall, J.; McNaughton, D.; Wood, B.R. In Vivo Prediction of the Nutrient Status of Individual Microalgal Cells Using Raman Microspectroscopy. FEMS Microbiol. Lett. 2007, 275, 24–30. [Google Scholar] [CrossRef]
- Wood, B.R.; Heraud, P.; Stojkovic, S.; Morrison, D.; Beardall, J.; McNaughton, D. A Portable Raman Acoustic Levitation Spectroscopic System for the Identification and Environmental Monitoring of Algal Cells. Anal. Chem. 2005, 77, 4955–4961. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zeng, H.; Larkum, A.W.D.; Cai, Z.-L. Raman Properties of Chlorophyll d, the Major Pigment of Acaryochloris Marina: Studies Using Both Raman Spectroscopy and Density Functional Theory. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2004, 60, 527–534. [Google Scholar] [CrossRef]
- Cannizzaro, C.; Rhiel, M.; Marison, I.; von Stockar, U. On-Line Monitoring of Phaffia Rhodozyma Fed-Batch Process with in Situ Dispersive Raman Spectroscopy. Biotechnol. Bioeng. 2003, 83, 668–680. [Google Scholar] [CrossRef]
- Marshall, C.P.; Leuko, S.; Coyle, C.M.; Walter, M.R.; Burns, B.P.; Neilan, B.A. Carotenoid Analysis of Halophilic Archaea by Resonance Raman Spectroscopy. Astrobiology 2007, 7, 631–643. [Google Scholar] [CrossRef]
- Parker, S.F.; Tavender, S.M.; Dixon, N.M.; Herman, H.; Williams, K.P.J.; Maddams, W.F. Raman Spectrum of β-Carotene Using Laser Lines from Green (514.5 Nm) to Near-Infrared (1064 Nm): Implications for the Characterization of Conjugated Polyenes. Appl. Spectrosc. 1999, 53, 86–91. [Google Scholar] [CrossRef]
- Mostaert, A.S.; Giordani, C.; Crockett, R.; Karsten, U.; Schumann, R.; Jarvis, S.P. Characterisation of Amyloid Nanostructures in the Natural Adhesive of Unicellular Subaerial Algae. J. Adhes. 2009, 85, 465–483. [Google Scholar] [CrossRef]
