LbCas12a-D156R Efficiently Edits LOB1 Effector Binding Elements to Generate Canker-Resistant Citrus Plants
Abstract
:1. Introduction
2. Materials and Methods
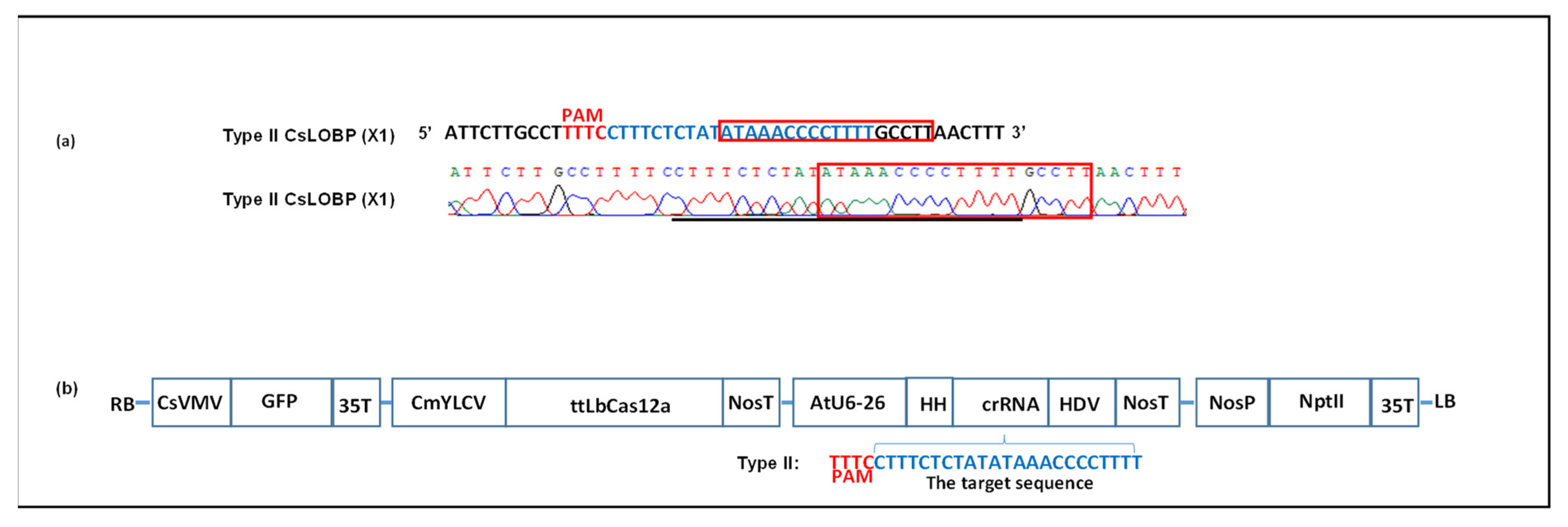
2.1. Plasmid Construction
2.2. Xcc-Facilitated Agroinfiltration in Pummelo
2.3. Agrobacterium-Mediated Pummelo Transformation
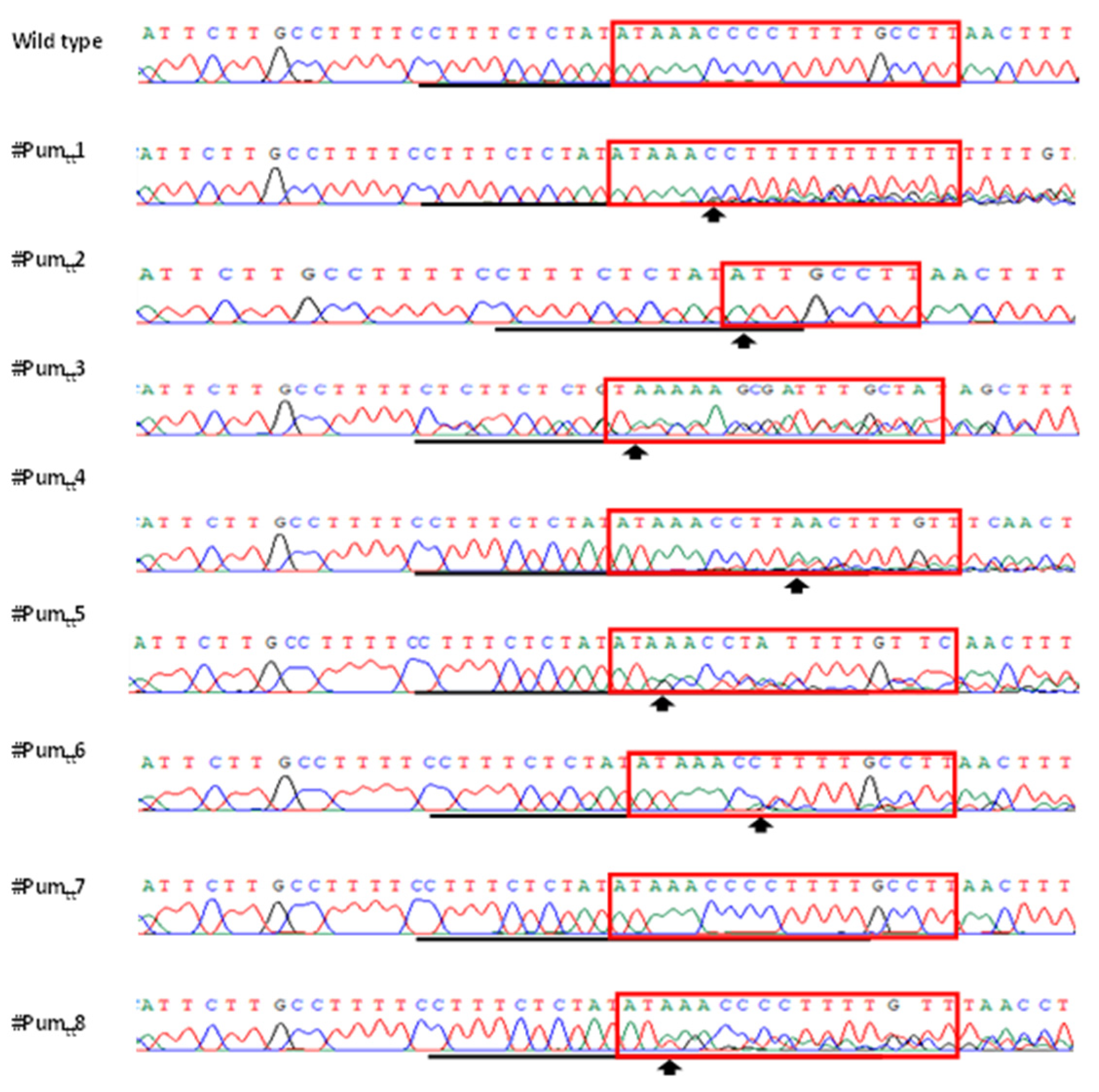
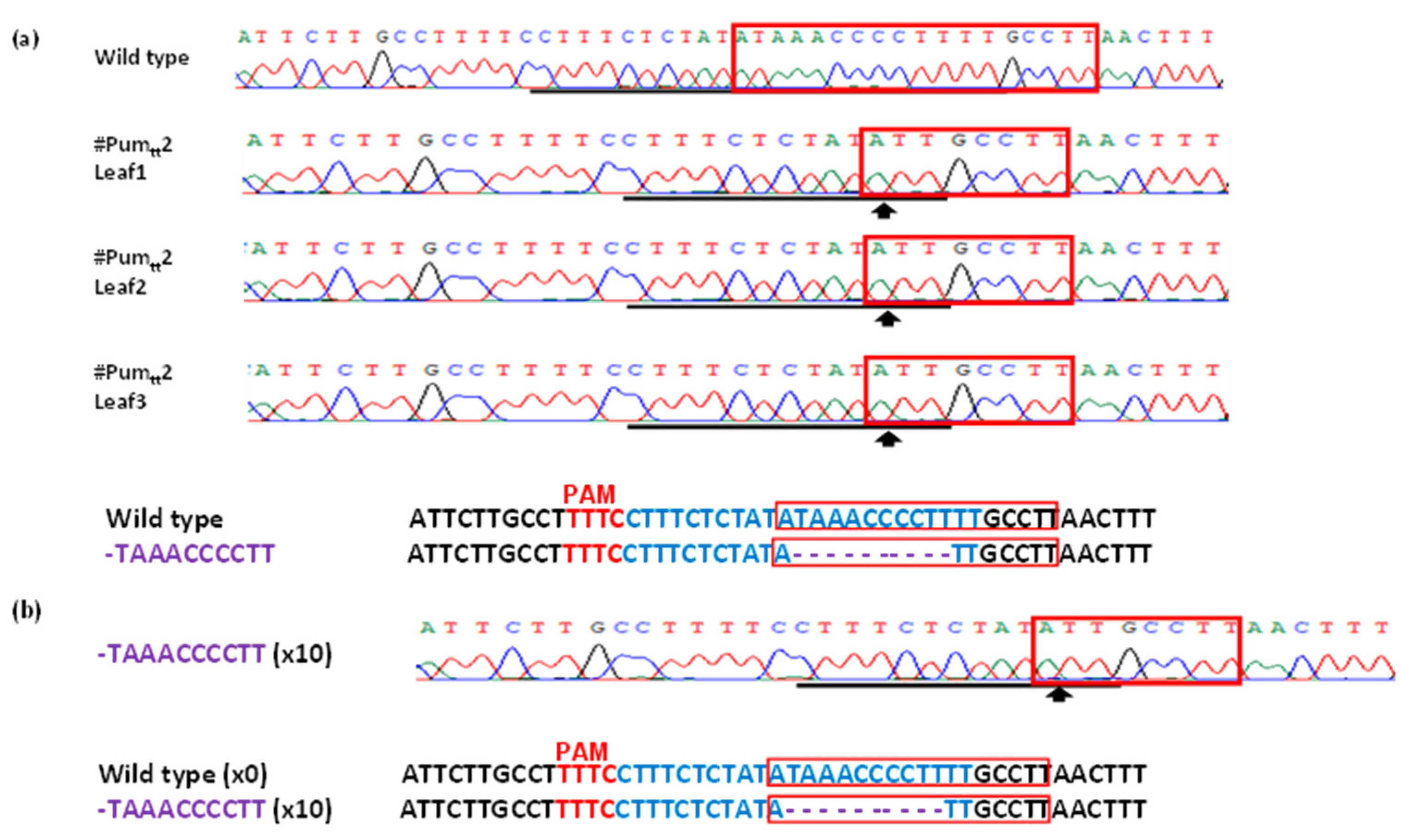
2.4. PCR Amplification of Mutagenized LOBP
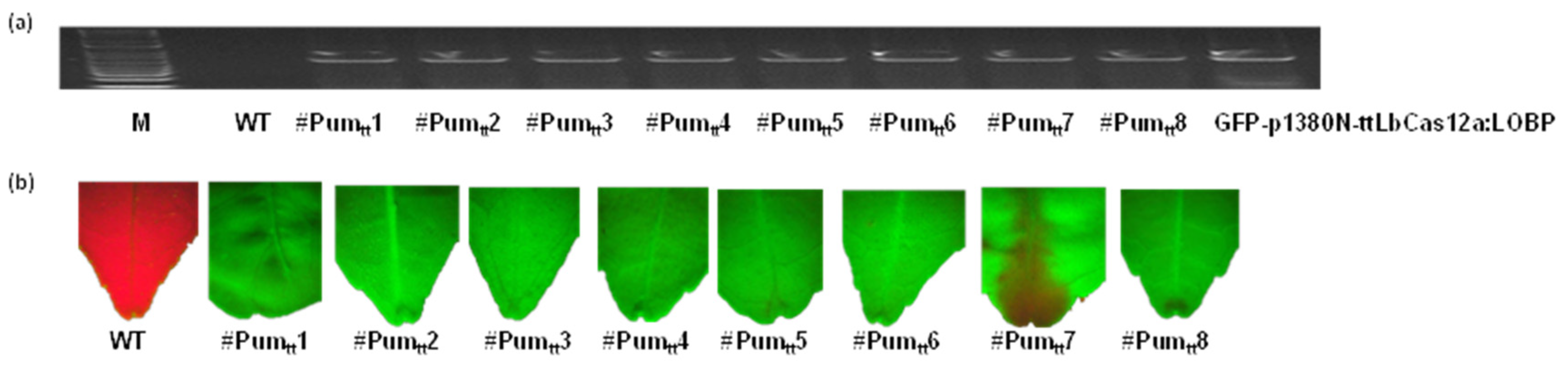
2.5. GFP Detection
2.6. Canker Symptom Assay in Citrus
3. Results
3.1. Transient Expression of ttLbCas12a to Edit Citrus Genome via Xcc-Facilitated Agroinfiltration
3.2. Transgenic Expression of ttLbCas12a in Pummelo
3.3. Mutation Genotypes of ttLbCpf1 in Transgenic Pummelo
3.4. Canker Resistance of ttLbCas12a-Transformed Pummelo Plants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balfagón, D.; Terán, F.; De Oliveira, T.; Santa-Catarina, C.; Gómez-Cadenas, A. Citrus rootstocks modify scion antioxidant system under drought and heat stress combination. Plant Cell Rep. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the under-standing of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [Green Version]
- Fernández, A.O.; Pinatti, C.A.O.; Peris, R.M.; Laguarda-Miró, N. Freeze-Damage Detection in Lemons Using Electrochemical Impedance Spectroscopy. Sensors 2019, 19, 4051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, Z.; Lopez-Climent, M.F.; Arbona, V.; Perez-Clemente, R.M.; Gomez-Cadenas, A. Modulation of the antioxidant system in citrus under waterlogging and subsequent drainage. J. Plant Physiol. 2009, 166, 1391–1404. [Google Scholar] [CrossRef]
