Plant Plasma Membrane Proton Pump: One Protein with Multiple Functions
Abstract
:1. Introduction
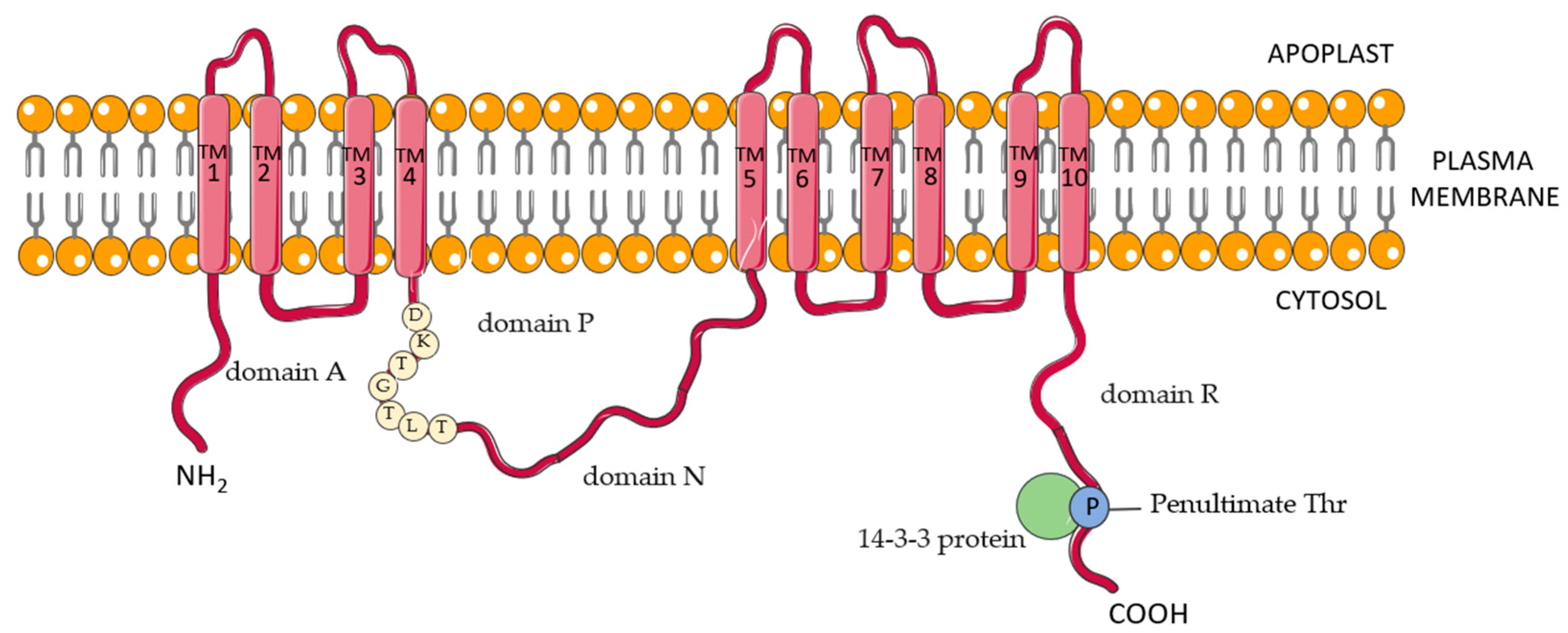
2. The Pleiotropic Effect of Plasma Membrane Proton Pump
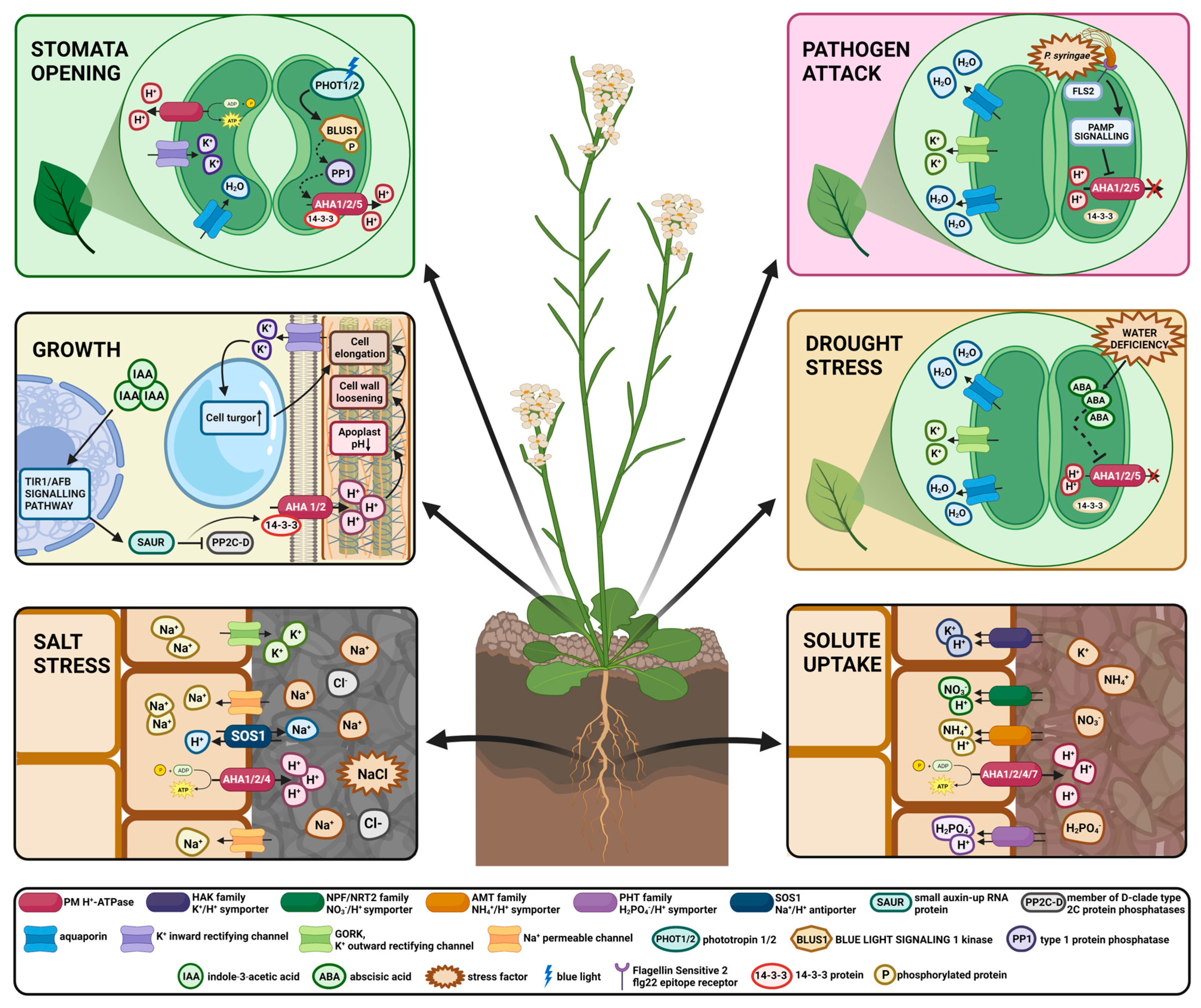
3. PM Proton Pump Engagement in Multiple Crucial Processes in Plants
3.1. Growth
3.2. Stomata Opening
3.3. Mineral Uptake
3.4. Cytosolic pH Homeostasis
3.5. Adaptation to Salt Stress
3.6. Adaptation to Drought Stress
3.7. Heavy Metals and Temperature Stresses—Poststress Responses for Homeostasis Maintenance
4. PM Proton Pump Involvement in Plant Pathogenesis
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Palmgren, M.G. PLANT PLASMA MEMBRANE H+-ATPases: Powerhouses for Nutrient Uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 817–845. [Google Scholar] [CrossRef] [Green Version]
- Haruta, M.; Gray, W.M.; Sussman, M.R. Regulation of the Plasma Membrane Proton Pump (H+-ATPase) by Phosphorylation. Curr. Opin. Plant Biol. 2015, 28, 68–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haruta, M.; Burch, H.L.; Nelson, R.B.; Barrett-Wilt, G.; Kline, K.G.; Mohsin, S.B.; Young, J.C.; Otegui, M.S.; Sussman, M.R. Molecular Characterization of Mutant Arabidopsis Plants with Reduced Plasma Membrane Proton Pump Activity. J. Biol. Chem. 2010, 285, 17918–17929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, S.; Kinoshita, T. Blue Light Regulation of Stomatal Opening and the Plasma Membrane H+-ATPase. Plant Physiol. 2017, 174, 531–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, R.D.; Olsen, L.I.; Ezike, C.V.; Pedersen, J.T.; Manstretta, R.; López-Marqués, R.L.; Palmgren, M. Roles of Plasma Membrane Proton ATPases AHA2 and AHA7 in Normal Growth of Roots and Root Hairs in Arabidopsis thaliana. Physiol. Plant. 2019, 166, 848–861. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.-Y.; Hao, D.-L.; Yang, G.-Z. Regulation of Cytosolic PH: The Contributions of Plant Plasma Membrane H+-ATPases and Multiple Transporters. IJMS 2021, 22, 12998. [Google Scholar] [CrossRef] [PubMed]
- Møller, J.V.; Juul, B.; le Maire, M. Structural Organization, Ion Transport, and Energy Transduction of P-Type ATPases. Biochim. Biophys. Acta—Biomembr. 1996, 1286, 1–51. [Google Scholar] [CrossRef]
- Jahn, T.; Dietrich, J.; Andersen, B.; Leidvik, B.; Otter, C.; Briving, C.; Kühlbrandt, W.; Palmgren, M.G. Large Scale Expression, Purification and 2D Crystallization of Recombinant Plant Plasma Membrane H+-ATPase. J. Mol. Biol. 2001, 309, 465–476. [Google Scholar] [CrossRef]
- Morsomme, P.; Boutry, M. The Plant Plasma Membrane H+-ATPase: Structure, Function and Regulation. Biochim. Biophys. Acta—Biomembr. 2000, 1465, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Duby, G.; Boutry, M. The Plant Plasma Membrane Proton Pump ATPase: A Highly Regulated P-Type ATPase with Multiple Physiological Roles. Pflug. Arch—Eur. J. Physiol. 2009, 457, 645–655. [Google Scholar] [CrossRef]
- Kanczewska, J.; Marco, S.; Vandermeeren, C.; Maudoux, O.; Rigaud, J.-L.; Boutry, M. Activation of the Plant Plasma Membrane H+-ATPase by Phosphorylation and Binding of 14-3-3 Proteins Converts a Dimer into a Hexamer. Proc. Natl. Acad. Sci. USA 2005, 102, 11675–11680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottmann, C.; Marco, S.; Jaspert, N.; Marcon, C.; Schauer, N.; Weyand, M.; Vandermeeren, C.; Duby, G.; Boutry, M.; Wittinghofer, A.; et al. Structure of a 14-3-3 Coordinated Hexamer of the Plant Plasma Membrane H+-ATPase by Combining X-ray Crystallography and Electron Cryomicroscopy. Mol. Cell 2007, 25, 427–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Guo, Y.; Yang, Y. The Molecular Mechanism of Plasma Membrane H+-ATPases in Plant Responses to Abiotic Stress. J. Genet. Genom. 2022, 49, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Gaxiola, R.A.; Palmgren, M.G.; Schumacher, K. Plant Proton Pumps. FEBS Lett. 2007, 581, 2204–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic Comparison of P-Type ATPase Ion Pumps in Arabidopsis and Rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef] [Green Version]
- Wdowikowska, A.; Klobus, G. The Plasma Membrane Proton Pump Gene Family in Cucumber. Acta Physiol. Plant. 2016, 38, 135. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Visconti, S.; Drumm, K.; Jahn, T.; Stensballe, A.; Mattei, B.; Jensen, O.N.; Aducci, P.; Palmgren, M.G. Binding of 14-3-3 Protein to the Plasma Membrane H+-ATPase AHA2 Involves the Three C-Terminal Residues Tyr946-Thr-Val and Requires Phosphorylation of Thr947. J. Biol. Chem. 1999, 274, 36774–36780. [Google Scholar] [CrossRef] [Green Version]
- Fuglsang, A.T.; Borch, J.; Bych, K.; Jahn, T.P.; Roepstorff, P.; Palmgren, M.G. The Binding Site for Regulatory 14-3-3 Protein in Plant Plasma Membrane H+-ATPase. J. Biol. Chem. 2003, 278, 42266–42272. [Google Scholar] [CrossRef] [Green Version]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S.; et al. Arabidopsis Protein Kinase PKS5 Inhibits the Plasma Membrane H+-ATPase by Preventing Interaction with 14-3-3 Protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef] [Green Version]
- Fuglsang, A.T.; Kristensen, A.; Cuin, T.A.; Schulze, W.X.; Persson, J.; Thuesen, K.H.; Ytting, C.K.; Oehlenschlaeger, C.B.; Mahmood, K.; Sondergaard, T.E.; et al. Receptor Kinase-Mediated Control of Primary Active Proton Pumping at the Plasma Membrane. Plant J. 2014, 80, 951–964. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.-K. The Arabidopsis Thaliana Salt Tolerance Gene SOS1 Encodes a Putative Na+/H+ Antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janicka-Russak, M.; Kabala, K.; Burzynski, M.; Klobus, G. Response of Plasma Membrane H+-ATPase to Heavy Metal Stress in Cucumis sativus Roots. J. Exp. Bot. 2008, 59, 3721–3728. [Google Scholar] [CrossRef] [Green Version]
- Janicka-Russak, M.; Kabała, K.; Wdowikowska, A.; Kłobus, G. Response of Plasma Membrane H+-ATPase to Low Temperature in Cucumber Roots. J. Plant Res. 2012, 125, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma Membrane H+-ATPase Regulation in the Center of Plant Physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Liu, W.; Zeng, F.; Chen, Z.; Zhang, G.; Wu, F. K+ Uptake, H+-ATPase Pumping Activity and Ca2+ Efflux Mechanism Are Involved in Drought Tolerance of Barley. Environ. Exp. Bot. 2016, 129, 57–66. [Google Scholar] [CrossRef]
- Haruta, M.; Sussman, M.R. The Effect of a Genetically Reduced Plasma Membrane Protonmotive Force on Vegetative Growth of Arabidopsis. Plant Physiol. 2012, 158, 1158–1171. [Google Scholar] [CrossRef] [Green Version]
- Gévaudant, F.; Duby, G.; von Stedingk, E.; Zhao, R.; Morsomme, P.; Boutry, M. Expression of a Constitutively Activated Plasma Membrane H+-ATPase Alters Plant Development and Increases Salt Tolerance. Plant Physiol. 2007, 144, 1763–1776. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shimazaki, K.; Kinoshita, T. Multiple Roles of the Plasma Membrane H+-ATPase and Its Regulation. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 2014; Volume 35, pp. 191–211. [Google Scholar] [CrossRef]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin Activates the Plasma Membrane H+-ATPase by Phosphorylation during Hypocotyl Elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef]
- Li, L.; Verstraeten, I.; Roosjen, M.; Takahashi, K.; Rodriguez, L.; Merrin, J.; Chen, J.; Shabala, L.; Smet, W.; Ren, H.; et al. Cell Surface and Intracellular Auxin Signalling for H+ Fluxes in Root Growth. Nature 2021, 599, 273–277. [Google Scholar] [CrossRef]
- Hager, A. Role of the Plasma Membrane H+-ATPase in Auxin-Induced Elongation Growth: Historical and New Aspects. J. Plant Res. 2003, 116, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Fendrych, M.; Leung, J.; Friml, J. TIR1/AFB-Aux/IAA Auxin Perception Mediates Rapid Cell Wall Acidification and Growth of Arabidopsis Hypocotyls. eLife 2016, 5, e19048. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T. Blue Light Activates the Plasma Membrane H+-ATPase by Phosphorylation of the C-Terminus in Stomatal Guard Cells. EMBO J. 1999, 18, 5548–5558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, T.; Shimazaki, K. Biochemical Evidence for the Requirement of 14-3-3 Protein Binding in Activation of the Guard-Cell Plasma Membrane H+-ATPase by Blue Light. Plant Cell Physiol. 2002, 43, 1359–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemiya, A.; Shimazaki, K. Arabidopsis Phot1 and Phot2 Phosphorylate BLUS1 Kinase with Different Efficiencies in Stomatal Opening. J. Plant Res. 2016, 129, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M.; Sugimoto, H.; Inoue, S.; Takahashi, Y.; Hayashi, M.; Hayashi, Y.; Mizutani, M.; Ogawa, T.; Kinoshita, D.; Ando, E.; et al. Type 2C Protein Phosphatase Clade D Family Members Dephosphorylate Guard Cell Plasma Membrane H+-ATPase. Plant Physiol. 2022, 188, 2228–2240. [Google Scholar] [CrossRef]
- Espen, L. Effect of NO3− Transport and Reduction on Intracellular PH: An in Vivo NMR Study in Maize Roots. J. Exp. Bot. 2004, 55, 2053–2061. [Google Scholar] [CrossRef]
- Ortiz-Ramirez, C.; Mora, S.I.; Trejo, J.; Pantoja, O. PvAMT1;1, a Highly Selective Ammonium Transporter That Functions as H+/NH4+ Symporter. J. Biol. Chem. 2011, 286, 31113–31122. [Google Scholar] [CrossRef] [Green Version]
- Młodzińska, E.; Zboińska, M. Phosphate Uptake and Allocation—A Closer Look at Arabidopsis Thaliana L. and Oryza Sativa L. Front. Plant Sci. 2016, 7, 1198. [Google Scholar] [CrossRef]
- Maathuis, F.J.M.; Sanders, D. Energization of Potassium Uptake in Arabidopsis Thaliana. Planta 1993, 191, 302–307. [Google Scholar] [CrossRef]
- Godwin, R.M.; Rae, A.L.; Carroll, B.J.; Smith, F.W. Cloning and Characterization of Two Genes Encoding Sulfate Transporters from Rice (Oryza Sativa L.) *. Plant Soil 2003, 257, 113–123. [Google Scholar] [CrossRef]
- Sze, H.; Chanroj, S. Plant Endomembrane Dynamics: Studies of K+/H+ Antiporters Provide Insights on the Effects of PH and Ion Homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef] [Green Version]
- Bassil, E.; Blumwald, E. The Ins and Outs of Intracellular Ion Homeostasis: NHX-Type Cation/H+ Transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wegner, L.H.; Li, X.; Zhang, J.; Yu, M.; Shabala, S.; Hao, Z. Biochemical and Biophysical PH Clamp Controlling Net H + Efflux across the Plasma Membrane of Plant Cells. New Phytol. 2021, 230, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Wegner, L.H.; Shabala, S. Biochemical PH Clamp: The Forgotten Resource in Membrane Bioenergetics. New Phytol. 2020, 225, 37–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janicka-Russak, M.; Kabała, K. The Role of Plasma Membrane H+-ATPase in Salinity Stress of Plants. In Progress in Botany; Lüttge, U., Beyschlag, W., Eds.; Progress in Botany; Springer International Publishing: Cham, Switzerland, 2015; Volume 76, pp. 77–92. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Y.; Ma, L.; Yang, Z.; Dong, Q.; Li, Q.; Ni, X.; Kudla, J.; Song, C.; Guo, Y. The Ca2+ Sensor SCaBP3/CBL7 Modulates Plasma Membrane H+-ATPase Activity and Promotes Alkali Tolerance in Arabidopsis. Plant Cell 2019, 31, 1367–1384. [Google Scholar] [CrossRef] [PubMed]
- Janicka-Russak, M.; Kabała, K.; Wdowikowska, A.; Kłobus, G. Modification of Plasma Membrane Proton Pumps in Cucumber Roots as an Adaptation Mechanism to Salt Stress. J. Plant Physiol. 2013, 170, 915–922. [Google Scholar] [CrossRef]
- Qiu, Q.-S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.-K. Regulation of SOS1, a Plasma Membrane Na+/H+ Exchanger in Arabidopsis Thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Chen, Z.; Pottosin, I.I.; Cuin, T.A.; Fuglsang, A.T.; Tester, M.; Jha, D.; Zepeda-Jazo, I.; Zhou, M.; Palmgren, M.G.; Newman, I.A.; et al. Root Plasma Membrane Transporters Controlling K+/Na+ Homeostasis in Salt-Stressed Barley. Plant Physiol. 2007, 145, 1714–1725. [Google Scholar] [CrossRef] [Green Version]
- Goh, C.-H.; Kinoshita, T.; Oku, T.; Shimazaki, K. Lnhibition of Blue Light-Dependent H+ Pumping by Abscisic Acid in Vicia Guard-Cell Protoplasts. Plant Physiol. 1996, 111, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, H.; Takemiya, A.; Song, C.; Kinoshita, T.; Shimazaki, K. Inhibition of Blue Light-Dependent H+ Pumping by Abscisic Acid through Hydrogen Peroxide-Induced Dephosphorylation of the Plasma Membrane H+-ATPase in Guard Cell Protoplasts. Plant Physiol. 2004, 136, 4150–4158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Adachi, Y.; Ye, W.; Hayashi, M.; Nakamura, Y.; Kinoshita, T.; Mori, I.C.; Murata, Y. Difference in Abscisic Acid Perception Mechanisms between Closure Induction and Opening Inhibition of Stomata. Plant Physiol. 2013, 163, 600–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muzi, C.; Camoni, L.; Visconti, S.; Aducci, P. Cold stress affects H+-ATPase and phospholipase D activity in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 328–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fodor, E.; Szabo-Nagy, A.; Erdei, L. The effects of cadmium on the fluidity and H+-ATPase activity of plasma membrane from sunflower and wheat roots. J. Plant Physiol. 1995, 147, 87–92. [Google Scholar] [CrossRef]
- Burzyński, M.; Kolano, E. In vivo and in vitro effects of copper and cadmium on the plasma membrane H+-ATPase from cucumber (Cucumis sativus L.) and maize (Zea mays L.) roots. Acta Physiol. Plant. 2003, 25, 39–45. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant Stomata Function in Innate Immunity against Bacterial Invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Fan, J.; Gao, Y.G.; Wang, Z.; Yang, P.; Liang, Y.; Opiyo, S.; Xia, Y. Arabidopsis Plasma Membrane ATPase AHA5 Is Negatively Involved in PAMP-Triggered Immunity. IJMS 2022, 23, 3857. [Google Scholar] [CrossRef]
- Liu, J.; Elmore, J.M.; Fuglsang, A.T.; Palmgren, M.G.; Staskawicz, B.J.; Coaker, G. RIN4 Functions with Plasma Membrane H+-ATPases to Regulate Stomatal Apertures during Pathogen Attack. PLoS Biol. 2009, 7, e1000139. [Google Scholar] [CrossRef]
- Kaundal, A.; Ramu, V.S.; Oh, S.; Lee, S.; Pant, B.; Lee, H.-K.; Rojas, C.M.; Senthil-Kumar, M.; Mysore, K.S. GENERAL CONTROL NONREPRESSIBLE4 Degrades 14-3-3 and the RIN4 Complex to Regulate Stomatal Aperture with Implications on Nonhost Disease Resistance and Drought Tolerance. Plant Cell 2017, 29, 2233–2248. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas Syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [Green Version]
- Arango, M.; Gévaudant, F.; Oufattole, M.; Boutry, M. The Plasma Membrane Proton Pump ATPase: The Significance of Gene Subfamilies. Planta 2003, 216, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in Action: Signalling, Transport and the Control of Plant Growth and Development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Quint, M.; Gray, W.M. Auxin Signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Rayle, D.L.; Cleland, R.E. The Acid Growth Theory of Auxin-Induced Cell Elongation Is Alive and Well. Plant Physiol. 1992, 99, 1271–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrot-Rechenmann, C. Cellular Responses to Auxin: Division versus Expansion. Cold Spring Harbor. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef] [PubMed]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in Plant Growth and Potential Applications in Crop Improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef] [Green Version]
- Philippar, K.; Ivashikina, N.; Ache, P.; Christian, M.; Lüthen, H.; Palme, K.; Hedrich, R. Auxin Activates KAT1 and KAT2, Two K+-Channel Genes Expressed in Seedlings of Arabidopsis Thaliana. Plant J. 2004, 37, 815–827. [Google Scholar] [CrossRef]
- Kutschera, U. The Current Status of the Acid-growth Hypothesis. New Phytol. 1994, 126, 549–569. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. In Auxin Molecular Biology; Perrot-Rechenmann, C., Hagen, G., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 373–385. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 Subfamily of SMALL AUXIN UP RNA Genes Promote Cell Expansion: SAUR Fusion Proteins Promote Cell Expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [Green Version]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Zhou, X.; Tang, W.; Takahashi, K.; Pan, X.; Dai, J.; Ren, H.; Zhu, X.; Pan, S.; Zheng, H.; et al. TMK-Based Cell-Surface Auxin Signalling Activates Cell-Wall Acidification. Nature 2021, 599, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, K.; Doi, M.; Assmann, S.M.; Kinoshita, T. Light Regulation of Stomatal Movement. Annu. Rev. Plant Biol. 2007, 58, 219–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, E.; Kinoshita, T. Red Light-Induced Phosphorylation of Plasma Membrane H+-ATPase in Stomatal Guard Cells. Plant Physiol. 2018, 178, 838–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, J.M. Phototropin Blue-Light Receptors. Annu. Rev. Plant Biol. 2007, 58, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Takemiya, A.; Kinoshita, T.; Asanuma, M.; Shimazaki, K. Protein Phosphatase 1 Positively Regulates Stomatal Opening in Response to Blue Light in Vicia Faba. Proc. Natl. Acad. Sci. USA 2006, 103, 13549–13554. [Google Scholar] [CrossRef] [Green Version]
- Takemiya, A.; Yamauchi, S.; Yano, T.; Ariyoshi, C.; Shimazaki, K. Identification of a Regulatory Subunit of Protein Phosphatase 1 Which Mediates Blue Light Signaling for Stomatal Opening. Plant Cell Physiol. 2013, 54, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Takemiya, A.; Sugiyama, N.; Fujimoto, H.; Tsutsumi, T.; Yamauchi, S.; Hiyama, A.; Tada, Y.; Christie, J.M.; Shimazaki, K. Phosphorylation of BLUS1 Kinase by Phototropins Is a Primary Step in Stomatal Opening. Nat. Commun. 2013, 4, 2094. [Google Scholar] [CrossRef]
- Kinoshita, T.; Ono, N.; Hayashi, Y.; Morimoto, S.; Nakamura, S.; Soda, M.; Kato, Y.; Ohnishi, M.; Nakano, T.; Inoue, S.; et al. FLOWERING LOCUS T Regulates Stomatal Opening. Curr. Biol. 2011, 21, 1232–1238. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Transporters Involved in Mineral Nutrient Uptake in Rice. J. Exp. Bot. 2016, 67, 3645–3653. [Google Scholar] [CrossRef] [Green Version]
- Muratore, C.; Espen, L.; Prinsi, B. Nitrogen Uptake in Plants: The Plasma Membrane Root Transport Systems from a Physiological and Proteomic Perspective. Plants 2021, 10, 681. [Google Scholar] [CrossRef]
- Tsay, Y.-F.; Chiu, C.-C.; Tsai, C.-B.; Ho, C.-H.; Hsu, P.-K. Nitrate Transporters and Peptide Transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, N.M.; Glass, A.D.M. Molecular and Physiological Aspects of Nitrate Uptake in Plants. Trends Plant Sci. 1998, 3, 389–395. [Google Scholar] [CrossRef]
- Søgaard, R.; Alsterfjord, M.; MacAulay, N.; Zeuthen, T. Ammonium Ion Transport by the AMT/Rh Homolog TaAMT1;1 Is Stimulated by Acidic PH. Pflug. Arch—Eur. J. Physiol. 2009, 458, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Preuss, C.P.; Huang, C.Y.; Gilliham, M.; Tyerman, S.D. Channel-Like Characteristics of the Low-Affinity Barley Phosphate Transporter PHT1;6 When Expressed in Xenopus Oocytes. Plant Physiol. 2010, 152, 1431–1441. [Google Scholar] [CrossRef] [Green Version]
- Coskun, D.; Britto, D.T.; Li, M.; Oh, S.; Kronzucker, H.J. Capacity and Plasticity of Potassium Channels and High-Affinity Transporters in Roots of Barley and Arabidopsis. Plant Physiol. 2013, 162, 496–511. [Google Scholar] [CrossRef] [Green Version]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. K+ Uptake in Plant Roots. The Systems Involved, Their Regulation and Parallels in Other Organisms. J. Plant Physiol. 2014, 171, 688–695. [Google Scholar] [CrossRef]
- Qi, Z.; Hampton, C.R.; Shin, R.; Barkla, B.J.; White, P.J.; Schachtman, D.P. The High Affinity K+ Transporter AtHAK5 Plays a Physiological Role in Planta at Very Low K + Concentrations and Provides a Caesium Uptake Pathway in Arabidopsis. J. Exp. Bot. 2008, 59, 595–607. [Google Scholar] [CrossRef]
- Véry, A.-A.; Nieves-Cordones, M.; Daly, M.; Khan, I.; Fizames, C.; Sentenac, H. Molecular Biology of K+ Transport across the Plant Cell Membrane: What Do We Learn from Comparison between Plant Species? J. Plant Physiol. 2014, 171, 748–769. [Google Scholar] [CrossRef]
- Maathuis, F.J.; Sanders, D. Mechanism of High-Affinity Potassium Uptake in Roots of Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 1994, 91, 9272–9276. [Google Scholar] [CrossRef] [Green Version]
- Walker, D.J.; Leigh, R.A.; Miller, A.J. Potassium Homeostasis in Vacuolate Plant Cells. Proc. Natl. Acad. Sci. USA 1996, 93, 10510–10514. [Google Scholar] [CrossRef] [Green Version]
- Rodr, A. Potassium Transport in Fungi and Plants. Biochim. Biophys. Acta 2000, 1469, 1–30. [Google Scholar] [CrossRef]
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The Roles of Three Functional Sulphate Transporters Involved in Uptake and Translocation of Sulphate in Arabidopsis Thaliana: Sulphate Transporters in Arabidopsis Thaliana. Plant J. 2000, 23, 171–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibagaki, N.; Rose, A.; McDermott, J.P.; Fujiwara, T.; Hayashi, H.; Yoneyama, T.; Davies, J.P. Selenate-Resistant Mutants of Arabidopsis Thaliana Identify Sultr1;2, a Sulfate Transporter Required for Efficient Transport of Sulfate into Roots. Plant J. 2002, 29, 475–486. [Google Scholar] [CrossRef]
- Zhang, B.; Pasini, R.; Dan, H.; Joshi, N.; Zhao, Y.; Leustek, T.; Zheng, Z.-L. Aberrant Gene Expression in the Arabidopsis SULTR1;2 Mutants Suggests a Possible Regulatory Role for This Sulfate Transporter in Response to Sulfur Nutrient Status. Plant J. 2014, 77, 185–197. [Google Scholar] [CrossRef]
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.-H. Evolutionary Relationships and Functional Diversity of Plant Sulfate Transporters. Front. Plant Sci. 2012, 2, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isayenkov, S.V.; Dabravolski, S.A.; Pan, T.; Shabala, S. Phylogenetic Diversity and Physiological Roles of Plant Monovalent Cation/H+ Antiporters. Front. Plant Sci. 2020, 11, 573564. [Google Scholar] [CrossRef]
- Davies, D.D. The Fine Control of Cytosolic PH. Physiol. Plant 1986, 67, 702–706. [Google Scholar] [CrossRef]
- Olivari, C.; Pugliarello, M.C.; Rasi-Caldogno, F.; De Michelis, M.I. Characteristics and Regulatory Properties of the H + -ATPase in a Plasma Membrane Fraction Purified from Arabidopsis Thaliana. Bot. Acta 1993, 106, 13–19. [Google Scholar] [CrossRef]
- Becker, D.; Zeilinger, C.; Lohse, G.; Depta, H.; Hedrich, R. Identification and Biochemical Characterization of the Plasma-Membrane H+-ATPase in Guard Cells of Vicia Faba L. Planta 1993, 190, 44–50. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Niu, X.; Damsz, B.; Bressan, R.A.; Hasegawa, P.M. NaCI-Lnduced Alterations in Both Cell Structure and Tissue- Specific Plasma Membrane H+-ATPase Gene Expression. Plant Physiol. 1996, 111, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Wu, H.; Bose, J. Salt Stress Sensing and Early Signalling Events in Plant Roots: Current Knowledge and Hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, X.; Lv, W.; Yang, Y. Molecular Mechanisms of Plant Responses to Salt Stress. Front. Plant Sci. 2022, 13, 934877. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qin, Y.; Xie, C.; Zhao, F.; Zhao, J.; Liu, D.; Chen, S.; Fuglsang, A.T.; Palmgren, M.G.; Schumaker, K.S.; et al. The Arabidopsis Chaperone J3 Regulates the Plasma Membrane H+-ATPase through Interaction with the PKS5 Kinase. Plant Cell 2010, 22, 1313–1332. [Google Scholar] [CrossRef] [Green Version]
- Hauser, F.; Horie, T. A Conserved Primary Salt Tolerance Mechanism Mediated by HKT Transporters: A Mechanism for Sodium Exclusion and Maintenance of High K +/Na + Ratio in Leaves during Salinity Stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Bose, J.; Hedrich, R. Salt Bladders: Do They Matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Anschütz, U.; Becker, D.; Shabala, S. Going beyond Nutrition: Regulation of Potassium Homoeostasis as a Common Denominator of Plant Adaptive Responses to Environment. J. Plant Physiol. 2014, 171, 670–687. [Google Scholar] [CrossRef]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium Transport in Plant Cells. Biochim. Biophys. Acta—Biomembr. 2000, 1465, 140–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Nguyen, N.H.; Jeong, C.Y.; Nguyen, N.T.; Hong, S.-W.; Lee, H. Loss of the R2R3 MYB, AtMyb73, Causes Hyper-Induction of the SOS1 and SOS3 Genes in Response to High Salinity in Arabidopsis. J. Plant Physiol. 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na +/H + Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Demidchik, V.; Shabala, L.; Cuin, T.A.; Smith, S.J.; Miller, A.J.; Davies, J.M.; Newman, I.A. Extracellular Ca2+ Ameliorates NaCl-Induced K+ Loss from Arabidopsis Root and Leaf Cells by Controlling Plasma Membrane K+-Permeable Channels. Plant Physiol. 2006, 141, 1653–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, J.I.; Raschke, K.; Neher, E. Voltage Dependence of K + Channels in Guard-Cell Protoplasts. Proc. Natl. Acad. Sci. USA 1987, 84, 4108–4112. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, J.I.; Allen, G.J.; Hugouvieux, V.; Kwak, J.M.; Waner, D. GUARD CELL SIGNAL TRANSDUCTION. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 627–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlot, S.; Leonhardt, N.; Fenzi, F.; Valon, C.; Costa, M.; Piette, L.; Vavasseur, A.; Genty, B.; Boivin, K.; Müller, A.; et al. Constitutive Activation of a Plasma Membrane H+-ATPase Prevents Abscisic Acid-Mediated Stomatal Closure. EMBO J. 2007, 26, 3216–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Inoue, S.; Takahashi, K.; Kinoshita, T. Immunohistochemical Detection of Blue Light-Induced Phosphorylation of the Plasma Membrane H+-ATPase in Stomatal Guard Cells. Plant Cell Physiol. 2011, 52, 1238–1248. [Google Scholar] [CrossRef] [Green Version]