- Shutova, V.V.; Tyutyaev, E.V.; Churin, A.A.; Ponomarev, V.Y.; Belyakova, G.A.; Maksimov, G.V. IR and Raman Spectroscopy in the Study of Carotenoids of Cladophora Rivularis Algae. Biophysics 2016, 61, 601–605. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Wyss, A. Carotenoids in Human Nutrition and Health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Chekanov, K.; Fedorenko, T.; Kublanovskaya, A.; Litvinov, D.; Lobakova, E. Diversity of Carotenogenic Microalgae in the White Sea Polar Region. FEMS Microbiol. Ecol. 2020, 96, fiz183. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.M.; Jones, H.D.T.; Han, D.; Hu, Q.; Beechem, T.E.; Timlin, J.A. Carotenoid Distribution in Living Cells of Haematococcus Pluvialis (Chlorophyceae). PLoS ONE 2011, 6, e24302. [Google Scholar] [CrossRef]
- Chiu, L.; Ho, S.-H.; Shimada, R.; Ren, N.-Q.; Ozawa, T. Rapid in Vivo Lipid/Carbohydrate Quantification of Single Microalgal Cell by Raman Spectral Imaging to Reveal Salinity-Induced Starch-to-Lipid Shift. Biotechnol. Biofuels 2017, 10, 9. [Google Scholar] [CrossRef]
- Mishra, K.B.; Vítek, P.; Mishra, A.; Hájek, J.; Barták, M. Chlorophyll a Fluorescence and Raman Spectroscopy Can Monitor Activation/Deactivation of Photosynthesis and Carotenoids in Antarctic Lichens. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 239, 118458. [Google Scholar] [CrossRef]
- Mirsafavi, R.; Moskovits, M.; Meinhart, C. Detection and Classification of Fentanyl and Its Precursors by Surface-Enhanced Raman Spectroscopy. Analyst 2020, 145, 3440–3446. [Google Scholar] [CrossRef]

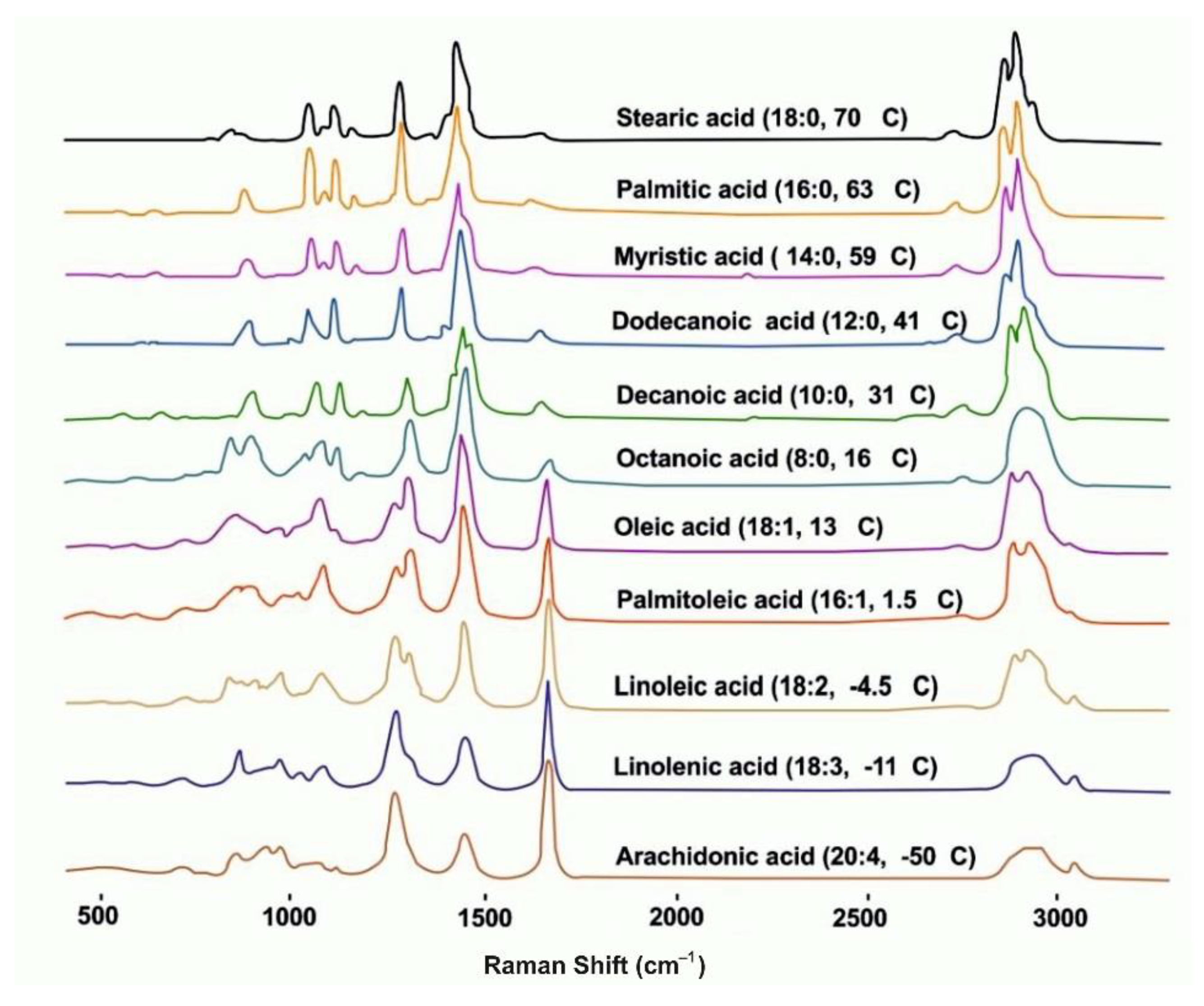

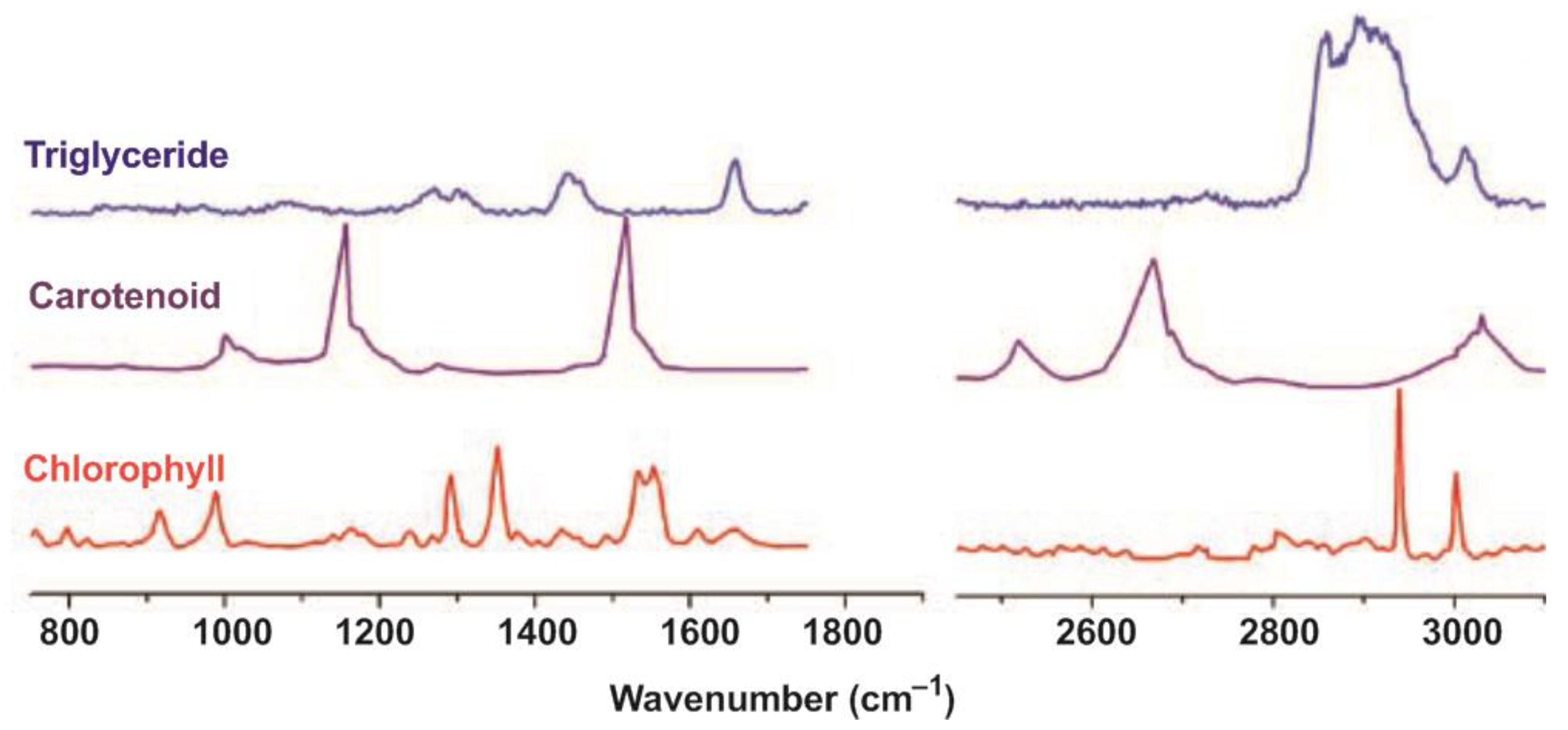
| Modification of Method | Object | Biomolecules | Reference/Link Number |
|---|---|---|---|
| Coherent anti-Stokes Raman scattering (CARS) and microscopy | microalgae | lipids, carotenoids | [4,15,16,17] |
| Confocal Raman microscopy | microalgae, algae | lipids | [18,19] |
| Raman micro spectroscopy | algae, animals | lipids, carotenoids | [2,5,20] |
| Resonance Raman spectroscopy (RRS) | bacteria, microalgae | carotenoids | [7,19,21] |
