Single-Cell RNAseq Resolve the Potential Effects of LanCL1 Gene in the Mouse Testis
Abstract
1. Introduction
2. Methods and Materials
2.1. Animals
2.2. Mouse Breeding Experiment
2.3. Semen Quality Analysis
2.4. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.5. Western Blotting
2.6. Immunofluorescence
2.7. Testosterone Level
2.8. Single Cell RNA Sequencing for the Testis Tissue
3. Data Processing
3.1. GO Annotations
3.2. Gene Set Enrichment Analysis (GSEA)
3.3. Cell Communication
4. Results
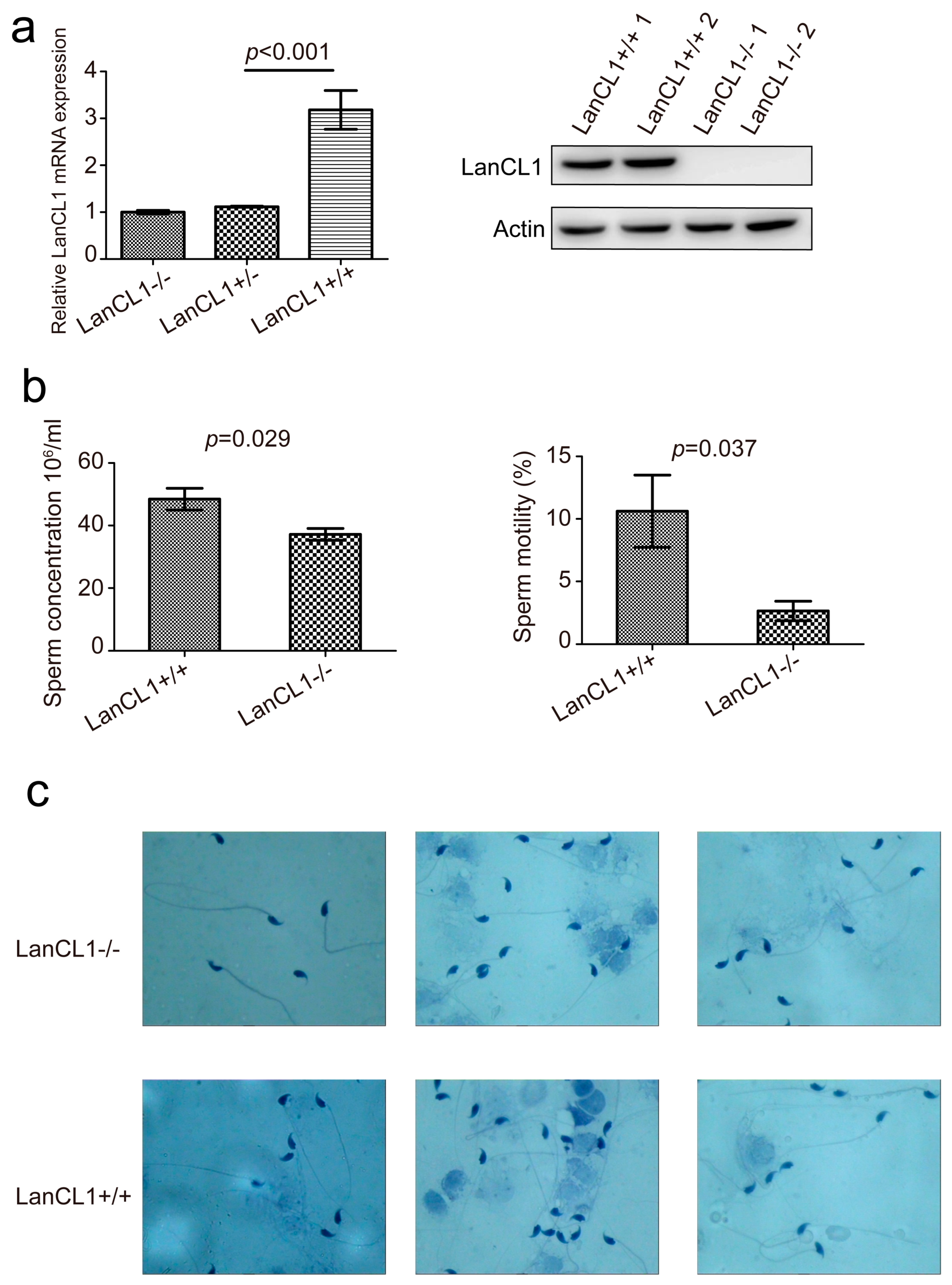
4.1. LanCL1 gene Associated with the Male Reproductive Ability
4.2. Poor Semen Quality Present in the LanCL1−/− Mice
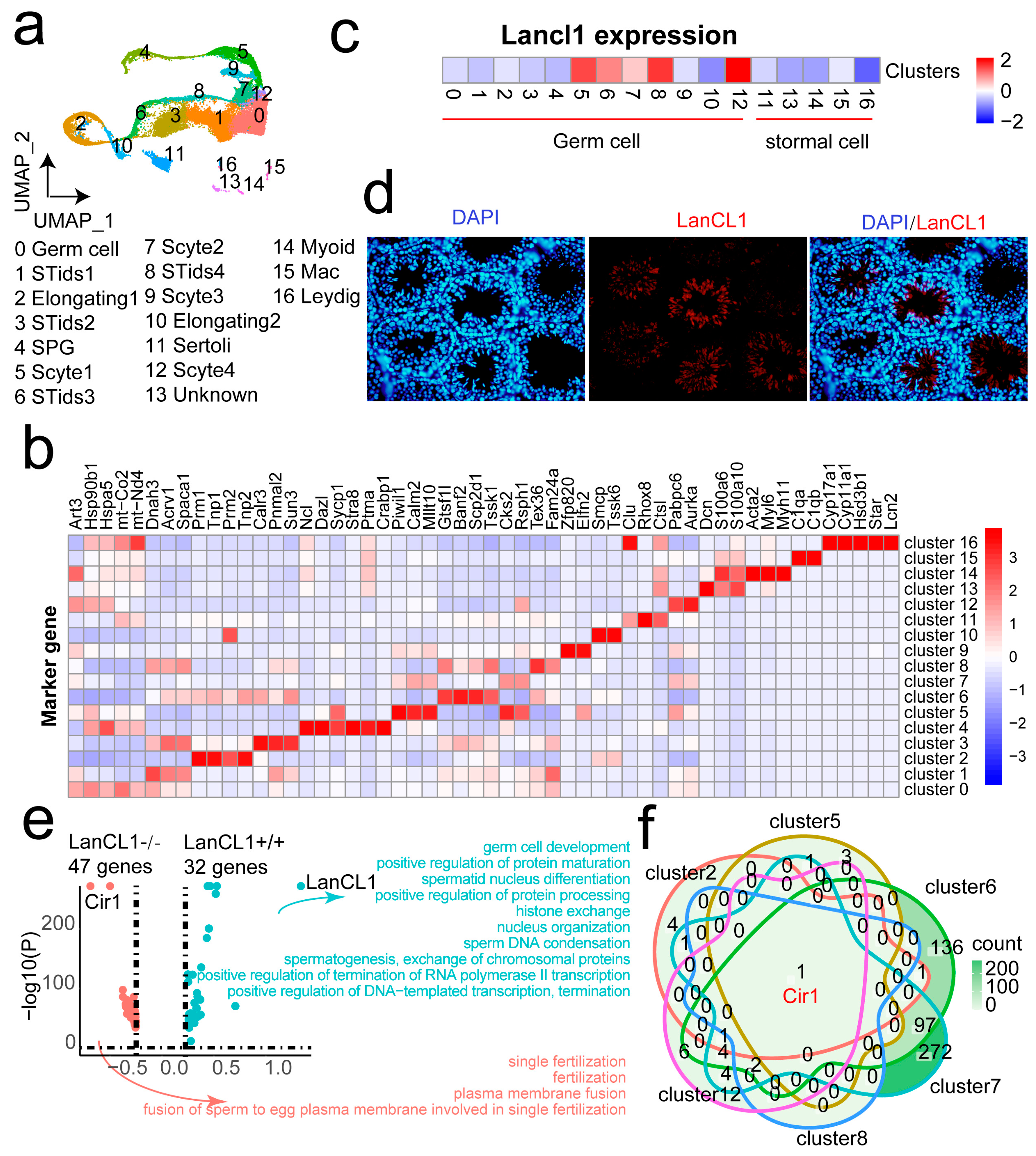
4.3. Single-Cell Analysis Discover the High Expression of Cir1 Gene in the LanCL1−/− Mice
4.4. LanCL1 gene Associate with the Normal Sperm Motility in Germ Cells
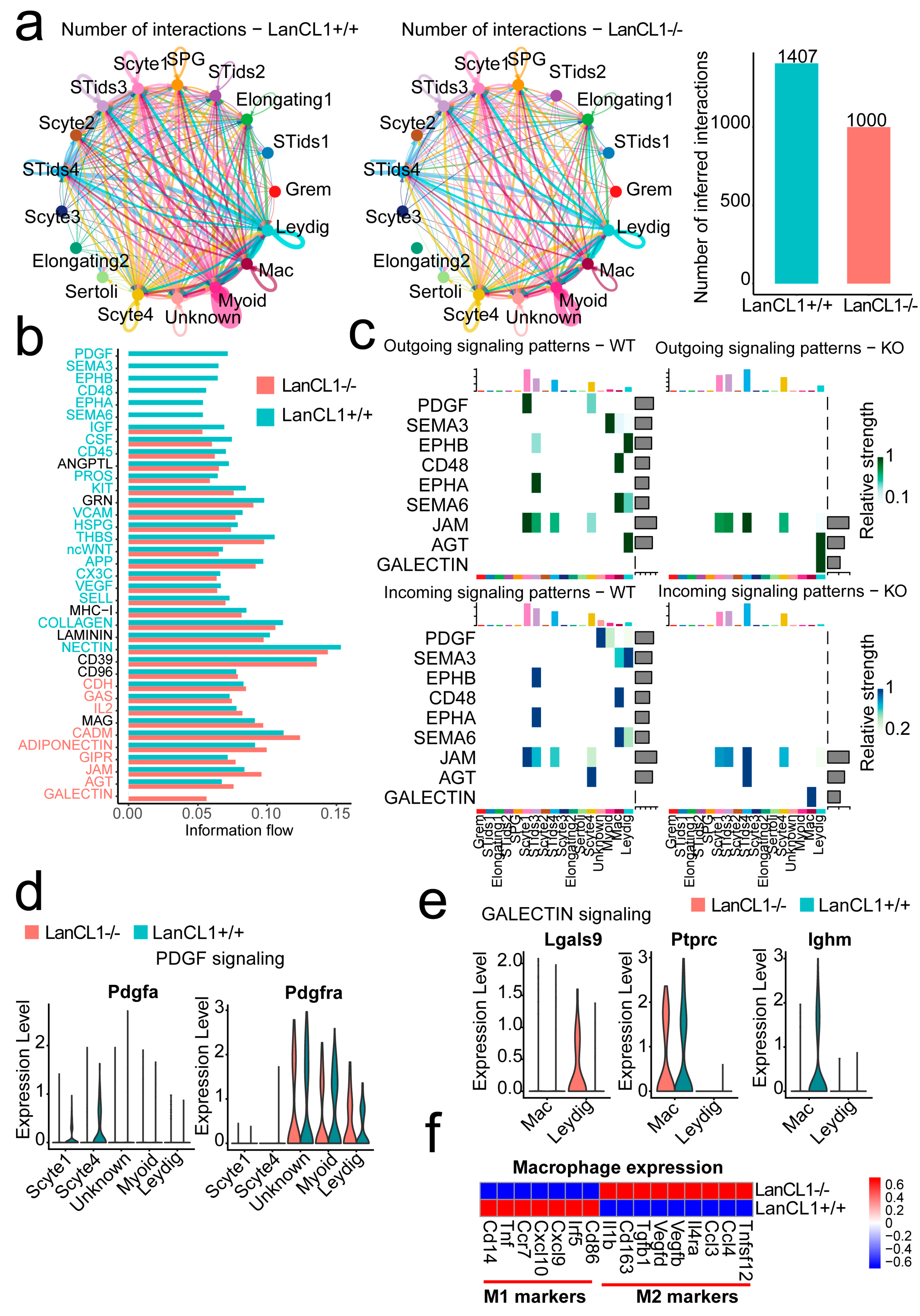
4.5. LanCL1 gene Influences the Cell Interactions Signaling within the Germ and Stromal Cells and Promotes the M2 Macrophage Polarity in Mouse Testis
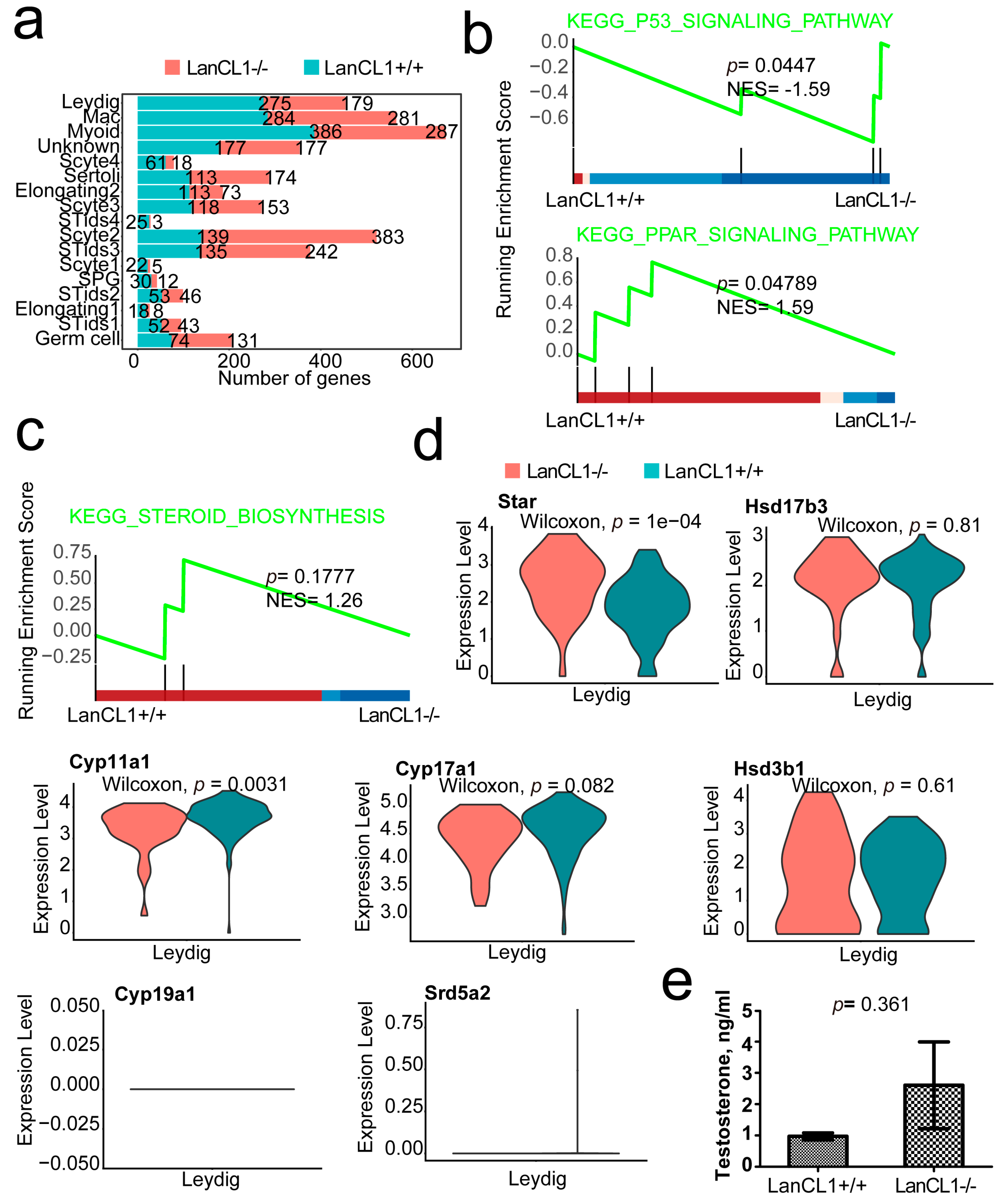
4.6. LanCL1 gene Regulated the P53 and PPAR Signaling Pathway Other Than the Gene Expressions of Testosterone Synthesis in the Leyding Cells
5. Discussion
6. Limitations
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duvuru, R.; Halabi, M.; Omolaoye, T.S.; Du Plessis, S.S. The genetic causes of male infertility: A Middle East and North Africa perspective. F1000Research 2022, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Vander Borght, M.