- Devi, S.R.; Prasad, M.N.V. Membrane Lipid Alterations in Heavy Metal Exposed Plants. In Heavy Metal Stress in Plants. From Molecules to Ecosystems; Prasad, M.N.V., Hagemeyer, J., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 99–116. [Google Scholar]
- Breckle, S.W.; Kahle, H. Ecological Geobotany/Autecology and Ecotoxicology. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 1991; Volume 52, pp. 391–406. [Google Scholar]
- Kennedy, C.D.; Gonsalves, F.A.N. The action of divalent Zn, Cd, Hg, Cu and Pb ions on the ATPase activity of plasma membranę fraction isolated from roots of Zea mays. Plant Soil 1989, 117, 167–175. [Google Scholar] [CrossRef]
- Janicka-Russak, M.; Kabała, K.; Burzyński, M. Different effect of cadmium and copper on H+-ATPase activity in plasma membrane vesicles from Cucumis sativus roots. J. Exp. Bot. 2012, 63, 4133–4142. [Google Scholar] [CrossRef]
- Ros, R.; Morales, A.; Segura, J.; Picazo, I. In vivo and in vitro effects of nickel and cadmium on the plasmalemma ATPase from rice (Oryza sativa L.) shoots and roots. Plant Sci. 1992, 83, 1–6. [Google Scholar] [CrossRef]
- Hippler, F.W.R.; Mttos, D., Jr.; Boaretto, R.M.; Williams, L.E. Copper excess reduces nitrate uptake by Arabidopsis roots with specific effects on gene expression. J. Plant Physiol. 2018, 228, 158–165. [Google Scholar] [CrossRef]
- Młodzińska, E.; Kłobus, G.; Christensen, M.D.; Fuglsang, A.T. The plasma membrane H+-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol. Plant. 2015, 154, 270–282. [Google Scholar] [CrossRef]
- Ram, H.; Kaur, A.; Gandass, N.; Singh, S.; Desmukh, R.; Sonah, H.; Sharma, T.R. Molecular characterization and expression dynamics of MTP genes under various spatiotemporal stages and metal stress conditions in rice. PLoS ONE 2019, 14, 0217360. [Google Scholar] [CrossRef] [PubMed]
- Talke, I.N.; Hanikenne, M.; Kramer, U. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 2006, 142, 148–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous Adaptation and the Regulation of Membrane Lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef] [Green Version]
- Los, D.A.; Murata, N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta. 2004, 1666, 142–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willing, R.P.; Leopold, A.C. Cellular expansion at low temperature as a cause of membrane lesions. Plant Physiol. 1983, 71, 118–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkindale, J.; Huang, B. Changes of lipid composition and saturation level in leaves and roots for heat-stress and heat-acclimated creeping bentgrass (Agrostis stolonifera). Environ. Exp. Bot. 2004, 51, 57–67. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Ponce-Pineda, I.G.; Carmona-Salazar, L.; Saucedo-García, M.; Cano-Ramírez, D.; Morales-Cedillo, F.; Peña-Moral, A.; Guevara-García, Á.A.; Sánchez-Nieto, S.; Gavilanes-Ruíz, M. MPK6 Kinase Regulates Plasma Membrane H+-ATPase Activity in Cold Acclimation. Int. J. Mol. Sci. 2021, 22, 6338. [Google Scholar] [CrossRef]
- Janicka-Russak, M.; Kabała, K. Abscisic acid and hydrogen peroxide induce modification of plasma membrane H(+)-ATPase from Cucumis sativus L. roots under heat shock. J. Plant Physiol. 2012, 169, 1607–1614. [Google Scholar] [CrossRef]
- Keinath, N.F.; Kierszniowska, S.; Lorek, J.; Bourdais, G.; Kessler, S.A.; Shimosato-Asano, H.; Grossniklaus, U.; Schulze, W.X.; Robatzek, S.; Panstruga, R. PAMP (Pathogen-Associated Molecular Pattern)-Induced Changes in Plasma Membrane Compartmentalization Reveal Novel Components of Plant Immunity. J. Biol. Chem. 2010, 285, 39140–39149. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Zhang, L.; Oblessuc, P.R.; He, S.Y. Stomatal Defense a Decade Later. Plant Physiol. 2017, 174, 561–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas Syringae: What It Takes to Be a Pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Jeworutzki, E.; Roelfsema, M.R.G.; Anschütz, U.; Krol, E.; Elzenga, J.T.M.; Felix, G.; Boller, T.; Hedrich, R.; Becker, D. Early Signaling through the Arabidopsis Pattern Recognition Receptors FLS2 and EFR Involves Ca2+-Associated Opening of Plasma Membrane Anion Channels: Flagellin-Activated Ion Channels. Plant J. 2010, 62, 367–378. [Google Scholar] [CrossRef]
