PARP1 Activation Controls Stress Granule Assembly after Oxidative Stress and DNA Damage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid and siRNA Transfection
2.2. Oxidative Stress Conditions and Drug Treatments
2.3. Immunofluorescence Staining and Image Analysis
2.4. RNA Hybridization In Situ
2.5. Preparation of Extracts and Polysome Fractionation Analysis
3. Results
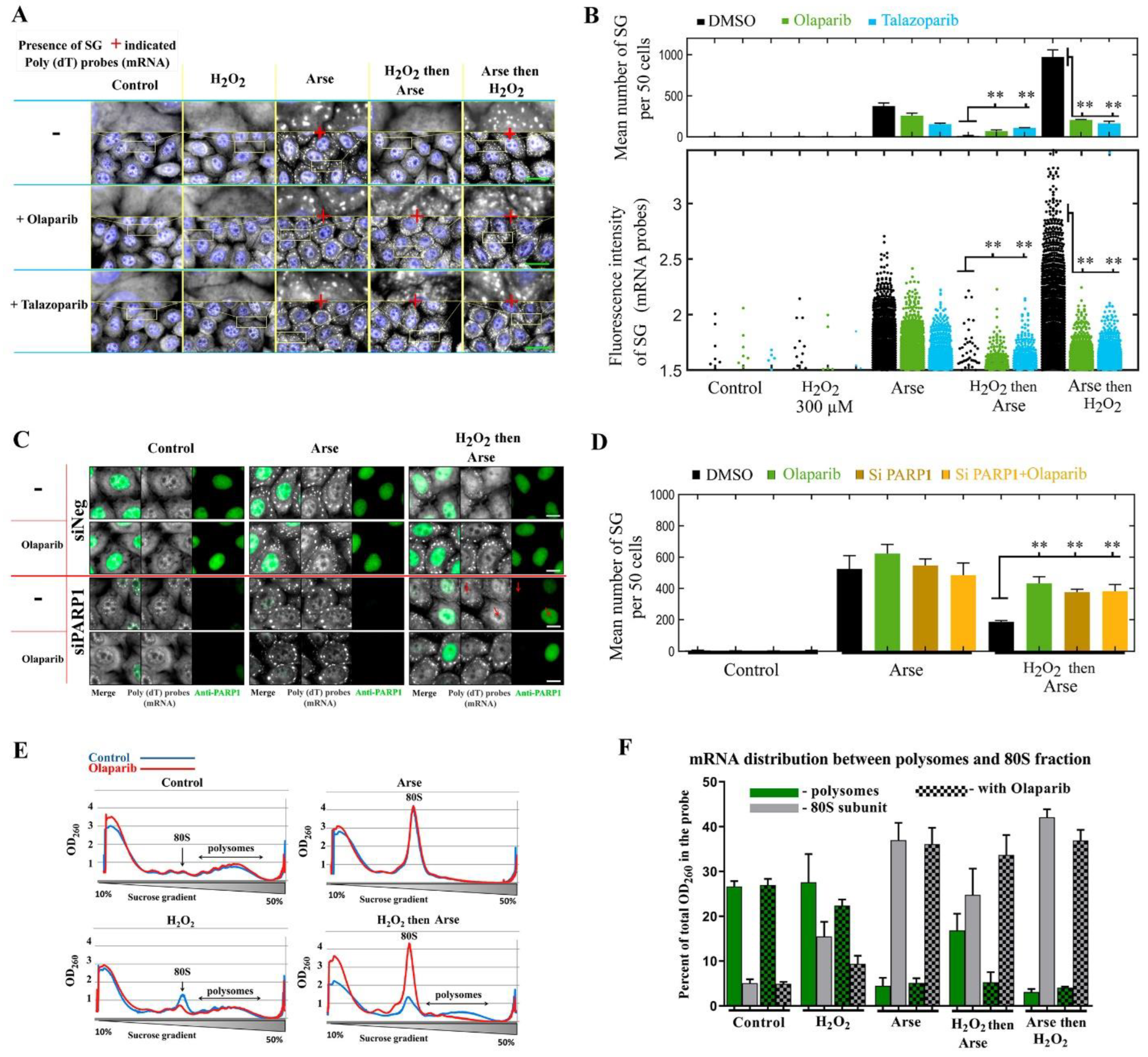
3.1. Activation of PARP1 following Oxidative DNA Damage Prevents Arsenite-Induced SG Formation and Influences Polysome Dissociation
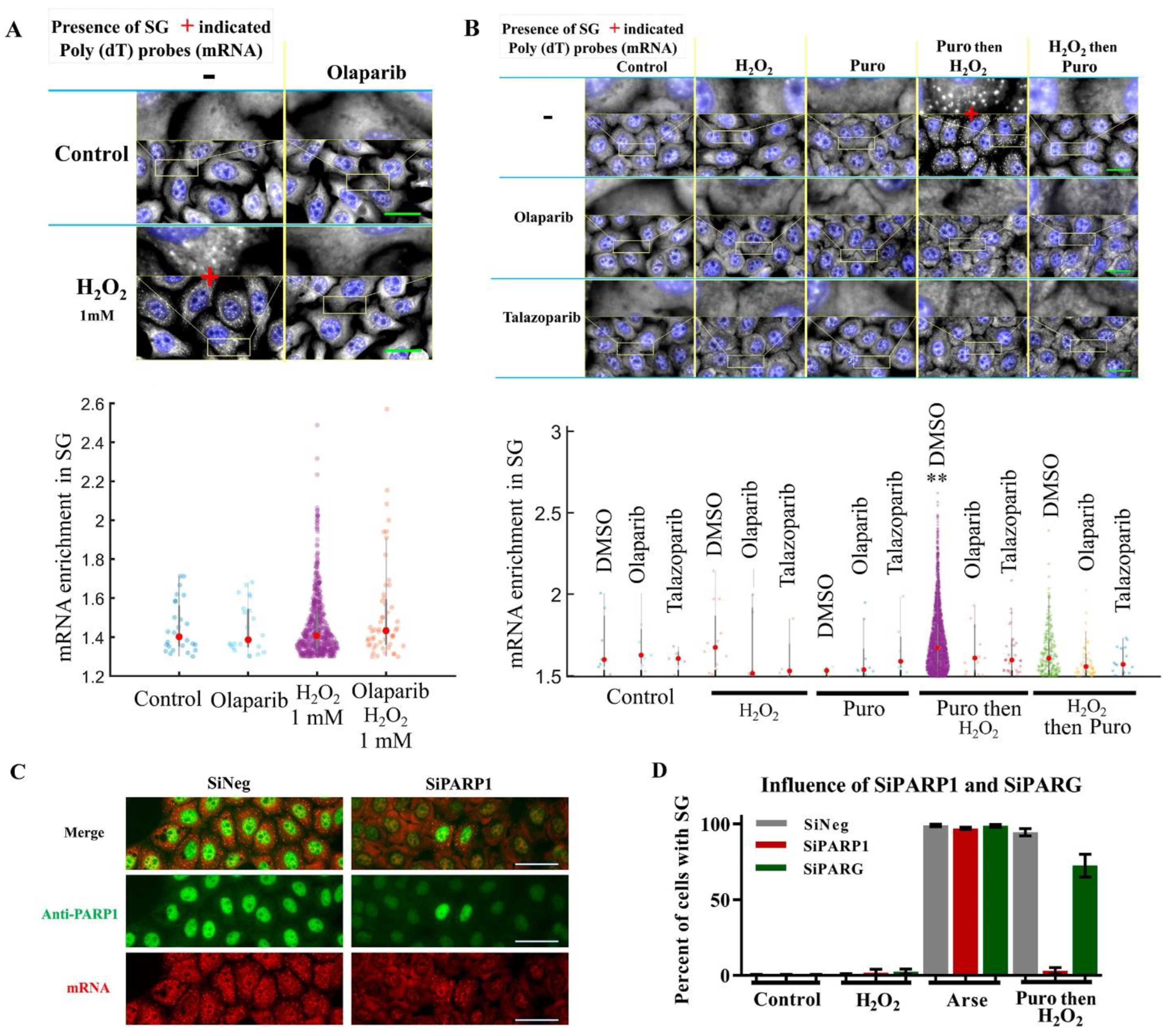
3.2. Activation of PARP1 Upregulates the Assembly of H2O2-Induced SGs
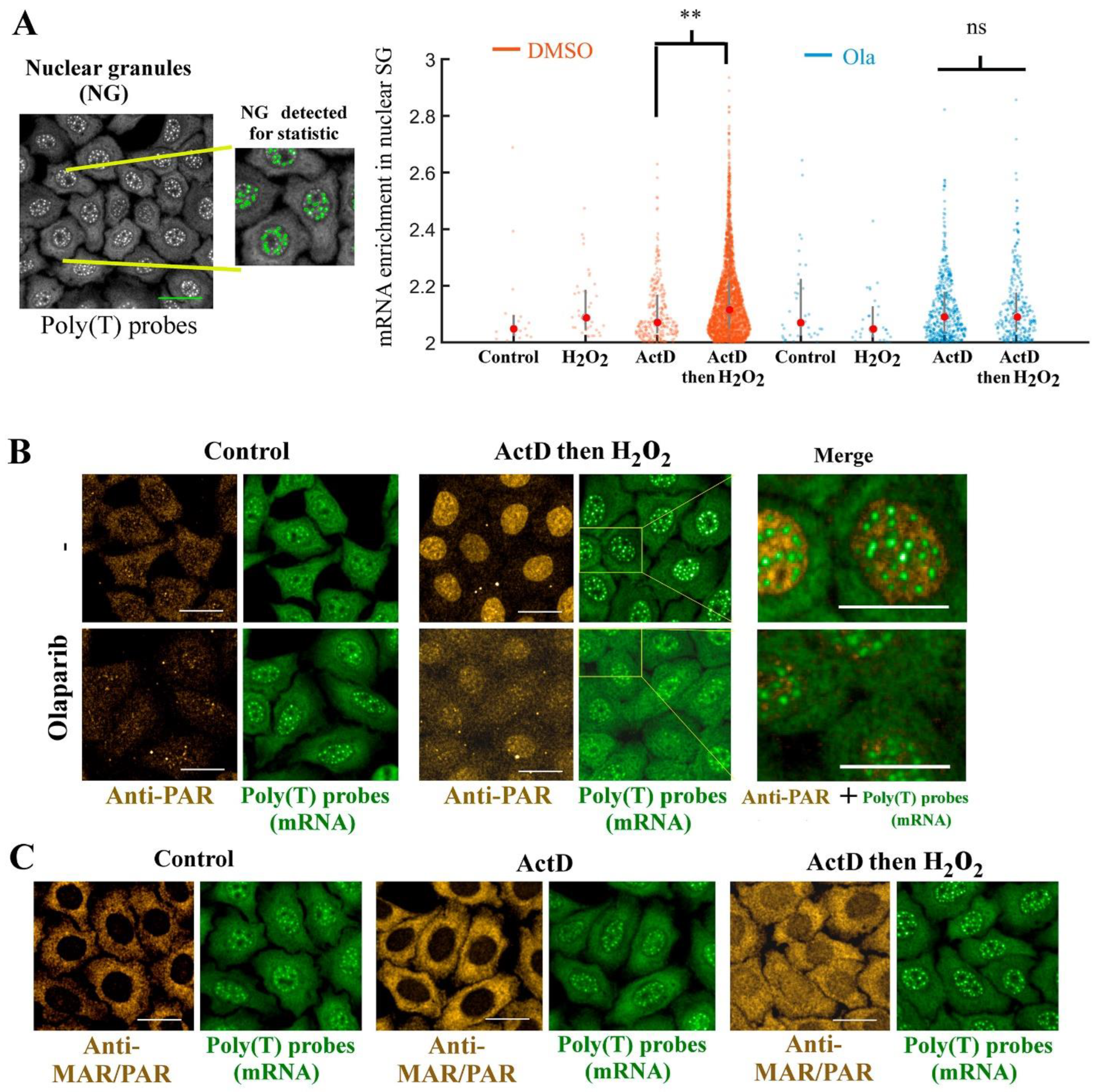
3.3. The Presence of PAR in SGs Is not Significantly Affected by PARP1 Activation Whether SGs Were Formed after Arsenite or H2O2 Treatment
3.4. No PAR Enrichment in TDP-43 and FUS-Rich Condensates in the Cytoplasm
3.5. The Translocation of Nuclear RBPs upon PARP1 Activation May Promote Stress Granule Assembly but FUS Translocation Is Not Enough
3.6. PARP1 Activation Promotes mRNA-rich Granule Assembly in the Nucleus without any Mixing with PAR
3.7. PARP1 Activation upon H2O2 Exposure Negatively Regulates SG Disassembly and Positively Regulates Preexisting TDP-43 and FUS-Rich Granules
4. Discussion
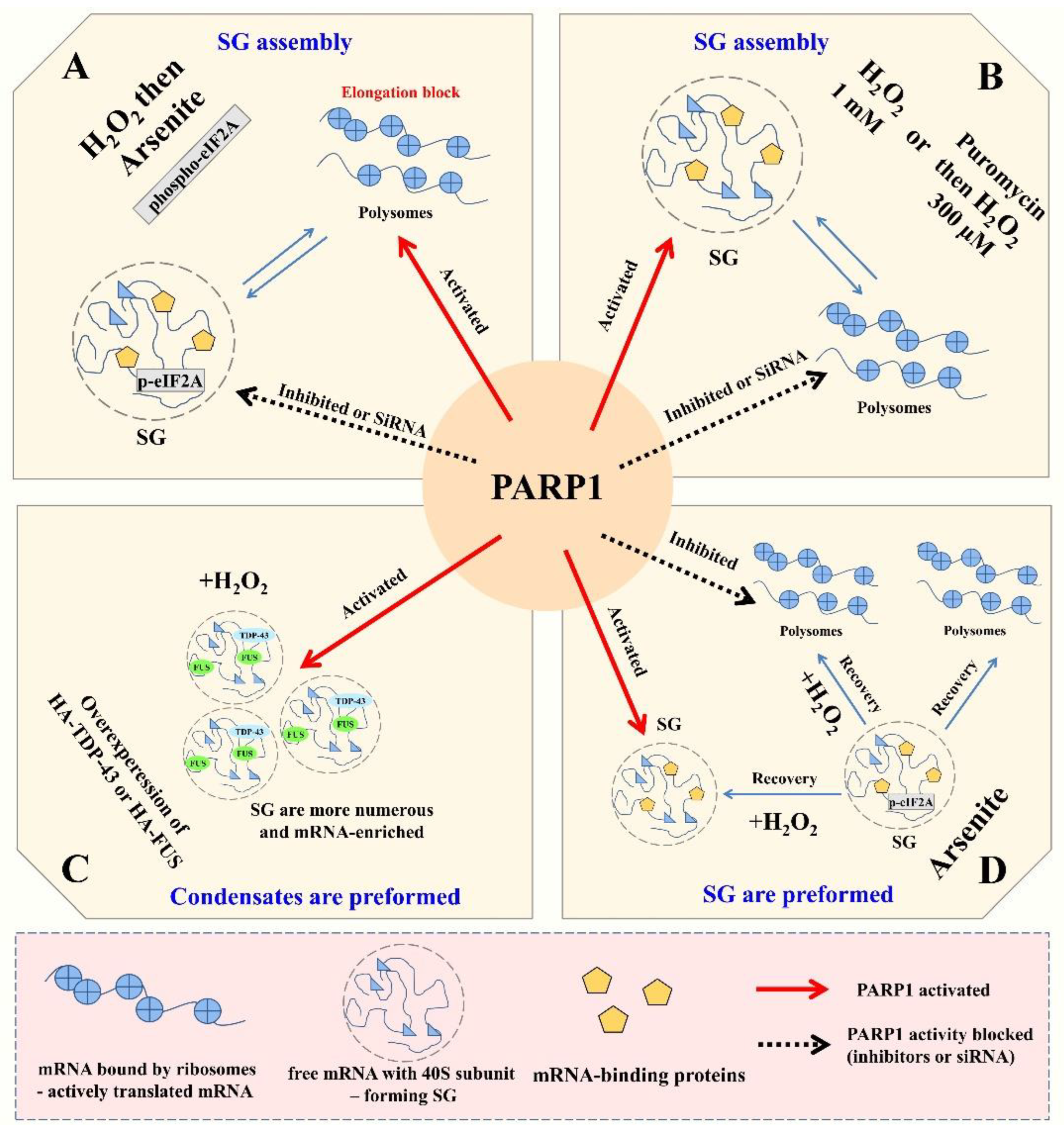
4.1. PARP1 Activation Prevents the Assembly of Arsenite-Induced SGs by Preventing Polysome Dissociation
4.2. PARP1 Activation Upregulates the Assembly of H2O2-Induced SGs
4.3. PARP1 Activation Positively Regulates Pre-Formed Nuclear and Cytoplasmic mRNA-Rich Granules but Does Not Induce PAR Accumulation in SGs
4.4. PARP1 Activation Promotes Assembly of Pre-Formed SGs and Prevents Their Dissociation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Aulas, A.; Fay, M.M.; Lyons, S.M.; Achorn, C.A.; Kedersha, N.; Anderson, P.; Ivanov, P. Stress-Specific Differences in Assembly and Composition of Stress Granules and Related Foci. J. Cell Sci. 2017, 130, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef] [PubMed]
- Bounedjah, O.; Desforges, B.; Wu, T.-D.; Pioche-Durieu, C.; Marco, S.; Hamon, L.; Curmi, P.A.; Guerquin-Kern, J.-L.; Piétrement, O.; Pastré, D. Free MRNA in Excess upon Polysome Dissociation Is a Scaffold for Protein Multimerization to Form Stress Granules. Nucleic Acids Res. 2014, 42, 8678–8691. [Google Scholar] [CrossRef]
- van Treeck, B.; Protter, D.S.W.; Matheny, T.; Khong, A.; Link, C.D.; Parker, R. RNA Self-Assembly Contributes to Stress Granule Formation and Defining the Stress Granule Transcriptome. Proc. Natl. Acad. Sci. USA 2018, 115, 2734–2739. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.R.E.A.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e17. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Kedersha, N.; Lee, D.S.W.; Strom, A.R.; Drake, V.; Riback, J.A.; Bracha, D.; Eeftens, J.M.; Iwanicki, A.; Wang, A.; et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020, 181, 306–324.e28. [Google Scholar] [CrossRef]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch That Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e28. [Google Scholar] [CrossRef]
- Duan, Y.; Du, A.; Gu, J.; Duan, G.; Wang, C.; Gui, X.; Ma, Z.; Qian, B.; Deng, X.; Zhang, K.; et al. PARylation Regulates Stress Granule Dynamics, Phase Separation, and Neurotoxicity of Disease-Related RNA-Binding Proteins. Cell Res. 2019, 29, 233–247. [Google Scholar] [CrossRef]
- Jin, X.; Cao, X.; Liu, S.; Liu, B. Functional Roles of Poly(ADP-Ribose) in Stress Granule Formation and Dynamics. Front. Cell Dev. Biol. 2021, 9, 671780. [Google Scholar] [CrossRef]
- Leung, A.K.L.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-Ribose) Regulates Stress Responses and MicroRNA Activity in the Cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar] [CrossRef]
- McGurk, L.; Gomes, E.; Guo, L.; Mojsilovic-Petrovic, J.; Tran, V.; Kalb, R.G.; Shorter, J.; Bonini, N.M. Poly(ADP-Ribose) Prevents Pathological Phase Separation of TDP-43 by Promoting Liquid Demixing and Stress Granule Localization. Mol. Cell 2018, 71, 703–717.e9. [Google Scholar] [CrossRef]
- Lüscher, B.; Ahel, I.; Altmeyer, M.; Ashworth, A.; Bai, P.; Chang, P.; Cohen, M.; Corda, D.; Dantzer, F.; Daugherty, M.D.; et al. ADP-ribosyltransferases, an Update on Function and Nomenclature. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.L. Poly(ADP-Ribose): A Dynamic Trigger for Biomolecular Condensate Formation. Trends Cell Biol. 2020, 30, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Altmeyer, M.; Neelsen, K.J.; Teloni, F.; Pozdnyakova, I.; Pellegrino, S.; Grøfte, M.; Rask, M.-B.D.; Streicher, W.; Jungmichel, S.; Nielsen, M.L.; et al. Liquid Demixing of Intrinsically Disordered Proteins Is Seeded by Poly(ADP-Ribose). Nat. Commun. 2015, 6, 8088. [Google Scholar] [CrossRef] [PubMed]
- Alemasova, E.E.; Pestryakov, P.E.; Sukhanova, M.v.; Kretov, D.A.; Moor, N.A.; Curmi, P.A.; Ovchinnikov, L.P.; Lavrik, O.I. Poly(ADP-Ribosyl)Ation as a New Posttranslational Modification of YB-1. Biochimie 2015, 119, 36–44. [Google Scholar] [CrossRef]
- Isabelle, M.; Gagné, J.-P.; Gallouzi, I.-E.; Poirier, G.G. Quantitative Proteomics and Dynamic Imaging Reveal That G3BP-Mediated Stress Granule Assembly Is Poly(ADP-Ribose)-Dependent Following Exposure to MNNG-Induced DNA Alkylation. J. Cell Sci. 2012, 125, 4555–4566. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, J.; Beeraka, N.M.; Tang, C.; Babayeva, Y.V.; Sinelnikov, M.Y.; Zhang, X.; Zhang, J.; Liu, J.; Reshetov, I.V.; et al. Advances in the Prevention and Treatment of Obesity-Driven Effects in Breast Cancers. Front. Oncol. 2022, 12, 2663. [Google Scholar] [CrossRef]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.R.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Mitochondrial Mutations and Mitoepigenetics: Focus on Regulation of Oxidative Stress-Induced Responses in Breast Cancers. Semin. Cancer Biol. 2022, 83, 556–569. [Google Scholar] [CrossRef]
- Walker, C.; El-Khamisy, S.F. Perturbed Autophagy and DNA Repair Converge to Promote Neurodegeneration in Amyotrophic Lateral Sclerosis and Dementia. Brain 2018, 141, 1247–1262. [Google Scholar] [CrossRef]
- Bai, P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol. Cell 2015, 58, 947–958. [Google Scholar] [CrossRef]
- Catara, G.; Grimaldi, G.; Schembri, L.; Spano, D.; Turacchio, G.; lo Monte, M.; Beccari, A.R.; Valente, C.; Corda, D. PARP1-Produced Poly-ADP-Ribose Causes the PARP12 Translocation to Stress Granules and Impairment of Golgi Complex Functions. Sci. Rep. 2017, 7, 14035. [Google Scholar] [CrossRef] [PubMed]
- Ayyappan, V.; Wat, R.; Barber, C.; Vivelo, C.A.; Gauch, K.; Visanpattanasin, P.; Cook, G.; Sazeides, C.; Leung, A.K.L. ADPriboDB 2.0: An Updated Database of ADP-Ribosylated Proteins. Nucleic Acids Res. 2021, 49, D261–D265. [Google Scholar] [CrossRef] [PubMed]
- Bonfiglio, J.J.; Fontana, P.; Zhang, Q.; Colby, T.; Gibbs-Seymour, I.; Atanassov, I.; Bartlett, E.; Zaja, R.; Ahel, I.; Matic, I. Serine ADP-Ribosylation Depends on HPF1. Mol. Cell 2017, 65, 932–940.e6. [Google Scholar] [CrossRef] [PubMed]
