UC-BSCs Exosomes Regulate Th17/Treg Balance in Patients with Systemic Lupus Erythematosus via miR-19b/KLF13
Abstract
:1. Introduction
2. Methods and Reagents
2.1. Research Object
2.2. Cell Sorting and Culture
2.3. Exosome Isolation
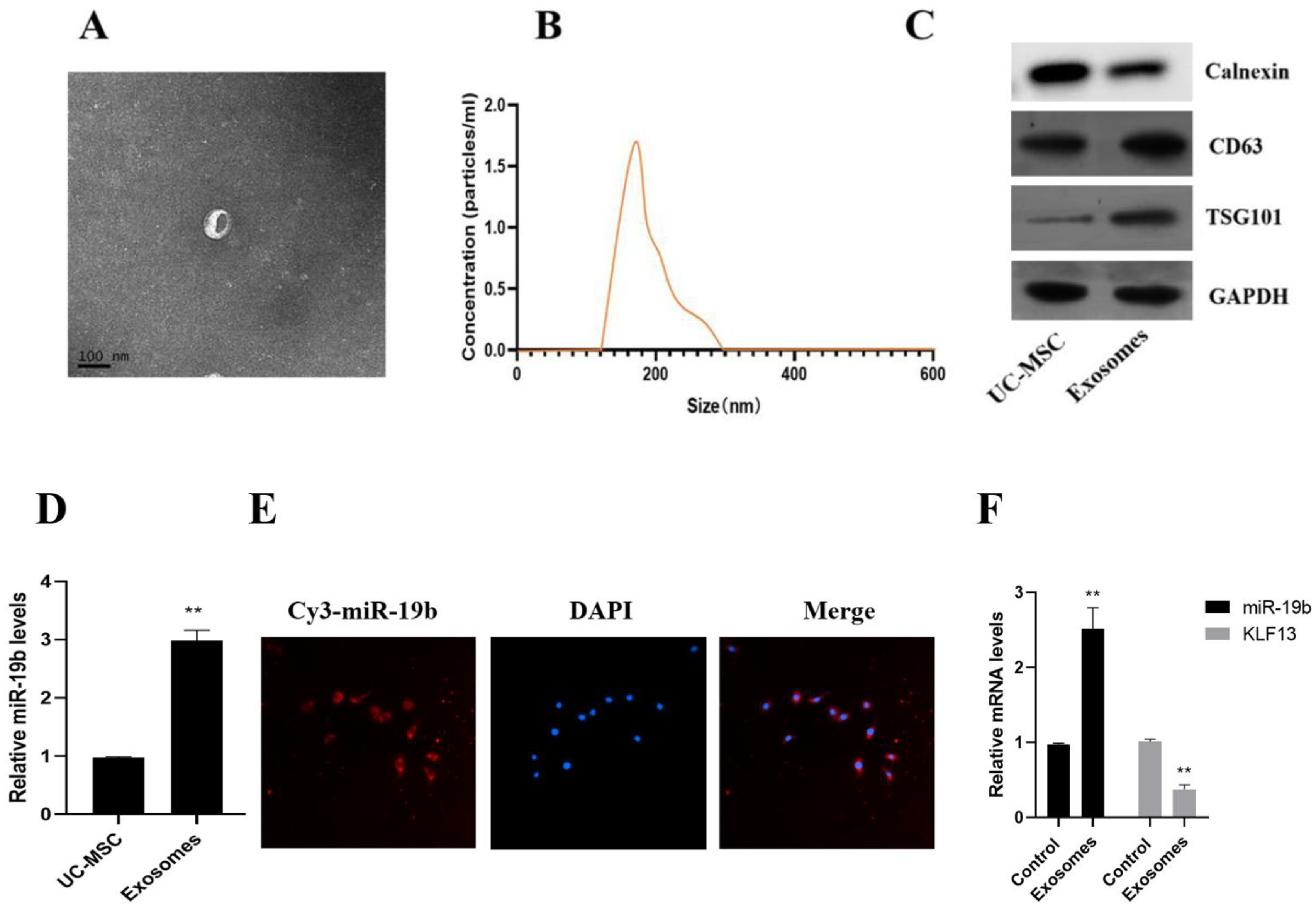
2.4. Exosome Characterization
2.5. Cellular Uptake of Exosomes
2.6. Detection of Th17/Treg Cell Content
2.7. RT-PCR
2.8. Cell Transfection
2.9. Dual Luciferase Reporter Gene Assay
2.10. Statistical Analysis
3. Results
3.1. Comparison of miR-19b Expression Levels
3.2. Balance of Th17 and Treg Cell Subsets
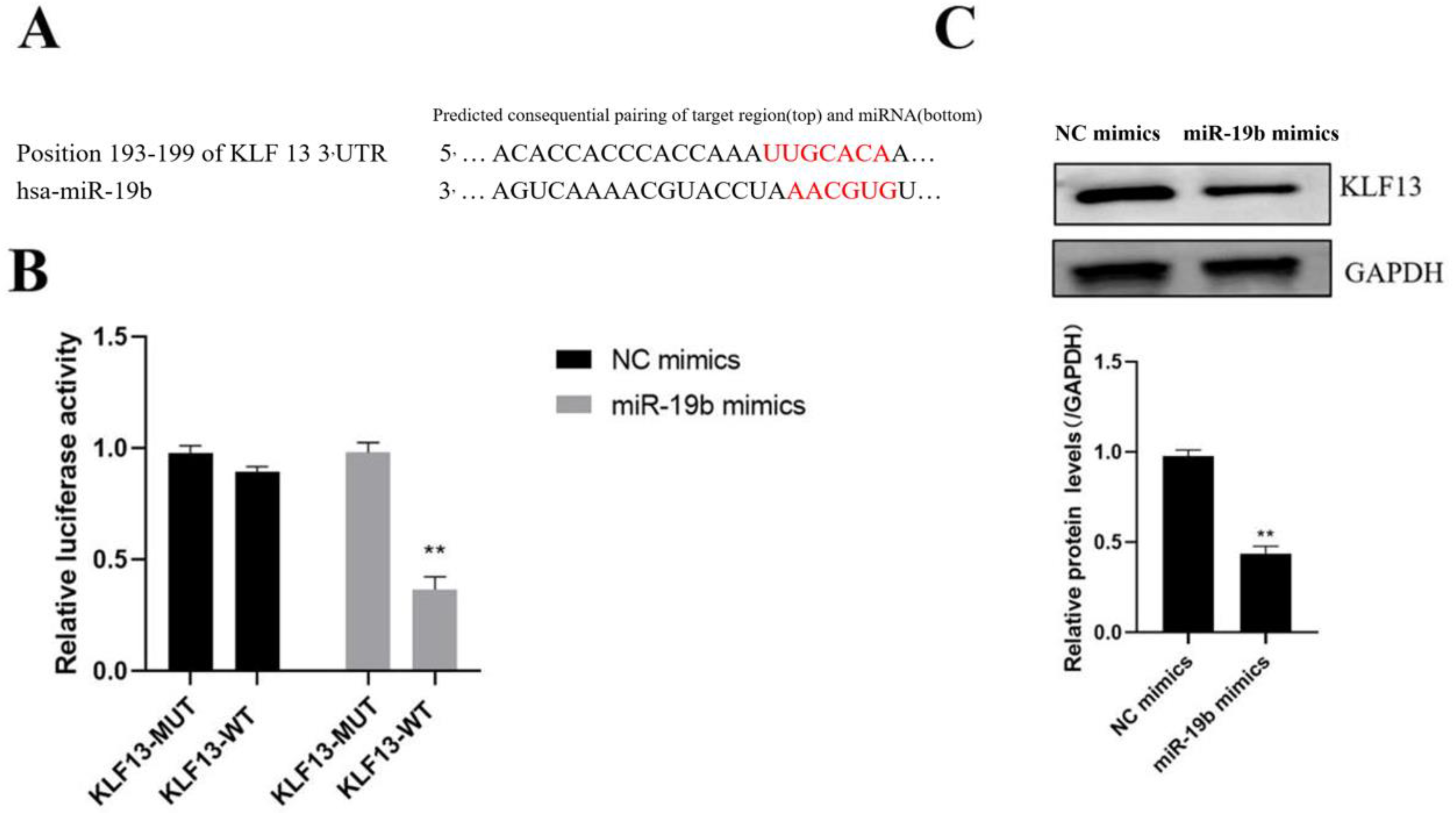
3.3. Identification of miR-19b Target Genes
3.4. Relationship of KLF13 and miR-19b Expression in SLE Patients
3.5. Uptake of UC-MSC Exosomes by PBMC Cells
3.6. Effects of UC-MSC Exosomes on Th17/Treg
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, S.; Long, H.; Lu, Q. New progress in systemic lupus erythematosus research in 2020. Chin. J. Dermatol. 2021, 54, 542–545. [Google Scholar]
- Kumar, H. Tools for fundamental understanding of systemic lupus erythematosus. Int. Rev. Immunol. 2020, 39, 151–152. [Google Scholar] [CrossRef]
- Zhan, H.; Li, Y. Advances in clinical application of biomarkers in systemic lupus erythematosus. Chin. J. Lab. Med. 2020, 43, 939–944. [Google Scholar]
- Cecchi, I.; Perez-Sanchez, C.; Sciascia, S.; Radin, M.; de la Rosa, I.A.; Puerto, N.B.; Lopez-Pedrera, C. Circulating microRNAs as potential biomarkers for monitoring the response to in vivo treatment with Rituximab in systemic lupus erythematosus patients. Autoimmun. Rev. 2020, 19, 128–132. [Google Scholar] [CrossRef]
- Dou, R.; Zhang, X.; Xu, X.; Wang, P.; Yan, B. Mesenchymal stem cell exosomal tsRNA-21109 alleviate systemic lupus erythematosus by inhibiting macrophage M1 polarization. Mol. Immunol. 2021, 139, 106–114. [Google Scholar] [CrossRef]