- Wang, N. The citrus huanglongbing crisis and potential solutions. Mol. Plant 2019, 12, 607–609. [Google Scholar] [CrossRef]
- Dutt, M.; Mou, Z.; Zhang, X.; Tanwir, S.E.; Grosser, J.W. Efficient CRISPR/Cas9 genome editing with Citrus embryogenic cell cultures. BMC Biotechnol. 2020, 20, 58. [Google Scholar] [CrossRef]
- Huang, X.; Wang, Y.; Xu, J.; Wang, N. Development of multiplex genome editing toolkits for citrus with high efficacy in biallelic and homozygous mutations. Plant Mol. Biol. 2020, 104, 297–307. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Targeted Genome Editing of Sweet Orange Using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Leblanc, C.; Irish, V.F.; Jacob, Y. Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 2017, 36, 1883–1887. [Google Scholar] [CrossRef]
- Huang, X.; Wang, N. Highly efficient generation of canker resistant sweet orange enabled by an improved CRISPR/Cas9 sys-tem. Front. Plant Sci. 2022. [Google Scholar] [CrossRef]
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 2016, 14, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Wang, N. Generation of homozygous canker-resistant citrus in the T0 generation using CRISPR-SpCas9p. Plant Biotechnol. J. 2020, 18, 1190–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Zheng, X.; Huang, Y.; Ye, J.; Chen, P.; Zhang, C.; Zhao, F.; Xie, Z.; Zhang, S.; Wang, N.; et al. Genome sequencing and CRISPR/Cas9 gene editing of an early flowering mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 2019, 17, 2199–2210. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Xu, J.; Orbović, V.; Zhang, Y.; Wang, N. Editing Citrus Genome via SaCas9/sgRNA System. Front. Plant Sci. 2017, 8, 2135. [Google Scholar] [CrossRef]