- Martello, R.; Leutert, M.; Jungmichel, S.; Bilan, V.; Larsen, S.C.; Young, C.; Hottiger, M.O.; Nielsen, M.L. Proteome-Wide Identification of the Endogenous ADP-Ribosylome of Mammalian Cells and Tissue. Nat. Commun. 2016, 7, 12917. [Google Scholar] [CrossRef] [PubMed]
- McGurk, L.; Rifai, O.M.; Bonini, N.M. Poly(ADP-Ribosylation) in Age-Related Neurological Disease. Trends Genet. 2019, 35, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Thapa, K.; Khan, H.; Sharma, U.; Grewal, A.K.; Singh, T.G. Poly (ADP-Ribose) Polymerase-1 as a Promising Drug Target for Neurodegenerative Diseases. Life Sci. 2021, 267, 118975. [Google Scholar] [CrossRef]
- Liu, C.; Fang, Y. New Insights of Poly(ADP-Ribosylation) in Neurodegenerative Diseases: A Focus on Protein Phase Separation and Pathologic Aggregation. Biochem. Pharm. 2019, 167, 58–63. [Google Scholar] [CrossRef]
- Park, H.; Kam, T.-I.; Dawson, T.M.; Dawson, V.L. Poly (ADP-Ribose) (PAR)-Dependent Cell Death in Neurodegenerative Diseases. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2020; Volume 353, pp. 1–29. ISBN 9780128201350. [Google Scholar]
- Singatulina, A.S.; Hamon, L.; Sukhanova, M.V.; Desforges, B.; Joshi, V.; Bouhss, A.; Lavrik, O.I.; Pastré, D. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 2019, 27, 1809–1821.e5. [Google Scholar] [CrossRef]
- da Cruz, S.; Cleveland, D.W. Understanding the Role of TDP-43 and FUS/TLS in ALS and Beyond. Curr. Opin. Neurobiol. 2011, 21, 904–919. [Google Scholar] [CrossRef]
- Dobra, I.; Pankivskyi, S.; Samsonova, A.; Pastre, D.; Hamon, L. Relation Between Stress Granules and Cytoplasmic Protein Aggregates Linked to Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2018, 18, 1–9. [Google Scholar] [CrossRef]
- Panda, A.C.; Martindale, J.L.; Gorospe, M. Polysome Fractionation to Analyze MRNA Distribution Profiles. Bio. Protoc. 2017, 7, e2126. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Chen, S.; Gilks, N.; Li, W.; Miller, I.J.; Stahl, J.; Anderson, P. Evidence That Ternary Complex (EIF2-GTP-TRNAiMet)-Deficient Preinitiation Complexes Are Core Constituents of Mammalian Stress Granules. Mol. Biol. Cell 2002, 13, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Erdélyi, K.; Bai, P.; Kovács, I.; Szabó, É.; Mocsár, G.; Kakuk, A.; Szabó, C.; Gergely, P.; Virág, L. Dual Role of Poly(ADP-ribose) Glycohydrolase in the Regulation of Cell Death in Oxidatively Stressed A549 Cells. FASEB J. 2009, 23, 3553–3563. [Google Scholar] [CrossRef]
- Emara, M.M.; Fujimura, K.; Sciaranghella, D.; Ivanova, V.; Ivanov, P.; Anderson, P. Hydrogen Peroxide Induces Stress Granule Formation Independent of EIF2α Phosphorylation. Biochem. Biophys. Res. Commun. 2012, 423, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Pothof, J.; Verkaik, N.S.; Hoeijmakers, J.H.; van Gent, D.C. Cell Cycle MicroRNA Responses and Stress Granule Formation Modulate the DNA Damage Response. Taylor Fr. 2009, 8, 3462–3468. [Google Scholar] [CrossRef]
- Wolf, A.; Krause-Gruszczynska, M.; Birkenmeier, O.; Ostareck-Lederer, A.; Hüttelmaier, S.; Hatzfeld, M. Plakophilin 1 Stimulates Translation by Promoting EIF4A1 Activity. J. Cell Biol. 2010, 188, 463–471. [Google Scholar] [CrossRef]
- Brown, J.A.L.; Roberts, T.L.; Richards, R.; Woods, R.; Birrell, G.; Lim, Y.C.; Ohno, S.; Yamashita, A.; Abraham, R.T.; Gueven, N.; et al. A Novel Role for HSMG-1 in Stress Granule Formation. Mol. Cell Biol. 2011, 31, 4417–4429. [Google Scholar] [CrossRef]
- Arimoto-Matsuzaki, K.; Saito, H.; Takekawa, M. TIA1 Oxidation Inhibits Stress Granule Assembly and Sensitizes Cells to Stress-Induced Apoptosis. Nat. Commun. 2016, 7, 10252. [Google Scholar] [CrossRef]
- Thedieck, K.; Holzwarth, B.; Prentzell, M.T.; Boehlke, C.; Kläsener, K.; Ruf, S.; Sonntag, A.G.; Maerz, L.; Grellscheid, S.N.; Kremmer, E.; et al. XInhibition of MTORC1 by Astrin and Stress Granules Prevents Apoptosis in Cancer Cells. Cell 2013, 154, 859–874. [Google Scholar] [CrossRef]
- Arimoto-Matsuzaki, K. Formation of Stress Granules Inhibits Apoptosis by Suppressing Stress-Responsive MAPK Pathways. Artic. Nat. Cell Biol. 2008, 10, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Higuchi, M.; Matsuki, H.; Yoshita, M.; Ohsawa, T.; Oie, M.; Fujii, M. Stress Granules Inhibit Apoptosis by Reducing Reactive Oxygen Species Production. Mol. Cell Biol. 2013, 33, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Thorsell, A.-G.; Ekblad, T.; Karlberg, T.; Löw, M.; Pinto, A.F.; Trésaugues, L.; Moche, M.; Cohen, M.S.; Schüler, H. Structural Basis for Potency and Promiscuity in Poly(ADP-Ribose) Polymerase (PARP) and Tankyrase Inhibitors. J. Med. Chem. 2017, 60, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Lin, Y.; Yang, C.-C.; McQuary, P.; Rosa Campos, A.; Aza Blanc, P.; Wolf, D.A. Cross Talk between EIF2α and EEF2 Phosphorylation Pathways Optimizes Translational Arrest in Response to Oxidative Stress. iScience 2019, 20, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic Shuttling of TIA-1 Accompanies the Recruitment of MRNA to Mammalian Stress Granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef]
- Beneke, S.; Meyer, K.; Holtz, A.; Hüttner, K.; Bürkle, A. Chromatin Composition Is Changed by Poly(ADP-Ribosyl)Ation during Chromatin Immunoprecipitation. PLoS ONE 2012, 7, e32914. [Google Scholar] [CrossRef][Green Version]
- Slade, D.; Dunstan, M.S.; Barkauskaite, E.; Weston, R.; Lafite, P.; Dixon, N.; Ahel, M.; Leys, D.; Ahel, I. The Structure and Catalytic Mechanism of a Poly(ADP-Ribose) Glycohydrolase. Nature 2011, 477, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Naumann, M.; Pal, A.; Goswami, A.; Lojewski, X.; Japtok, J.; Vehlow, A.; Naujock, M.; Günther, R.; Jin, M.; Stanslowsky, N.; et al. Impaired DNA Damage Response Signaling by FUS-NLS Mutations Leads to Neurodegeneration and FUS Aggregate Formation. Nat. Commun. 2018, 9, 335. [Google Scholar] [CrossRef]
- Bosco, D.A.; Lemay, N.; Ko, H.K.; Zhou, H.; Burke, C.; Kwiatkowski, T.J.; Sapp, P.; McKenna-Yasek, D.; Brown, R.H.; Hayward, L.J. Mutant FUS Proteins That Cause Amyotrophic Lateral Sclerosis Incorporate into Stress Granules. Hum. Mol. Genet. 2010, 19, 4160–4175. [Google Scholar] [CrossRef] [PubMed]
- Colombrita, C.; Zennaro, E.; Fallini, C.; Weber, M.; Sommacal, A.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 Is Recruited to Stress Granules in Conditions of Oxidative Insult. J. Neurochem. 2009, 111, 1051–1061. [Google Scholar] [CrossRef]
- Fang, M.Y.; Markmiller, S.; Vu, A.Q.; Javaherian, A.; Dowdle, W.E.; Jolivet, P.; Bushway, P.J.; Castello, N.A.; Baral, A.; Chan, M.Y.; et al. Small-Molecule Modulation of TDP-43 Recruitment to Stress Granules Prevents Persistent TDP-43 Accumulation in ALS/FTD. Neuron 2019, 103, 802–819.e11. [Google Scholar] [CrossRef] [PubMed]
- Gal, J.; Zhang, J.; Kwinter, D.M.; Zhai, J.; Jia, H.; Jia, J.; Zhu, H. Nuclear Localization Sequence of FUS and Induction of Stress Granules by ALS Mutants. Neurobiol. Aging 2011, 32, 2323.e27–2323.e40. [Google Scholar] [CrossRef]
- Rengifo-Gonzalez, J.C.; el Hage, K.; Clément, M.-J.; Steiner, E.; Joshi, V.; Craveur, P.; Durand, D.; Pastré, D.; Bouhss, A. The Cooperative Binding of TDP-43 to GU-Rich RNA Repeats Antagonizes TDP-43 Aggregation. Elife 2021, 10, e67605. [Google Scholar] [CrossRef]
- Wheeler, E.C.; Vu, A.Q.; Einstein, J.M.; DiSalvo, M.; Ahmed, N.; van Nostrand, E.L.; Shishkin, A.A.; Jin, W.; Allbritton, N.L.; Yeo, G.W. Pooled CRISPR Screens with Imaging on Microraft Arrays Reveals Stress Granule-Regulatory Factors. Nat. Methods 2020, 17, 636–642. [Google Scholar] [CrossRef]
- Youn, J.-Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of MRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef] [PubMed]
- Bley, N.; Lederer, M.; Pfalz, B.; Reinke, C.; Fuchs, T.; Glaß, M.; Möller, B.; Hüttelmaier, S. Stress Granules Are Dispensable for MRNA Stabilization during Cellular Stress. Nucleic Acids Res. 2015, 43, e26. [Google Scholar] [CrossRef] [PubMed]
- Páhi, Z.G.; Borsos, B.N.; Pantazi, V.; Ujfaludi, Z.; Pankotai, T. PARylation During Transcription: Insights into the Fine-Tuning Mechanism and Regulation. Cancers 2020, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Child, J.R.; Chen, Q.; Reid, D.W.; Jagannathan, S.; Nicchitta, C.V. Recruitment of Endoplasmic Reticulum-Targeted and Cytosolic MRNAs into Membrane-Associated Stress Granules. RNA 2021, 27, 1241–1256. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct Stages in Stress Granule Assembly and Disassembly. Elife 2016, 5, e18413. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.; Tedim Ferreira, M.; Gagné, J.-P.; Sharma, A.K.; Hendzel, M.J.; Masson, J.-Y.; Poirier, G.G. Emerging Roles of Eraser Enzymes in the Dynamic Control of Protein ADP-Ribosylation. Nat. Commun. 2019, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Lagueux, J.; Shah, G.M.; Menard, L.; Thomassin, H.; Duchaine, C.; Hengartner, C.; Poirier, G.G. Poly(ADP-Ribose) Catabolism in Mammalian Cells. Mol. Cell Biochem. 1994, 138, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jin, X.; Liu, B. The Involvement of Stress Granules in Aging and Aging-Associated Diseases. Aging Cell 2020, 19, e13136. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, B. Relationships between Stress Granules, Oxidative Stress, and Neurodegenerative Diseases. Oxid. Med. Cell Longev. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; King, O.D.; Shorter, J.; Gitler, A.D. Stress Granules as Crucibles of ALS Pathogenesis. J. Cell Biol. 2013, 201, 361–372. [Google Scholar] [CrossRef]