| Single-cell Raman spectroscopy (SCRS) | microalgae | lipids | [22,23] |
| Surface-enhanced Raman spectroscopy (SERS) | animals, bacteria, microalgae | lipids, carotenoids, proteins | [1,2,6,10,14,24] |
| Bioanalyte/Disease | RS Substrate | Reference |
|---|---|---|
| Cancer (blood plasma protein) | Ag NPs | [51] |
| Quantification of hepatitis B DNA | Ag NPs | [52] |
| Breast cancer tissue | Ag NPs | [53] |
| Sjogren’s syndrome from saliva | Cl-Ag NPs | [54] |
| Human tear uric acid | SiO2 and Au | [55] |
| Creatinine | Nano-Au | [56] |
| Mouse IgG | Au NPs | [57] |
| Single prostate cancer cells | Au NPs | [58] |
| Plasmodium falciparum DNA | Magnetic beads | [59] |
| HeLa cells | Au NPs | [60] |
| Gastritis | Au NPs | [61] |
| Bioactive Compounds | Microalgal Strain Name | Type of RS | Wavenumber | Ref. |
|---|---|---|---|---|
| α-helix protein | Arthrospira platensis | Macro-Raman spectrometry | 1574 cm−1 | [107] |
| Amide bonds | Arthrospira platensis | Macro-Raman spectrometry | 1400 cm−1 | [107] |
| Antioxidant protein enzyme | Arthrospira platensis | Macro-Raman spectrometry | 1030 and 1120 cm−1 | [107] |
| Polyphosphates | Phaeodactylum tricornutum | Single-cell micro-Raman spectrometry | 1160 cm−1 | [107] |
| ß-carotene | Phaeodactylum tricornutum | Single-cell micro-Raman spectrometry | 1522 cm−1 | [107] |
| ß-carotene | Dunaliella tertiolecta | Resonance Raman spectrometry | 1158 and 1527 cm−1 | [108] |
| Triglyceride | Chlorella sorokiniana | Micro-Raman spectroscopy | 2800 and 3000 cm−1 | [14] |
| ß-carotenoid | Neochloris oleoabundans | Micro-Raman spectroscopy | 1505 and 1535 cm−1 | |
| ß-carotene | Chlorella sorokiniana | Raman micro-spectroscopy | 1515 and 1157 cm−1 | [109] |
| Astaxanthin | Chlainomonas sp. | Micro-Raman spectroscopy | 1520 and 1156 cm−1 | [20] |
| Astaxanthin | Chlamydomonas nivalis | Micro-Raman spectroscopy | 1520 and 1156 cm−1 | |
| Violaxanthin | Chloromonas nivalis | Micro-Raman spectroscopy | 1525 cm−1 | |
