MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of Melanoma Cell Lines, Fibroblasts and Melanocytes
2.2. SiRNA-Mediated Knockdown (KD)
2.3. Western Blot
2.4. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
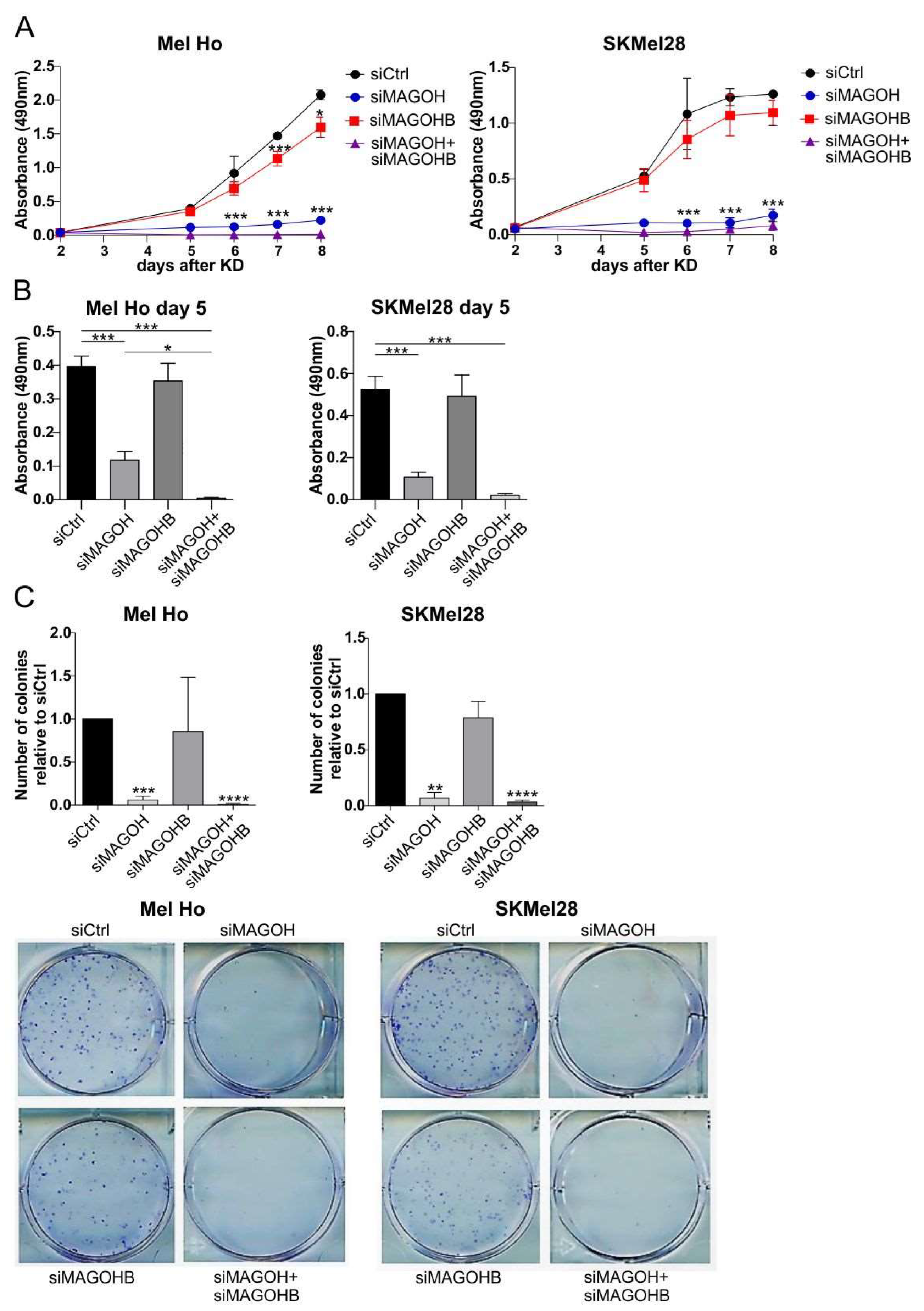
2.5. Cell Proliferation Assay
2.6. Clonogenic Assay
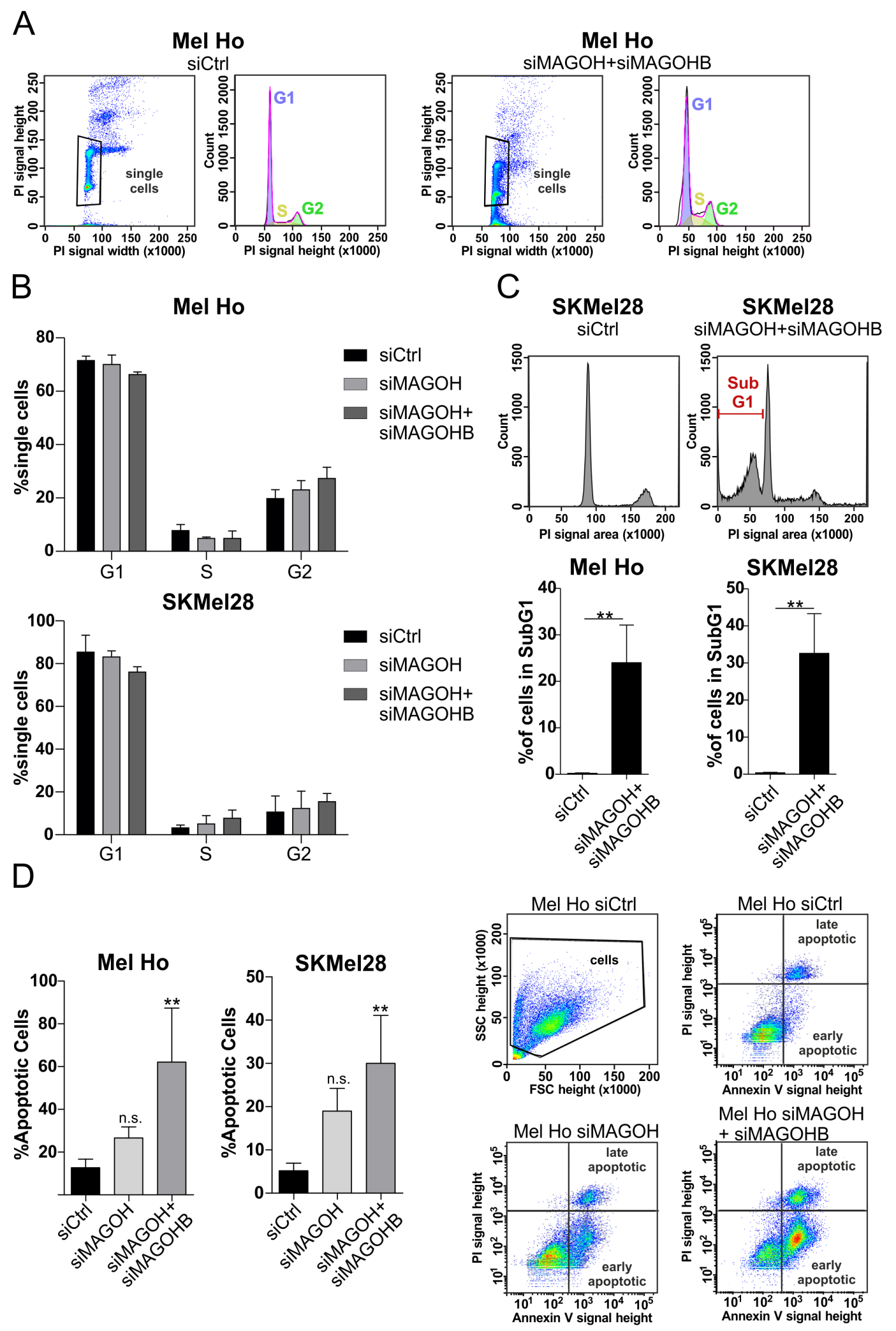
2.7. Cell Cycle Analysis
2.8. Apoptosis Assay
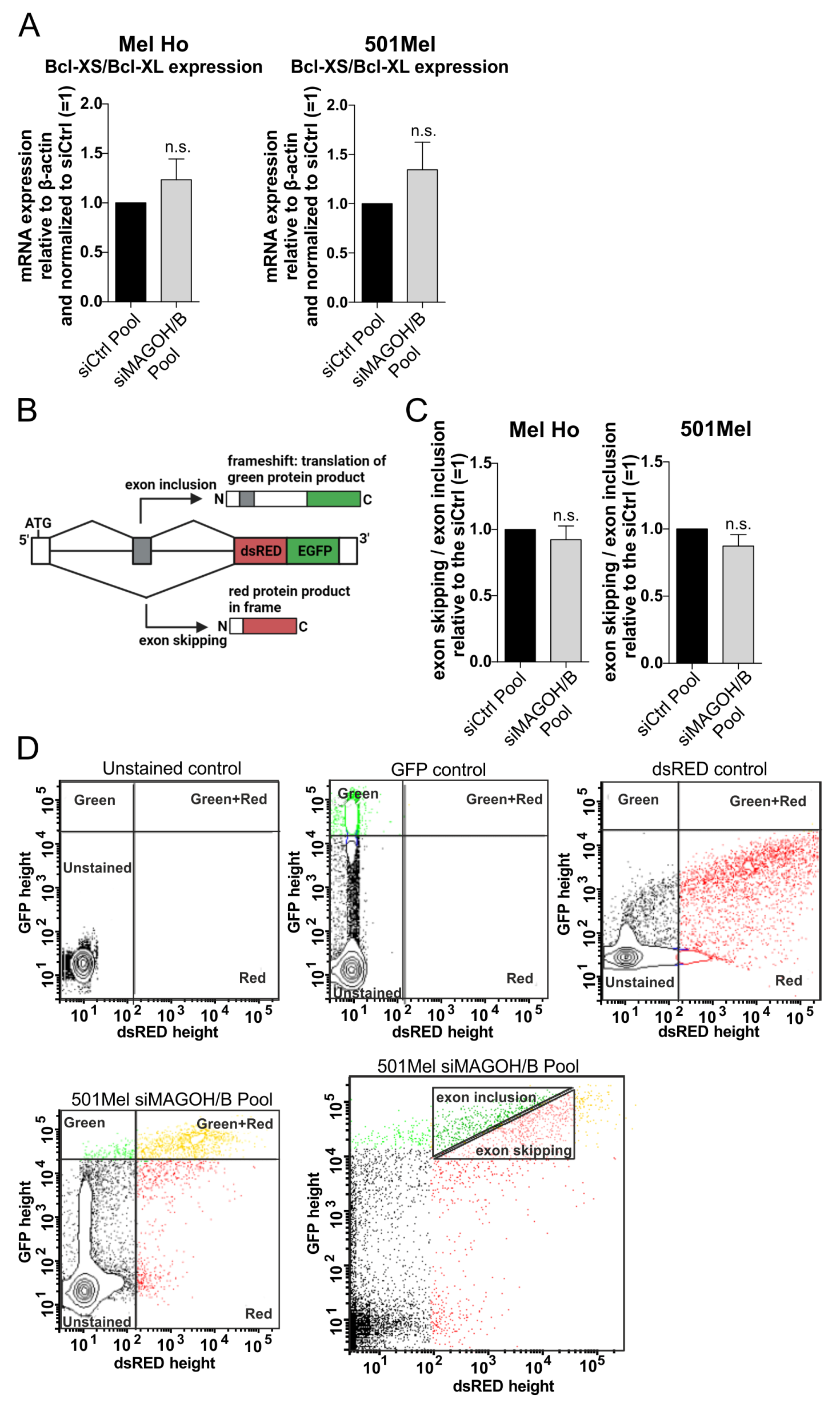
2.9. Analysis of Exon Skipping
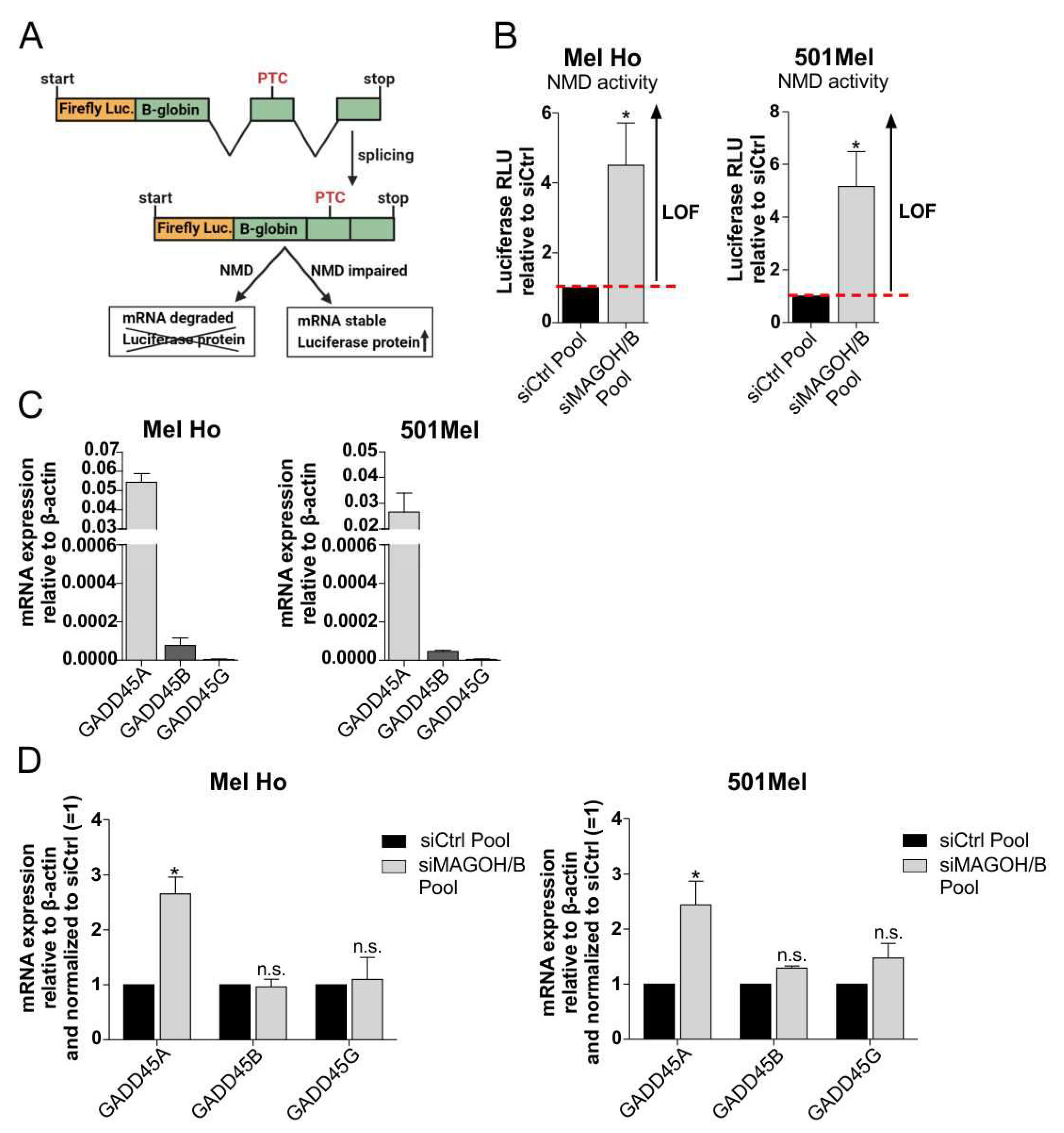
2.10. Analysis of Nonsense-Mediated Decay (NMD) Activity