- Wolozin, B.; Ivanov, P. Stress Granules and Neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666. [Google Scholar] [CrossRef]
- Rhine, K.; Dasovich, M.; Yoniles, J.; Badiee, M.; Skanchy, S.; Ganser, L.R.; Ge, Y.; Fare, C.M.; Shorter, J.; Leung, A.K.L.; et al. Poly(ADP-Ribose) Drives Condensation of FUS via a Transient Interaction. Mol. Cell 2022, 82, 969–985.e11. [Google Scholar] [CrossRef]
- Ederle, H.; Dormann, D. TDP-43 and FUS En Route from the Nucleus to the Cytoplasm. FEBS Lett. 2017, 591, 1489–1507. [Google Scholar] [CrossRef]
- Gasset-Rosa, F.; Lu, S.; Yu, H.; Chen, C.; Melamed, Z.; Guo, L.; Shorter, J.; da Cruz, S.; Cleveland, D.W. Cytoplasmic TDP-43 De-Mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron 2019, 102, 339–357.e7. [Google Scholar] [CrossRef]
- Jungmichel, S.; Rosenthal, F.; Altmeyer, M.; Lukas, J.; Hottiger, M.O.; Nielsen, M.L. Proteome-Wide Identification of Poly(ADP-Ribosyl)Ation Targets in Different Genotoxic Stress Responses. Mol. Cell 2013, 52, 272–285. [Google Scholar] [CrossRef]
- Éthier, C.; Tardif, M.; Arul, L.; Poirier, G.G. PARP-1 Modulation of MTOR Signaling in Response to a DNA Alkylating Agent. PLoS ONE 2012, 7, e47978. [Google Scholar] [CrossRef]
- Cohen-Armon, M. PARP-1 Activation in the ERK Signaling Pathway. Trends Pharmacol. Sci. 2007, 28, 556–560. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singatulina, A.S.; Sukhanova, M.V.; Desforges, B.; Joshi, V.; Pastré, D.; Lavrik, O.I. PARP1 Activation Controls Stress Granule Assembly after Oxidative Stress and DNA Damage. Cells 2022, 11, 3932. https://doi.org/10.3390/cells11233932
Singatulina AS, Sukhanova MV, Desforges B, Joshi V, Pastré D, Lavrik OI. PARP1 Activation Controls Stress Granule Assembly after Oxidative Stress and DNA Damage. Cells. 2022; 11(23):3932. https://doi.org/10.3390/cells11233932
Chicago/Turabian StyleSingatulina, Anastasia S., Maria V. Sukhanova, Bénédicte Desforges, Vandana Joshi, David Pastré, and Olga I. Lavrik. 2022. "PARP1 Activation Controls Stress Granule Assembly after Oxidative Stress and DNA Damage" Cells 11, no. 23: 3932. https://doi.org/10.3390/cells11233932
APA StyleSingatulina, A. S., Sukhanova, M. V., Desforges, B., Joshi, V., Pastré, D., & Lavrik, O. I. (2022). PARP1 Activation Controls Stress Granule Assembly after Oxidative Stress and DNA Damage. Cells, 11(23), 3932. https://doi.org/10.3390/cells11233932