- Lindeberg, M.; Cunnac, S.; Collmer, A. Pseudomonas Syringae Type III Effector Repertoires: Last Words in Endless Arguments. Trends Microbiol. 2012, 20, 199–208. [Google Scholar] [CrossRef]
- Ueno, K.; Kinoshita, T.; Inoue, S.; Emi, T.; Shimazaki, K. Biochemical Characterization of Plasma Membrane H+-ATPase Activation in Guard Cell Protoplasts of Arabidopsis Thaliana in Response to Blue Light. Plant Cell Physiol. 2005, 46, 955–963. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wang, Y.; Chen, X.; Xu, F.; Ding, M.; Ye, W.; Kawai, Y.; Toda, Y.; Hayashi, Y.; Suzuki, T.; et al. Plasma membrane H+-ATPase overexpression increases rice yield via simultaneous enhancement of nutrient uptake and photosynthesis. Nat. Commun. 2021, 12, 735. [Google Scholar] [CrossRef]
- Ding, M.; Zhang, M.; Zeng, H.; Hayashi, Y.; Zhu, Y.; Kinoshita, T. Molecular basis of plasma membrane H+-ATPase function and potential application in the agricultural production. Plant Physiol. Biochem. 2021, 168, 10–16. [Google Scholar] [CrossRef]
- Zandonadi, D.B.; Santos, M.P.; Caixeta, L.S.; Marinho, E.B.; Peres, L.E.P.; Façanha, A.R. Plant Proton Pumps as Markers of Biostimulant Action. Sci. Agric. 2016, 73, 24–28. [Google Scholar] [CrossRef] [Green Version]
- Havshøi, N.W.; Fuglsang, A.T. A Critical Review on Natural Compounds Interacting with the Plant Plasma Membrane H+-ATPase and Their Potential as Biologicals in Agriculture. J. Integr. Plant Biol. 2022, 64, jipb.13221. [Google Scholar] [CrossRef]
| Process | References |
|---|---|
| Development | Haruta et al. 2010 [3], Hoffmann et al. 2019 [5], Haruta et al. 2012 [27], Gévundant et al. 2007 [28], Wang et al. 2014 [29] |
| Growth of root and shoot | Takahashi et al. 2012 [30], Li et al. 2021 [31], Hager 2003 [32], Fendrych et al. 2016 [33] |
| Physiological Processes | |
| Stomata opening | Kinoshita et al. 1999 [34], Kinoshita et al. 2002 [35], Talemiya et al. 2016 [36], Akiyama et al. 2022 [37] |
| Mineral compounds uptake | Espen 2004 [38], Ortiz-Ramirez et al. 2011 [39], Młodzińska and Zboińska 2016 [40], Maathius et al. 1993 [41], Godwin et al. 2003 [42] |
| pH homeostasis maintenance | Sze and Charnoj 2018 [43], Bassil and Blumwald 2014 [44], Wegner et al. 2021 [45], Wegner and Shabala 2020 [46] |
| Adaptation to Abiotic Stresses | |
| Salt stress | Janicka-Russak and Kabała 2015 [47], Yang et al. 2019 [48], Janicka-Russak et al. 2013 [49], Qiu et al. 2002 [50], Chen et al. 2007 [51] |
| Drought | Goh et al. 1996 [52], Zhang et al. 2004 [53], Yin et al. 2013 [54] |
| Low temperature | Janicka-Russak et al. 2012 [24], Muzi et al. 2016 [55] |
| Heavy metals | Janicka-Russak et al. 2008 [23], Fodor et al. 1995 [56], Burzyński and Kolano 2003 [57] |
| Pathogenesis | |
| Plant defense mechanism | Falhof et al. 2016 [25], Melotto et al. 2006 [58], Zhao et al. 2022 [59] |
| Pathogen attack | Liu et al. 2009 [60], Kundal et al. 2017 [61], Zhou et al. 2015 [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalak, A.; Wdowikowska, A.; Janicka, M. Plant Plasma Membrane Proton Pump: One Protein with Multiple Functions. Cells 2022, 11, 4052. https://doi.org/10.3390/cells11244052
Michalak A, Wdowikowska A, Janicka M. Plant Plasma Membrane Proton Pump: One Protein with Multiple Functions. Cells. 2022; 11(24):4052. https://doi.org/10.3390/cells11244052
Chicago/Turabian StyleMichalak, Adrianna, Anna Wdowikowska, and Małgorzata Janicka. 2022. "Plant Plasma Membrane Proton Pump: One Protein with Multiple Functions" Cells 11, no. 24: 4052. https://doi.org/10.3390/cells11244052
APA StyleMichalak, A., Wdowikowska, A., & Janicka, M. (2022). Plant Plasma Membrane Proton Pump: One Protein with Multiple Functions. Cells, 11(24), 4052. https://doi.org/10.3390/cells11244052