2.11. Immunohistochemistry
2.12. Statistical Analysis
3. Results
3.1. MAGOH mRNA Shows Higher Expression Than MAGOHB in Cutaneous Melanoma
3.2. MAGOH/MAGOHB Protein Expression Is Upregulated in Cutaneous Melanoma Cell Lines and Tissue Samples
3.3. An siRNA-Mediated Knockdown (KD) of MAGOH and MAGOHB Proves MAGOH to Be the Predominatnly Expressed Protein of Both Homologues in Cutaneous Melanoma Cells
3.4. MAGOH Expression Is Vital for Melanoma Cell Proliferation
3.5. Knockdown of MAGOH and MAGOHB Does Not Affect Cell Cycle Progression but Induces Apoptosis
3.6. MAGOH and MAGOHB Do Not Regulate Alternative Splicing of the Pro-Apoptotic Gene Bcl-XS or Exon Skipping in Cutaneous Melanoma
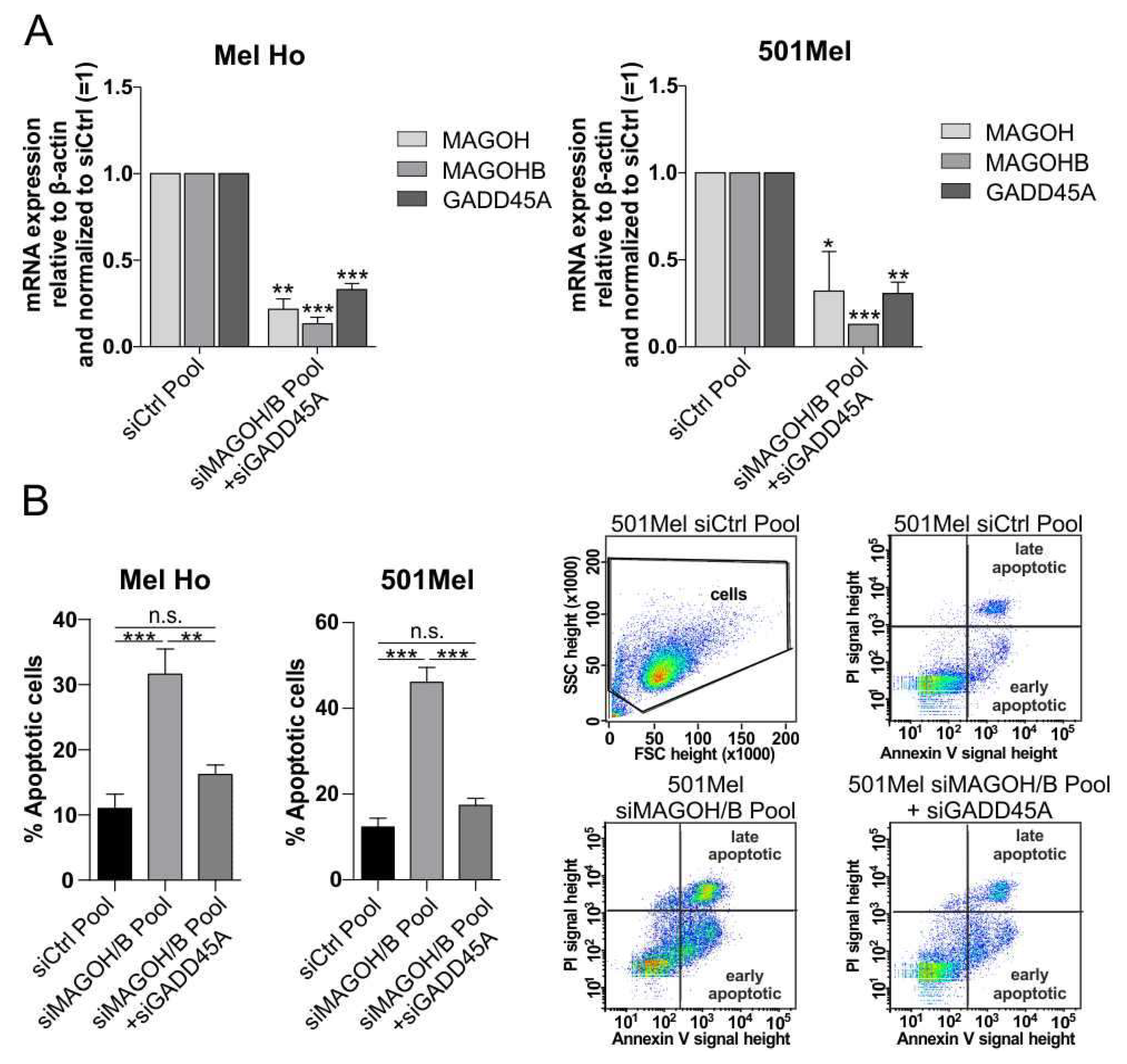
3.7. MAGOH and MAGOHB Knockdown Decreases NMD Activity and Promotes Apoptosis via Upregulation of GADD45A
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Šitum, M.; Buljan, M.; Kolić, M.; Vučić, M. Melanoma-clinical, dermatoscopical, and histopathological morphological characteristics. Acta Dermatovenerol. Croat. 2014, 22, 1–12. [Google Scholar] [PubMed]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar]