- Jia, H.; Orbović, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019, 17, 1928–1937. [Google Scholar] [CrossRef] [Green Version]
- Duan, S.; Jia, H.; Pang, Z.; Teper, D.; White, F.; Jones, J.; Zhou, C.; Wang, N. Functional characterization of the citrus canker susceptibility gene CsLOB1. Mol. Plant Pathol. 2018, 19, 1908–1916. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Carlos Huguet Tapia, J.; Hu, Y.; Jones, J.; Wang, N.; Liu, S.; White, F.F. Homologs of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol. Plant Pathol. 2016, 18, 798–810. [Google Scholar] [CrossRef]
- Jia, H.; Omar, A.; Orbović, V.; Wang, N. Biallelic editing of the LOB1 promoter via CRISPR/Cas9 creates canker-resistant ‘Duncan’ grapefruit. Phytopathology 2021. [Google Scholar] [CrossRef] [PubMed]
- Teper, D.; Wang, N. Consequences of adaptation of TAL effectors on host susceptibility to Xanthomonas. PLoS Genet. 2021, 17, e1009310. [Google Scholar] [CrossRef]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017, 7, 11606. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.-T.; Ryu, J.; Kang, B.-C.; Kim, J.-S.; Kim, S.-G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017, 8, 14406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Ren, Q.; Tang, X.; Liu, S.; Malzahn, A.A.; Zhou, J.; Wang, J.; Yin, D.; Pan, C.; Yuan, M.; et al. Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 2021, 12, 1944. [Google Scholar] [CrossRef]
- Tang, X.; Ren, Q.; Yang, L.; Bao, Y.; Zhong, Z.; He, Y.; Liu, S.; Qi, C.; Liu, B.; Wang, Y.; et al. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol. J. 2019, 17, 1431–1445. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Wang, C.; Liu, Q.; Fu, Y.; Wang, K. Targeted mutagenesis in rice using CRISPR-Cpf1 system. J. Genet. Genom. 2017, 44, 71–73. [Google Scholar] [CrossRef]
- Lee, K.; Zhang, Y.; Kleinstiver, B.P.; Guo, J.A.; Aryee, M.J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.K.; et al. Activities and specificities of CRISPR /Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2018, 17, 362–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malzahn, A.A.; Tang, X.; Lee, K.; Ren, Q.; Sretenovic, S.; Zhang, Y.; Chen, H.; Kang, M.; Bao, Y.; Zheng, X.; et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019, 17, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17018. [Google Scholar] [CrossRef]
- Wang, M.; Mao, Y.; Lu, Y.; Tao, X.; Zhu, J.-K. Multiplex Gene Editing in Rice Using the CRISPR-Cpf1 System. Mol. Plant 2017, 10, 1011–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Qin, R.; Li, H.; Li, D.; Li, L.; Wei, P.; Yang, J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 2017, 15, 713–717. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.-C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting as a stomatal developmental gene EPL9 in rice. Plant Cell. Rep. 2017, 36, 745–757. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhang, Y.; You, Q.; Tang, X.; Ren, Q.; Liu, S.; Yang, L.; Wang, Y.; Liu, X.; Liu, B.; et al. Plant Genome Editing Using FnCpf1 and LbCpf1 Nucleases at Redefined and Altered PAM Sites. Mol. Plant 2018, 11, 999–1002. [Google Scholar] [CrossRef] [Green Version]
- Bernabé-Orts, J.M.; Casas-Rodrigo, I.; Minguet, E.G.; Landolfi, V.; Garcia-Carpintero, V.; Gianoglio, S.; Vázquez-Vilar, M.; Granell, A.; Orzaez, D. Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol. J. 2019, 17, 1971–1984. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhang, J.; Sun, L.; Ma, Y.; Xu, J.; Liang, S.; Deng, J.; Tan, J.; Zhang, Q.; Tu, L.; et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018, 16, 137–150. [Google Scholar] [CrossRef] [Green Version]
- Joung, J.K.; Kleinstiver, B.; Sousa, A. Variants of CPF1 (Cas12a) with Altered PAM Specificity. U.S. Patent US20190010481A1, 25 October 2018. [Google Scholar]
- Schindele, P.; Puchta, H. Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 2020, 18, 1118–1120. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.K.; Armstrong, B.; Schindele, P.; Puchta, H. Efficient gene targeting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a. Plant Biotechnol. J. 2021, 19, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wang, N. Xcc-facilitated agroinfiltration of citrus leaves: A tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 2014, 33, 1993–2001. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wang, N. High-Throughput Screening and Analysis of Genes of Xanthomonas citri subsp. citri Involved in Citrus Canker Symptom Development. Mol. Plant Microbe Interact. 2012, 25, 69–84. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Zou, X.; Orbovic, V.; Wang, N. Genome Editing in Citrus Tree with CRISPR/Cas9. Methods Mol. Biol. 2019, 235–241. [Google Scholar]
- Duan, K.; Cheng, Y.; Ji, J.; Wang, C.; Wei, Y.; Wang, Y. Large chromosomal segment deletions by CRISPR/LbCpf1-mediated multiplex gene editing in soybean. J. Integr. Plant Biol. 2021, 63, 1620–1631. [Google Scholar] [CrossRef]