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Leisegang, K.; Majzoub, A.; Henkel, R.; Finelli, R.; Panner Selvam, M.K.; Tadros, N.; Parekh, N.; Ko, E.Y.; Cho, C.L.; et al. Utility of Antioxidants in the Treatment of Male Infertility: Clinical Guidelines Based on a Systematic Review and Analysis of Evidence. World J. Men’s Health. 2021, 39, 233–290. [Google Scholar] [CrossRef] [PubMed]
- Carlsen, E.; Giwercman, A.; Keiding, N.; Skakkebaek, N.E. Evidence for decreasing quality of semen during past 50 years. BMJ 1992, 305, 609–613. [Google Scholar] [CrossRef]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Jolles, M.; Pinotti, R.; Swan, S.H. Temporal trends in sperm count: A systematic review and meta-regression analysis of samples collected globally in the 20th and 21st centuries. Hum. Reprod. Update 2022, dmac035. [Google Scholar] [CrossRef]
- Bisht, S.; Faiq, M.; Tolahunase, M.; Dada, R. Oxidative stress and male infertility. Nat. Rev. Urol. 2017, 14, 470–485. [Google Scholar] [CrossRef]
- Mayer, H.; Breuss, J.; Ziegler, S.; Prohaska, R. Molecular characterization and tissue-specific expression of a murine putative G-protein-coupled receptor. Biochim. Biophys. Acta 1998, 1399, 51–56. [Google Scholar] [CrossRef]
- Mayer, H.; Salzer, U.; Breuss, J.; Ziegler, S.; Marchler-Bauer, A.; Prohaska, R. Isolation, molecular characterization, and tissue-specific expression of a novel putative G protein-coupled receptor. Biochim. Biophys. Acta 1998, 1395, 301–308. [Google Scholar] [CrossRef]