- Liu, F.; Chen, H.; Chen, T.; Lau, C.-S.; Yu, F.-X.; Chen, K.; Chen, H.-P.; Pan, R.-S.; Chan, G.C.-F.; Zhang, X.-Y.; et al. Immunotherapeutic effects of allogeneic mesenchymal stem cells on systemic lupus erythematosus. Lupus 2020, 29, 872–883. [Google Scholar] [CrossRef]
- Li, W.; Chen, W.; Sun, L. An Update for Mesenchymal Stem Cell Therapy in Lupus Nephritis. Kidney Dis. 2021, 7, 79–89. [Google Scholar] [CrossRef]
- Xu, J.; Chen, J.; Li, W.; Lian, W.; Huang, J.; Lai, B.; Li, L.; Huang, Z. Additive Therapeutic Effects of Mesenchymal Stem Cells and IL-37 for Systemic Lupus Erythematosus. J. Am. Soc. Nephrol. 2020, 31, 54–65. [Google Scholar] [CrossRef]
- Ding, N.; Li, E.; Ouyang, X.; Guo, J.; Wei, B. The Therapeutic Potential of Bone Marrow Mesenchymal Stem Cells for Articular Cartilage Regeneration in Osteoarthritis. Curr. Stem. Cell Res. Ther. 2021, 16, 840–847. [Google Scholar] [CrossRef]
- Liu, H.; Jin, M.; Ji, M.; Zhang, W.; Liu, A.; Wang, T. Hypoxic pretreatment of adipose-derived stem cell exosomes improved cognition by delivery of circ-Epc1 and shifting microglial M1/M2 polarization in an Alzheimer’s disease mice model. Aging 2022, 14, 3070. [Google Scholar] [CrossRef]
- Zhang, S.; Wong, K.L.; Ren, X.; Teo, K.Y.W.; Afizah, H.; Choo, A.B.H.; Toh, W.S. Mesenchymal Stem Cell Exosomes Promote Functional Osteochondral Repair in a Clinically Relevant Porcine Model. Am. J. Sports Med. 2022, 50, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tang, Y.; Qu, B.; Cui, H.; Wang, S.; Wang, L.; Luo, X.; Huang, X.; Li, J.; Chen, S.; et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010, 62, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Sato, R.; Imaizumi, T.; Aizawa, T.; Watanabe, S.; Tsugawa, K.; Kawaguchi, S.; Seya, K.; Matsumiya, T.; Tanaka, H. Inhibitory effect of anti-malarial agents on the expression of proinflammatory chemokines via Toll-like receptor 3 signaling in human glomerular endothelial cells. Ren. Fail. 2021, 43, 643–650. [Google Scholar] [CrossRef]