- Hu, X.; Meng, X.; Li, J.; Wang, K.; Yu, H. Improving the efficiency of the CRISPR-Cas12a system with tRNA-crRNA arrays. Crop J. 2019, 8, 403–407. [Google Scholar] [CrossRef]
- Campa, C.C.; Weisbach, N.R.; Santinha, A.J.; Incarnato, D.; Platt, R.J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods 2019, 16, 887–893. [Google Scholar] [CrossRef]
- Gao, L.; Cox, D.B.T.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef]
- Yu, K.; Liu, Z.; Gui, H.; Geng, L.; Wei, J.; Liang, D.; Lv, J.; Xu, J.; Chen, X. Highly efficient generation of bacterial leaf blight-resistant and transgene-free rice using a genome editing and multiplexed selection system. BMC Plant Biol. 2021, 21, 197. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Wang, Y.; Su, H.; Huang, X.; Wang, N. LbCas12a-D156R Efficiently Edits LOB1 Effector Binding Elements to Generate Canker-Resistant Citrus Plants. Cells 2022, 11, 315. https://doi.org/10.3390/cells11030315
Jia H, Wang Y, Su H, Huang X, Wang N. LbCas12a-D156R Efficiently Edits LOB1 Effector Binding Elements to Generate Canker-Resistant Citrus Plants. Cells. 2022; 11(3):315. https://doi.org/10.3390/cells11030315
Chicago/Turabian StyleJia, Hongge, Yuanchun Wang, Hang Su, Xiaoen Huang, and Nian Wang. 2022. "LbCas12a-D156R Efficiently Edits LOB1 Effector Binding Elements to Generate Canker-Resistant Citrus Plants" Cells 11, no. 3: 315. https://doi.org/10.3390/cells11030315
APA StyleJia, H., Wang, Y., Su, H., Huang, X., & Wang, N. (2022). LbCas12a-D156R Efficiently Edits LOB1 Effector Binding Elements to Generate Canker-Resistant Citrus Plants. Cells, 11(3), 315. https://doi.org/10.3390/cells11030315