- Mayer, H.; Bauer, H.; Breuss, J.; Ziegler, S.; Prohaska, R. Characterization of rat LANCL1, a novel member of the lanthionine synthetase C-like protein family, highly expressed in testis and brain. Gene 2001, 269, 73–80. [Google Scholar] [CrossRef]
- Nielsen, J.E.; Hansen, M.A.; Jørgensen, M.; Tanaka, M.; Almstrup, K.; Skakkebaek, N.E.; Leffers, H. Germ cell differentiation-dependent and stage-specific expression of LANCL1 in rodent testis. Eur. J. Histochem. 2003, 47, 215–222. [Google Scholar] [CrossRef][Green Version]
- Huang, C.; Chen, M.; Pang, D.; Bi, D.; Zou, Y.; Xia, X.; Yang, W.; Luo, L.; Deng, R.; Tan, H.; et al. Developmental and activity-dependent expression of LanCL1 confers antioxidant activity required for neuronal survival. Dev. Cell 2014, 30, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Adeoye, O.; Olawumi, J.; Opeyemi, A.; Christiania, O. Review on the role of glutathione on oxidative stress and infertility. JBRA Assist. Reprod. 2018, 22, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiao, Q.; Chen, X.; Tong, S.; Sun, J.; Lv, R.; Wang, S.; Gou, Y.; Tan, L.; Xu, J.; et al. LanCL1 protects prostate cancer cells from oxidative stress via suppression of JNK pathway. Cell Death Dis. 2018, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Qi, N.; Zeng, Y.; Bao, M.; Chen, Y.; Liao, J.; Wei, L.; Cao, D.; Huang, S.; Luo, Q.; et al. The Endogenous Alterations of the Gut Microbiota and Feces Metabolites Alleviate Oxidative Damage in the Brain of LanCL1 Knockout Mice. Front. Microbiol. 2020, 11, 557342. [Google Scholar] [CrossRef]
- Huang, C.; Yang, C.; Pang, D.; Li, C.; Gong, H.; Cao, X.; He, X.; Chen, X.; Mu, B.; Cui, Y.; et al. Animal models of male subfertility targeted on LanCL1-regulated spermatogenic redox homeostasis. Lab Anim. 2022, 51, 133–145. [Google Scholar] [CrossRef]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667. [Google Scholar] [CrossRef]
- Bauer, H.; Mayer, H.; Marchler-Bauer, A.; Salzer, U.; Prohaska, R. Characterization of p40/GPR69A as a peripheral membrane protein related to the lantibiotic synthetase component C. Biochem. Biophys. Res. Commun. 2000, 275, 69–74. [Google Scholar] [CrossRef]
- Chung, C.H.; Kurien, B.T.; Mehta, P.; Mhatre, M.; Mou, S.; Pye, Q.N.; Stewart, C.; West, M.; Williamson, K.S.; Post, J.; et al. Identification of lanthionine synthase C-like protein-1 as a prominent glutathione binding protein expressed in the mammalian central nervous system. Biochemistry 2007, 46, 3262–3269. [Google Scholar] [CrossRef]
- Zhong, W.X.; Wang, Y.B.; Peng, L.; Ge, X.Z.; Zhang, J.; Liu, S.S.; Zhang, X.N.; Xu, Z.H.; Chen, Z.; Luo, J.H. Lanthionine synthetase C-like protein 1 interacts with and inhibits cystathionine β-synthase: A target for neuronal antioxidant defense. J. Biol. Chem. 2012, 287, 34189–34201. [Google Scholar] [CrossRef]
- Kimble, J.; Crittenden, S.L. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 2007, 23, 405–433. [Google Scholar] [CrossRef]
- Morichika, K.; Kataoka, K.; Terayama, K.; Tazaki, A.; Kinoshita, T.; Watanabe, K.; Mochii, M. Perturbation of Notch/Suppressor of Hairless pathway disturbs migration of primordial germ cells in Xenopus embryo. Dev. Growth Differ. 2010, 52, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.L.; Qian, Y.; Schulz, C. Notch and Delta are required for survival of the germline stem cell lineage in testes of Drosophila melanogaster. PLoS ONE 2019, 14, e0222471. [Google Scholar] [CrossRef] [PubMed]
- Garcia, T.X.; DeFalco, T.; Capel, B.; Hofmann, M.C. Constitutive activation of NOTCH1 signaling in Sertoli cells causes gonocyte exit from quiescence. Dev. Biol. 2013, 377, 188–201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, H.; Brennan, J.; Karl, J.; Hamada, Y.; Raetzman, L.; Capel, B. Notch signaling maintains Leydig progenitor cells in the mouse testis. Development 2008, 135, 3745–3753. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Kageyama, Y.; Ishizaka, K.; Xia, G.; Kihara, K.; Oshima, H. Requirement of Notch 1 and its ligand jagged 2 expressions for spermatogenesis in rat and human testes. J. Androl. 2001, 22, 999–1011. [Google Scholar] [CrossRef]
- Murta, D.; Batista, M.; Trindade, A.; Silva, E.; Henrique, D.; Duarte, A.; Lopes-da-Costa, L. In vivo notch signaling blockade induces abnormal spermatogenesis in the mouse. PLoS ONE 2014, 9, e113365. [Google Scholar] [CrossRef]
- Murta, D.; Batista, M.; Silva, E.; Trindade, A.; Henrique, D.; Duarte, A.; Lopes-da-Costa, L. Notch signaling in the epididymal epithelium regulates sperm motility and is transferred at a distance within epididymosomes. Andrology 2016, 4, 314–327. [Google Scholar] [CrossRef]
- Gnessi, L.; Basciani, S.; Mariani, S.; Arizzi, M.; Spera, G.; Wang, C.; Bondjers, C.; Karlsson, L.; Betsholtz, C. Leydig cell loss and spermatogenic arrest in platelet-derived growth factor (PDGF)-A-deficient mice. J. Cell Biol. 2000, 149, 1019–1026. [Google Scholar] [CrossRef]
- Lv, R.; Bao, Q.; Li, Y. Regulation of M1-type and M2-type macrophage polarization in RAW264.7 cells by Galectin-9. Mol. Med. Rep. 2017, 16, 9111–9119. [Google Scholar] [CrossRef]
- Santoro, M.; Guido, C.; De Amicis, F.; Sisci, D.; Vizza, D.; Gervasi, S.; Carpino, A.; Aquila, S. Sperm metabolism in pigs: A role for peroxisome proliferator-activated receptor gamma (PPARγ). J. Exp. Biol. 2013, 216, 1085–1092. [Google Scholar] [CrossRef]
- Aquila, S.; Bonofiglio, D.; Gentile, M.; Middea, E.; Gabriele, S.; Belmonte, M.; Catalano, S.; Pellegrino, M.; Andò, S. Peroxisome proliferator-activated receptor (PPAR) gamma is expressed by human spermatozoa: Its potential role on the sperm physiology. J. Cell. Physiol. 2006, 209, 977–986. [Google Scholar] [CrossRef]
- Yin, Y.; Stahl, B.C.; DeWolf, W.C.; Morgentaler, A. p53-mediated germ cell quality control in spermatogenesis. Dev. Biol. 1998, 204, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Ghandehari-Alavijeh, R.; Zohrabi, D.; Tavalaee, M.; Nasr-Esfahani, M.H. Association between expression of TNF-α, P53 and HIF1α with asthenozoospermia. Hum. Fertil. 2019, 22, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.N.; Karimi, J.; Khodadadi, I.; Amiri, I.; Karami, M.; Saidijam, M.; Vatannejad, A.; Tavilani, H. Evaluation of the p53 and Thioredoxin reductase in sperm from asthenozoospermic males in comparison to normozoospermic males. Free Radic. Biol. Med. 2018, 116, 123–128. [Google Scholar] [CrossRef] [PubMed]