- Tsao, H.; Chin, L.; Garraway, L.A.; Fisher, D.E. Melanoma: From mutations to medicine. Genes Dev. 2012, 26, 1131. [Google Scholar] [CrossRef]
- Roh, M.R.; Eliades, P.; Gupta, S.; Tsao, H. Genetics of melanocytic nevi. Pigment Cell Melanoma Res. 2015, 28, 661–672. [Google Scholar] [CrossRef]
- Magnoni, C.; Tenedini, E.; Ferrari, F.; Benassi, L.; Bernardi, C.; Gualdi, G.; Bertazzoni, G.; Roncaglia, E.; Fantoni, L.; Manfredini, R.; et al. Transcriptional profiles in melanocytes from clinically unaffected skin distinguish the neoplastic growth pattern in patients with melanoma. Br. J. Dermatol. 2007, 156, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Matthews, N.H.; Li, W.-Q.; Qureshi, A.A.; Weinstock, M.A.; Cho, E. Epidemiology of Melanoma. In Cutaneous Melanoma: Etiology and Therapy; Codon Publications: Brisbane, Australia, 2017; pp. 3–22. ISBN 978-0-9944381-4-0. [Google Scholar]
- Europe. The Cancer Atlas. Available online: https://canceratlas.cancer.org/the-burden/europe/ (accessed on 18 November 2022).
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Nagore, E.; Martorell-Calatayud, A.; Botella-Estrada, R.; Guillén, C. Growth rate as an independent prognostic factor in localized invasive cutaneous melanoma. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 618–620. [Google Scholar] [CrossRef]
- Gualdi, G.; Panarese, F.; Meogrossi, G.; Marchioni, M.; De Tursi, M.; Di Marino, P.; Angelucci, D.; Amatetti, M.; Proietto, G.; Di Nicola, M.; et al. Speed rate (SR) as a new dynamic index of melanoma behavior. Pigment Cell Melanoma Res. 2020, 33, 709–718. [Google Scholar] [CrossRef]
- Melanoma Survival Rates. Melanoma Survival Statistics. Available online: https://www.cancer.org/cancer/melanoma-skin-cancer/detection-diagnosis-staging/survival-rates-for-melanoma-skin-cancer-by-stage.html (accessed on 5 October 2022).
- Orthaber, K.; Pristovnik, M.; Skok, K.; Perić, B.; Maver, U. Skin Cancer and Its Treatment: Novel Treatment Approaches with Emphasis on Nanotechnology. J. Nanomater. 2017, 2017, 1–20. [Google Scholar] [CrossRef]
- Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866. [Google Scholar] [CrossRef]
- Moore, M.J.; Proudfoot, N.J. Pre-mRNA Processing Reaches Back toTranscription and Ahead to Translation. Cell 2009, 136, 688–700. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Supek, F.; Lehner, B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef]
- Shkreta, L.; Cloutier, A.; Toutant, J.; Rendos, H.V.; Chabot, B. Regulation of alternative splicing and the case of Bcl-x. Pak. J. Biochem. Mol. Biol. 2015, 48, 27–38. [Google Scholar]
- Blazquez, L.; Emmett, W.; Faraway, R.; Pineda, J.M.B.; Bajew, S.; Gohr, A.; Haberman, N.; Sibley, C.R.; Bradley, R.K.; Irimia, M.; et al. Exon Junction Complex Shapes the Transcriptome by Repressing Recursive Splicing. Mol. Cell 2018, 72, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Murigneux, V.; Le Hir, H. Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol. 2014, 15, 551. [Google Scholar] [CrossRef]
- Singh, K.K.; Wachsmuth, L.; Kulozik, A.E.; Gehring, N.H. Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay. RNA Biol. 2013, 10, 1291–1298. [Google Scholar] [CrossRef]
- Ishigaki, Y.; Nakamura, Y.; Tatsuno, T.; Hashimoto, M.; Shimasaki, T.; Iwabuchi, K.; Tomosugi, N. Depletion of RNA-binding protein RBM8A (Y14) causes cell cycle deficiency and apoptosis in human cells. Exp. Biol. Med. 2013, 238, 889–897. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Nogueira, M.F.; Buss, C.G.; Krill-Burger, J.M.; Wawer, M.J.; Malolepsza, E.; Berger, A.C.; Choi, P.S.; Shih, J.; Taylor, A.M.; et al. Genome-scale analysis identifies paralog lethality as a vulnerability of chromosome 1p loss in cancer. Nat. Genet. 2018, 50, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Otani, Y.; Fujita, K.I.; Kameyama, T.; Mayeda, A. The Exon Junction Complex Core Represses Cancer-Specific Mature mRNA Re-splicing: A Potential Key Role in Terminating Splicing. Int. J. Mol. Sci. 2021, 22, 6519. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, Z.; Wu, X.; Tou, L.; Zheng, J.; Zhou, D. MAGOH/MAGOHB Inhibits the Tumorigenesis of Gastric Cancer via Inactivation of b-RAF/MEK/ERK Signaling. