CHRDL1 Regulates Stemness in Glioma Stem-like Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Cell Culture
2.2. Lentiviral Transduction of GSCs
2.3. Cell-Based Assays
2.3.1. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Assay
2.3.2. Limiting Dilution Assay
2.3.3. Sphere Formation Assays
2.3.4. Cell Cycle Analysis
2.4. SDS-PAGE and Western Blot
2.5. Irradiation Procedures
2.6. Immunofluorescence Microscopy/Foci Assay
2.7. Taqman-Based qRT-PCR
2.8. Interrogation of the Human Protein Atlas
2.9. Statistics
3. Results
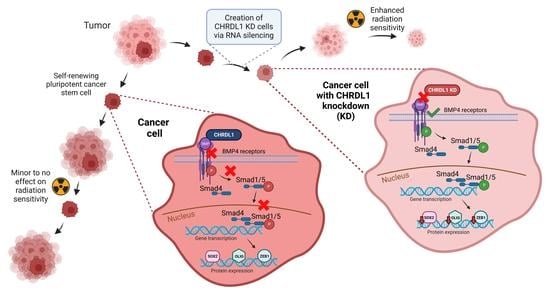
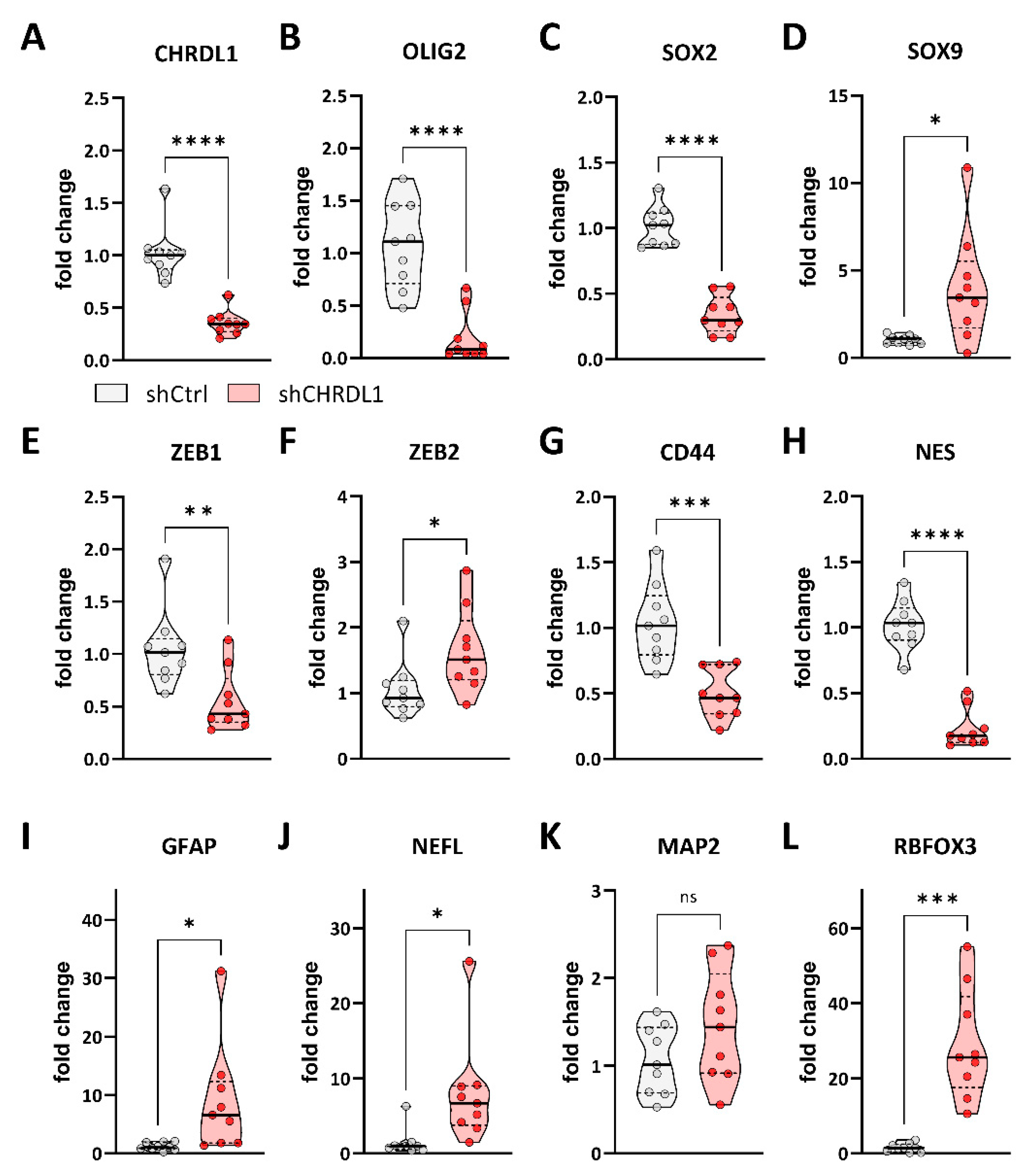
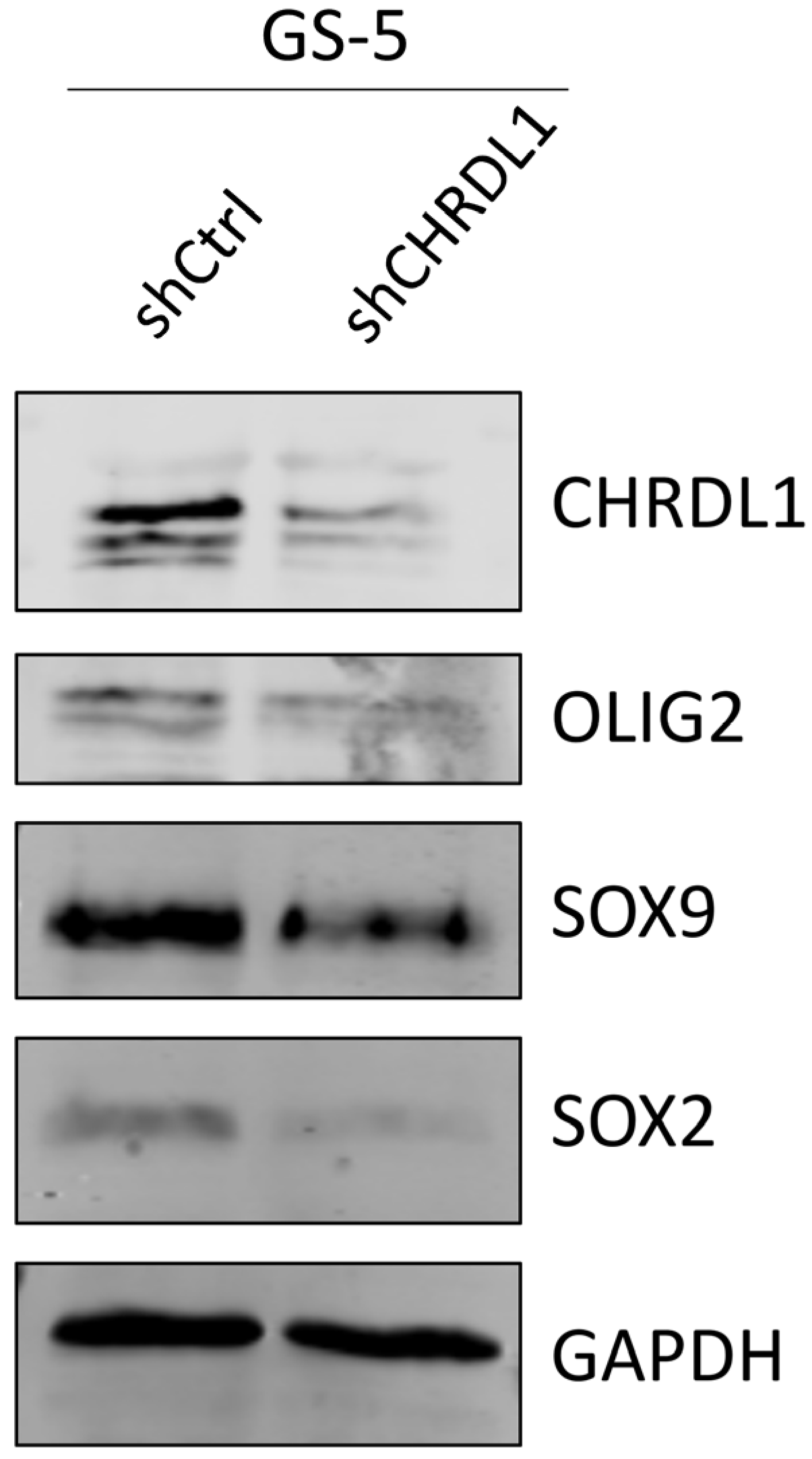
3.1. CHRDL1 Depletion Reduces Stemness of GSCs
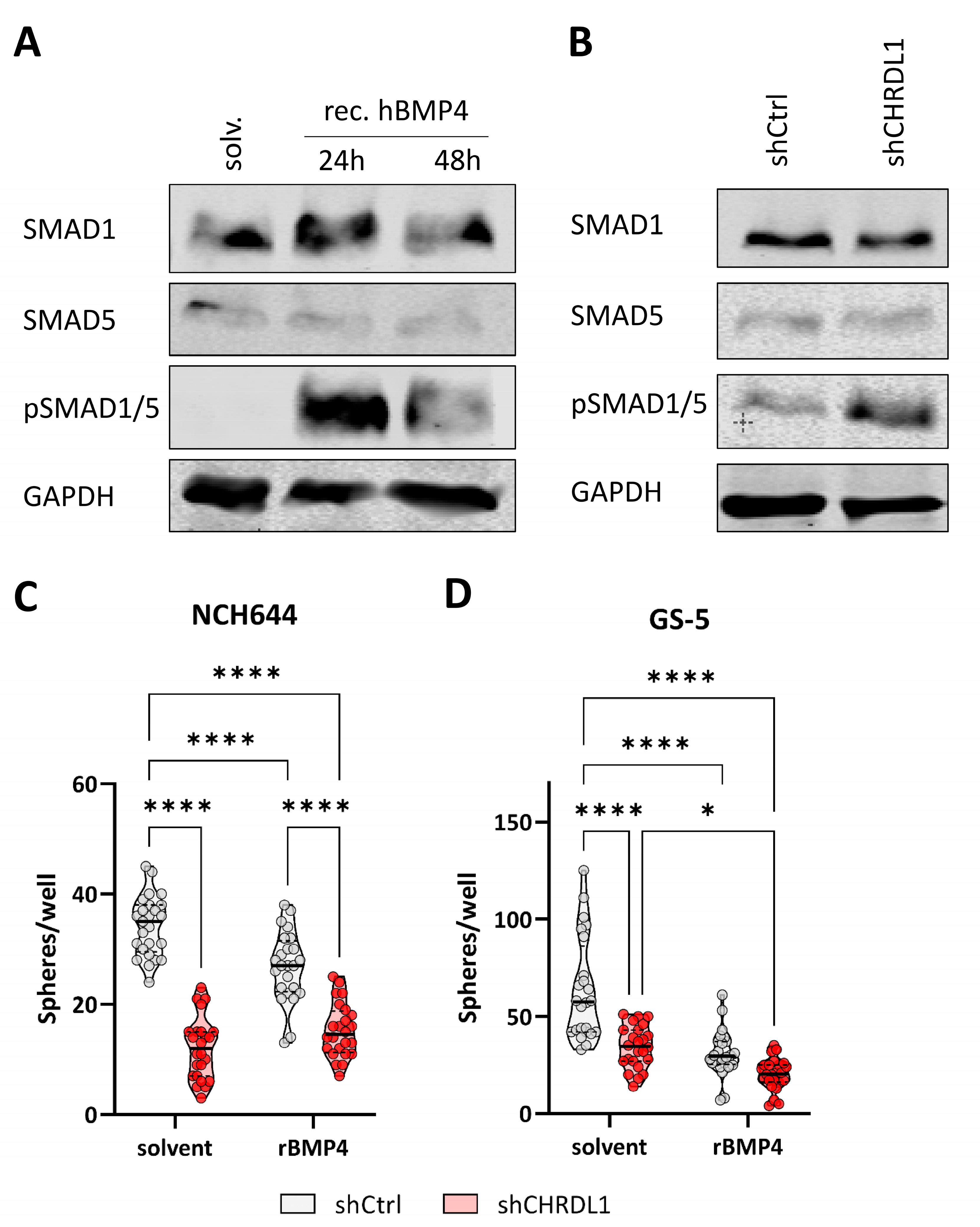
3.2. CHRDL1 Depletion Re-Activates BMP4-Signaling in GSCs
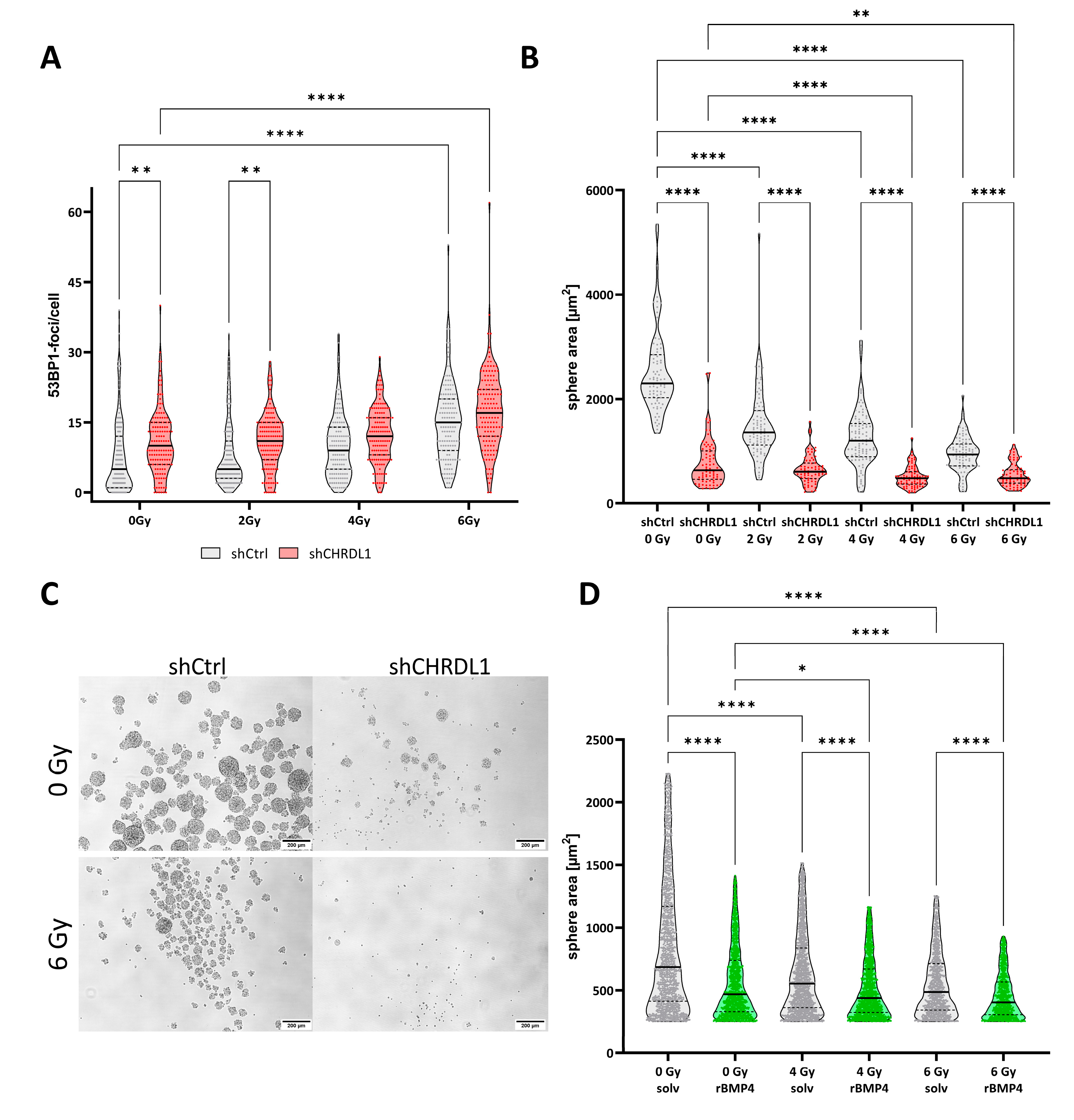
3.3. CHRDL1 Depletion Sensitizes GSCs towards Radiation Treatment
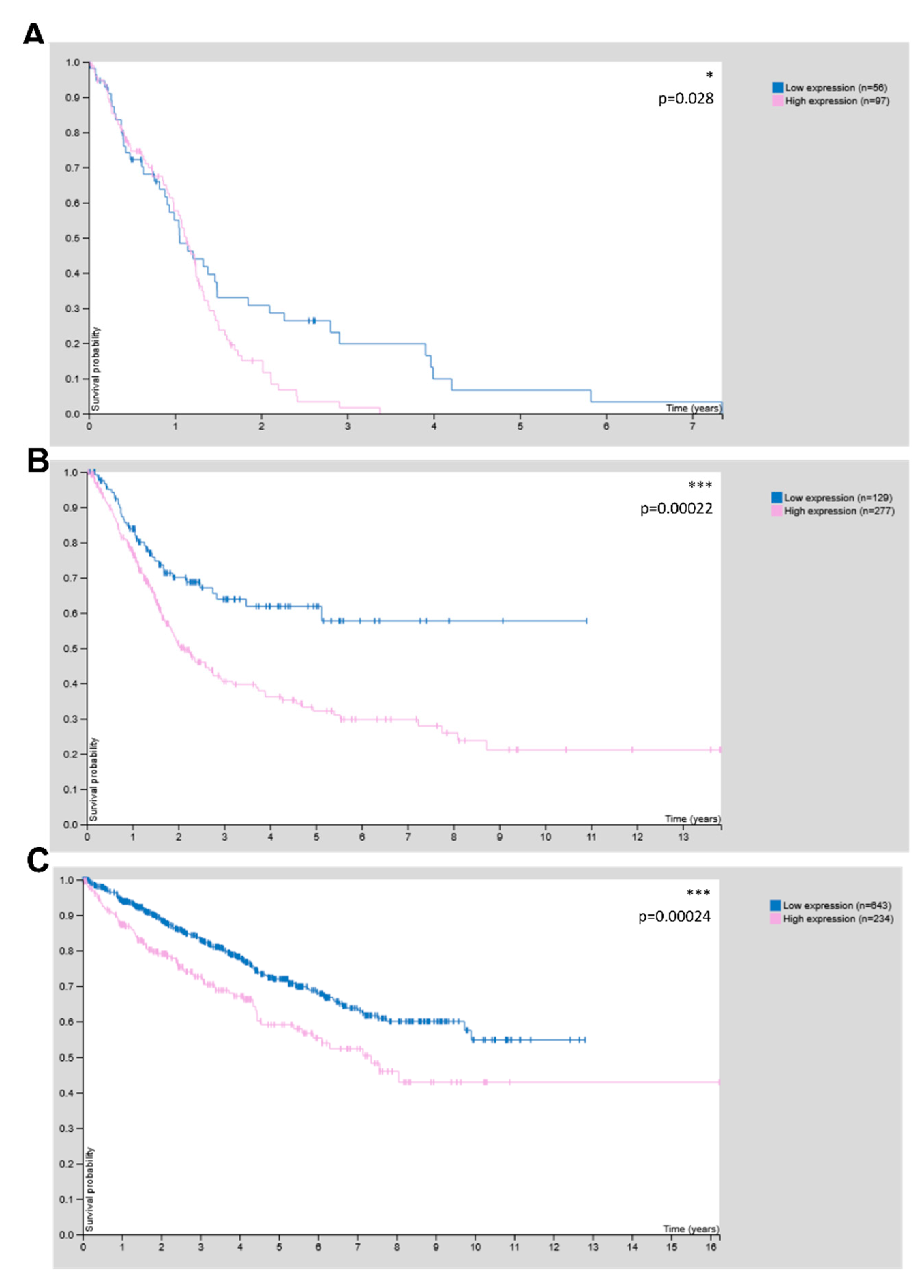
3.4. CHRDL1 Is Associated with Poor Prognosis in Glioma and Other Cancers
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Stupp, R.; Hegi, M.E.; Gilbert, M.R.; Chakravarti, A. Chemoradiotherapy in malignant glioma: Standard of care and future directions. J. Clin. Oncol. 2007, 25, 4127–4136. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Luwor, R.B.; Baradaran, B.; Taylor, L.E.; Iaria, J.; Nheu, T.V.; Amiry, N.; Hovens, C.M.; Wang, B.; Kaye, A.H.; Zhu, H.J. Targeting Stat3 and Smad7 to restore TGF-beta cytostatic regulation of tumor cells in vitro and in vivo. Oncogene 2013, 32, 2433–2441. [Google Scholar] [CrossRef]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef]
- Tirosh, I.; Venteicher, A.S.; Hebert, C.; Escalante, L.E.; Patel, A.P.; Yizhak, K.; Fisher, J.M.; Rodman, C.; Mount, C.; Filbin, M.G.; et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016, 539, 309–313. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef]
- Dirkse, A.; Golebiewska, A.; Buder, T.; Nazarov, P.V.; Muller, A.; Poovathingal, S.; Brons, N.H.C.; Leite, S.; Sauvageot, N.; Sarkisjan, D.; et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 2019, 10, 1787. [Google Scholar] [CrossRef]
- Krakstad, C.; Chekenya, M. Survival signalling and apoptosis resistance in glioblastomas: Opportunities for targeted therapeutics. Mol. Cancer 2010, 9, 135. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front. Surg. 2016, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Schiffer, D.; Mellai, M.; Annovazzi, L.; Caldera, V.; Piazzi, A.; Denysenko, T.; Melcarne, A. Stem cell niches in glioblastoma: A neuropathological view. Biomed. Res. Int. 2014, 2014, 725921. [Google Scholar] [CrossRef] [PubMed]
- Signore, M.; Ricci-Vitiani, L.; De Maria, R. Targeting apoptosis pathways in cancer stem cells. Cancer Lett. 2013, 332, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, X.; Chen, L.-C.; Du, W.-Z.; Cui, Y.-Q.; Piao, X.-Y.; Li, Y.-L.; Jiang, C.-L. Targeting glioma stem cells via the Hedgehog signaling pathway. Neuroimmunol. Neuroinflamm. 2014, 1, 51–59. [Google Scholar] [CrossRef]
- Gilbert, C.A.; Ross, A.H. Cancer stem cells: Cell culture, markers, and targets for new therapies. J. Cell Biochem. 2009, 108, 1031–1038. [Google Scholar] [CrossRef]
- Linder, B.; Wehle, A.; Hehlgans, S.; Bonn, F.; Dikic, I.; Rodel, F.; Seifert, V.; Kogel, D. Arsenic Trioxide and (-)-Gossypol Synergistically Target Glioma Stem-Like Cells via Inhibition of Hedgehog and Notch Signaling. Cancers 2019, 11, 350. [Google Scholar] [CrossRef]
- Kane, R.; Godson, C.; O’Brien, C. Chordin-like 1, a bone morphogenetic protein-4 antagonist, is upregulated by hypoxia in human retinal pericytes and plays a role in regulating angiogenesis. Mol. Vis. 2008, 14, 1138–1148. [Google Scholar]
- Piccirillo, S.G.; Reynolds, B.A.; Zanetti, N.; Lamorte, G.; Binda, E.; Broggi, G.; Brem, H.; Olivi, A.; Dimeco, F.; Vescovi, A.L. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 2006, 444, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Koguchi, M.; Nakahara, Y.; Ito, H.; Wakamiya, T.; Yoshioka, F.; Ogata, A.; Inoue, K.; Masuoka, J.; Izumi, H.; Abe, T. BMP4 induces asymmetric cell division in human glioma stem-like cells. Oncol. Lett. 2020, 19, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, R.; Wu, M.; Johnson, K.; Kim, H.; Celebre, A.; Shahzad, U.; Graham, M.S.; Kessler, J.A.; Chuang, J.H.; Karamchandani, J.; et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019, 9, 14569. [Google Scholar] [CrossRef]
- Itoh, N.; Ohta, H. Secreted bone morphogenetic protein antagonists of the Chordin family. Biomol. Concepts 2010, 1, 297–304. [Google Scholar] [CrossRef]
- Cyr-Depauw, C.; Northey, J.J.; Tabaries, S.; Annis, M.G.; Dong, Z.; Cory, S.; Hallett, M.; Rennhack, J.P.; Andrechek, E.R.; Siegel, P.M. Chordin-Like 1 Suppresses Bone Morphogenetic Protein 4-Induced Breast Cancer Cell Migration and Invasion. Mol. Cell. Biol. 2016, 36, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.L.; Zhang, S.Q.; Zhang, H.; Wan, B.; Yin, Z.S. Chordin-like protein 1 promotes neuronal differentiation by inhibiting bone morphogenetic protein-4 in neural stem cells. Mol. Med. Rep. 2013, 7, 1143–1148. [Google Scholar] [CrossRef]
- Campos, B.; Wan, F.; Farhadi, M.; Ernst, A.; Zeppernick, F.; Tagscherer, K.E.; Ahmadi, R.; Lohr, J.; Dictus, C.; Gdynia, G.; et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin. Cancer Res. 2010, 16, 2715–2728. [Google Scholar] [CrossRef]
- Gunther, H.S.; Schmidt, N.O.; Phillips, H.S.; Kemming, D.; Kharbanda, S.; Soriano, R.; Modrusan, Z.; Meissner, H.; Westphal, M.; Lamszus, K. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene 2008, 27, 2897–2909. [Google Scholar] [CrossRef]
- Gerstmeier, J.; Possmayer, A.L.; Bozkurt, S.; Hoffmann, M.E.; Dikic, I.; Herold-Mende, C.; Burger, M.C.; Munch, C.; Kogel, D.; Linder, B. Calcitriol Promotes Differentiation of Glioma Stem-Like Cells and Increases Their Susceptibility to Temozolomide. Cancers 2021, 13, 3577. [Google Scholar] [CrossRef]
- Remy, J.; Linder, B.; Weirauch, U.; Day, B.W.; Stringer, B.W.; Herold-Mende, C.; Aigner, A.; Krohn, K.; Kogel, D. STAT3 Enhances Sensitivity of Glioblastoma to Drug-Induced Autophagy-Dependent Cell Death. Cancers 2022, 14, 339. [Google Scholar] [CrossRef]
- Linder, B.; Kohler, L.H.F.; Reisbeck, L.; Menger, D.; Subramaniam, D.; Herold-Mende, C.; Anant, S.; Schobert, R.; Biersack, B.; Kogel, D. A New Pentafluorothio-Substituted Curcuminoid with Superior Antitumor Activity. Biomolecules 2021, 11, 947. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Reyner, K.; Deleyrolle, L.; Millette, S.; Azari, H.; Day, B.W.; Stringer, B.W.; Boyd, A.W.; Johns, T.G.; Blot, V.; et al. Neurosphere and adherent culture conditions are equivalent for malignant glioma stem cell lines. Anat. Cell Biol. 2015, 48, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.A.; Daou, M.C.; Moser, R.P.; Ross, A.H. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010, 70, 6870–6879. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Antonietti, P.; Linder, B.; Hehlgans, S.; Mildenberger, I.C.; Burger, M.C.; Fulda, S.; Steinbach, J.P.; Gessler, F.; Rodel, F.; Mittelbronn, M.; et al. Interference with the HSF1/HSP70/BAG3 Pathway Primes Glioma Cells to Matrix Detachment and BH3 Mimetic-Induced Apoptosis. Mol. Cancer Ther. 2017, 16, 156–168. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Ghisolfi, L.; Keates, A.C.; Hu, X.; Lee, D.K.; Li, C.J. Ionizing radiation induces stemness in cancer cells. PLoS ONE 2012, 7, e43628. [Google Scholar] [CrossRef]
- Park, H.R.; Choi, Y.J.; Kim, J.Y.; Kim, I.G.; Jung, U. Repeated Irradiation with gamma-Ray Induces Cancer Stemness through TGF-beta-DLX2 Signaling in the A549 Human Lung Cancer Cell Line. Int. J. Mol. Sci. 2021, 22, 4284. [Google Scholar] [CrossRef]
- Sun, Y.; Yan, K.; Wang, Y.; Xu, C.; Wang, D.; Zhou, W.; Guo, S.; Han, Y.; Tang, L.; Shao, Y.; et al. Context-dependent tumor-suppressive BMP signaling in diffuse intrinsic pontine glioma regulates stemness through epigenetic regulation of CXXC5. Nat. Cancer 2022, 3, 1105–1122. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Q.; Tian, D.; Wu, L.; Dong, H.; Wang, J.; Ji, B.; Zhu, X.; Cai, Q.; Wang, L.; et al. BMP4 reverses multidrug resistance through modulation of BCL-2 and GDNF in glioblastoma. Brain Res. 2013, 1507, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Azari, H.; Deleyrolle, L.; Millette, S.; Zeng, H.; Reynolds, B.A. Controlling tumor invasion: Bevacizumab and BMP4 for glioblastoma. Future Oncol. 2013, 9, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Xi, G.; Best, B.; Mania-Farnell, B.; James, C.D.; Tomita, T. Therapeutic Potential for Bone Morphogenetic Protein 4 in Human Malignant Glioma. Neoplasia 2017, 19, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e46. [Google Scholar] [CrossRef]
- Podergajs, N.; Motaln, H.; Rajcevic, U.; Verbovsek, U.; Korsic, M.; Obad, N.; Espedal, H.; Vittori, M.; Herold-Mende, C.; Miletic, H.; et al. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget 2016, 7, 593–609. [Google Scholar] [CrossRef]
- Han, Y.; Xia, L.; Wang, X.; Xiong, H.; Zeng, L.; Wang, Z.; Zhang, T.; Xia, K.; Hu, X.; Su, T. Study on the expression and function of chordin-like 1 in oral squamous cell carcinoma. Oral Dis. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Wu, Q.; Zheng, Z.; Zhang, J.; Piao, Z.; Xin, M.; Xiang, X.; Wu, A.; Zhao, T.; Huang, S.; Qiao, Y.; et al. Chordin-Like 1 Regulates Epithelial-to-Mesenchymal Transition and Metastasis via the MAPK Signaling Pathway in Oral Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 862751. [Google Scholar] [CrossRef]
- Blanco-Suarez, E.; Allen, N.J. Astrocyte-secreted chordin-like 1 regulates spine density after ischemic injury. Sci. Rep. 2022, 12, 4176. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berglar, I.; Hehlgans, S.; Wehle, A.; Roth, C.; Herold-Mende, C.; Rödel, F.; Kögel, D.; Linder, B. CHRDL1 Regulates Stemness in Glioma Stem-like Cells. Cells 2022, 11, 3917. https://doi.org/10.3390/cells11233917
Berglar I, Hehlgans S, Wehle A, Roth C, Herold-Mende C, Rödel F, Kögel D, Linder B. CHRDL1 Regulates Stemness in Glioma Stem-like Cells. Cells. 2022; 11(23):3917. https://doi.org/10.3390/cells11233917
Chicago/Turabian StyleBerglar, Inka, Stephanie Hehlgans, Andrej Wehle, Caterina Roth, Christel Herold-Mende, Franz Rödel, Donat Kögel, and Benedikt Linder. 2022. "CHRDL1 Regulates Stemness in Glioma Stem-like Cells" Cells 11, no. 23: 3917. https://doi.org/10.3390/cells11233917
APA StyleBerglar, I., Hehlgans, S., Wehle, A., Roth, C., Herold-Mende, C., Rödel, F., Kögel, D., & Linder, B. (2022). CHRDL1 Regulates Stemness in Glioma Stem-like Cells. Cells, 11(23), 3917. https://doi.org/10.3390/cells11233917