| Genotype of Mice | No. Male Mice | No. Female Mice | Pregnant Rate | Time of Vaginal Plug b | p c | Litter Size | p c | No. Neonatal Male Mice | p c | No. Neonatal Female Mice | p c | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | |||||||||||
| (+/+) | (+/+) | 6 | 24 | 91.67% (22/24) | 2.33 ± 0.16 a | 0.084 | 7.73 ± 0.41 | 0.027 | 3.81 ± 0.34 | 0.210 | 3.75 ± 0.52 | 0.467 |
| (−/−) | (+/+) | 6 | 24 | 91.67% (22/24) | 3.00 ± 0.35 | 6.59 ± 0.28 | 3.30 ± 0.23 | 3.35 ± 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Liao, J.; Qin, M.; Li, H.; Zhang, Q.; Chen, Y.; Cheng, J. Single-Cell RNAseq Resolve the Potential Effects of LanCL1 Gene in the Mouse Testis. Cells 2022, 11, 4135. https://doi.org/10.3390/cells11244135
Lu J, Liao J, Qin M, Li H, Zhang Q, Chen Y, Cheng J. Single-Cell RNAseq Resolve the Potential Effects of LanCL1 Gene in the Mouse Testis. Cells. 2022; 11(24):4135. https://doi.org/10.3390/cells11244135
Chicago/Turabian StyleLu, Jiangting, Jinling Liao, Min Qin, Hui Li, Qingyuan Zhang, Yang Chen, and Jiwen Cheng. 2022. "Single-Cell RNAseq Resolve the Potential Effects of LanCL1 Gene in the Mouse Testis" Cells 11, no. 24: 4135. https://doi.org/10.3390/cells11244135
APA StyleLu, J., Liao, J., Qin, M., Li, H., Zhang, Q., Chen, Y., & Cheng, J. (2022). Single-Cell RNAseq Resolve the Potential Effects of LanCL1 Gene in the Mouse Testis. Cells, 11(24), 4135. https://doi.org/10.3390/cells11244135