| Antheraxanthin | Chloromonas nivalis | Micro-Raman spectroscopy | 1523 cm−1 | |
| Myxoxanthophyll | Botrydiopsis alpina | Micro-Raman spectroscopy | 1527 cm−1 | [110] |
| Neoxanthin | Dunaliella parva | Micro-Raman spectroscopy | 1525 and 1530 cm−1 | |
| Chlorophyll c | Dunaliella tertiolecta | Micro-Raman spectroscopy | 1670 cm−1 | |
| Lipid | Botryococcus brauniiis | Micro-Raman spectroscopy | 1640 and 1674 cm−1 | [111] |
| FAME | Scenedemus quadricauda | Surface-enhanced Raman spectroscopy | 1430, 1157, 1544, 1257, 1307, 961 and 596 cm−1 | [112] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allakhverdiev, E.S.; Khabatova, V.V.; Kossalbayev, B.D.; Zadneprovskaya, E.V.; Rodnenkov, O.V.; Martynyuk, T.V.; Maksimov, G.V.; Alwasel, S.; Tomo, T.; Allakhverdiev, S.I. Raman Spectroscopy and Its Modifications Applied to Biological and Medical Research. Cells 2022, 11, 386. https://doi.org/10.3390/cells11030386
Allakhverdiev ES, Khabatova VV, Kossalbayev BD, Zadneprovskaya EV, Rodnenkov OV, Martynyuk TV, Maksimov GV, Alwasel S, Tomo T, Allakhverdiev SI. Raman Spectroscopy and Its Modifications Applied to Biological and Medical Research. Cells. 2022; 11(3):386. https://doi.org/10.3390/cells11030386
Chicago/Turabian StyleAllakhverdiev, Elvin S., Venera V. Khabatova, Bekzhan D. Kossalbayev, Elena V. Zadneprovskaya, Oleg V. Rodnenkov, Tamila V. Martynyuk, Georgy V. Maksimov, Saleh Alwasel, Tatsuya Tomo, and Suleyman I. Allakhverdiev. 2022. "Raman Spectroscopy and Its Modifications Applied to Biological and Medical Research" Cells 11, no. 3: 386. https://doi.org/10.3390/cells11030386
APA StyleAllakhverdiev, E. S., Khabatova, V. V., Kossalbayev, B. D., Zadneprovskaya, E. V., Rodnenkov, O. V., Martynyuk, T. V., Maksimov, G. V., Alwasel, S., Tomo, T., & Allakhverdiev, S. I. (2022). Raman Spectroscopy and Its Modifications Applied to Biological and Medical Research. Cells, 11(3), 386. https://doi.org/10.3390/cells11030386