- Wang, L.; Wu, J.; Ye, N.; Li, F.; Zhan, H.; Chen, S.; Xu, J. Plasma-Derived Exosome MiR-19b Acts as a Diagnostic Marker for Pancreatic Cancer. Front. Oncol. 2021, 11, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Pi, L.; Fang, B.R.; Meng, X.X. Adipose mesenchymal stem cell-derived exosomes accelerate skin wound healing via the lncRNA H19/miR-19b/SOX9 axis. Lab. Investig. 2021, 101, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xu, Y.; Feng, S.; He, P.; Sheng, B.; Ni, J. miR-19b enhances osteogenic differentiation of mesenchymal stem cells and promotes fracture healing through the WWP1/Smurf2-mediated KLF5/β-catenin signaling pathway. Exp. Mol. Med. 2021, 53, 159–162. [Google Scholar] [CrossRef]
- Zeng, Q.; Zhu, Z.; Song, L.; He, Z. Transferred by Exosomes-Derived MiR-19b-3p Targets PTEN to Regulate Esophageal Cancer Cell Apoptosis, Migration and Invasion. Biosci. Rep. 2020, 20, 78–91. [Google Scholar] [CrossRef]
- Price, S. Retraction: Cajanonic acid A regulates the ratio of Th17/Treg via inhibition of expression of IL-6 and TGF-β in insulin-resistant HepG2 cells. Biosci. Rep. 2022, 42, 319–321. [Google Scholar]
- Lambert, S.; Nair, R.; Wang, S.; Elder, J.T. 078 The effect of LPS on neutrophil extracellular trap-induced Th17 polarization is IL-6 and STAT3-Dependent. J. Investig. Dermatol. 2020, 140, 473–476. [Google Scholar] [CrossRef]
- Tang, Z.; Wang, Y.; Xing, R.; Zeng, S.; Di, J.; Xing, F. Deltex-1 is indispensible for the IL-6 and TGF-β treatment-triggered differentiation of Th17 cells. Cell. Immunol. 2020, 94, 138–146. [Google Scholar] [CrossRef]
- Ortega, A.; Martinez-Arroyo, O.; Forner, M.J.; Cortes, R. Exosomes as Drug Delivery Systems: Endogenous Nanovehicles for Treatment of Systemic Lupus Erythematosus. Pharmaceutics 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, Y.; Park, J.; Lee, E.; Song, Y. CD47 Potentiates Inflammatory Response in Systemic Lupus Erythematosus. Cells 2021, 10, 1151. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.C.; Cardoso, R.M.S.; Freire, P.C.; Gomes, C.F.; Duarte, F.V.; das Neves, R.P.; Simões-Correia, J. Immunomodulatory Properties of Umbilical Cord Blood-Derived Small Extracellular Vesicles and Their Therapeutic Potential for Inflammatory Skin Disorders. Int. J. Mol. Sci. 2021, 22, 9797. [Google Scholar] [CrossRef]
- Li, H.; Guan, Y.; Sun, B.; Dou, X.; Liu, X.; Xue, F.; Fu, R.; Zhang, L.; Yang, R. Role of bone marrow-derived mesenchymal stem cell defects in CD8+ CD28- suppressor T-lymphocyte induction in patients with immune thrombocytopenia and associated mechanisms. Br. J. Haematol. 2020, 191, 852–862. [Google Scholar] [CrossRef] [PubMed]




| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| IL-6 | CCG GTT CTG TTC TGC CAG CG | CTG CGA CAC TCA CCT CTG CA |
| IL-17 | CTC CTG TGA TGG CCC ACC TG | CTC CTG GAT CTG AGA CGG AT |
| IL-10 | CGG TCA AGA TCG CCA CTG | ACG GCT GAC TCT AGT AGG GC |
| TGF-β | AAC GAA CTG GCT AGG TGC | CCT AGG CTC ATT AGG CTT AG |
| MiR-19b | CAA GTG GAA GTG GGC AGA G | |
| GAPDH | CAAGAGCCAACGAGCCAAGTC | GCCCAACGTGGACTCCAATTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, J.; Zheng, N.; Mao, C.; Liu, S.; Zhang, H.; Sun, L. UC-BSCs Exosomes Regulate Th17/Treg Balance in Patients with Systemic Lupus Erythematosus via miR-19b/KLF13. Cells 2022, 11, 4123. https://doi.org/10.3390/cells11244123
Tu J, Zheng N, Mao C, Liu S, Zhang H, Sun L. UC-BSCs Exosomes Regulate Th17/Treg Balance in Patients with Systemic Lupus Erythematosus via miR-19b/KLF13. Cells. 2022; 11(24):4123. https://doi.org/10.3390/cells11244123
Chicago/Turabian StyleTu, Jianxin, Nan Zheng, Chentong Mao, Shan Liu, Hongxing Zhang, and Li Sun. 2022. "UC-BSCs Exosomes Regulate Th17/Treg Balance in Patients with Systemic Lupus Erythematosus via miR-19b/KLF13" Cells 11, no. 24: 4123. https://doi.org/10.3390/cells11244123
APA StyleTu, J., Zheng, N., Mao, C., Liu, S., Zhang, H., & Sun, L. (2022). UC-BSCs Exosomes Regulate Th17/Treg Balance in Patients with Systemic Lupus Erythematosus via miR-19b/KLF13. Cells, 11(24), 4123. https://doi.org/10.3390/cells11244123






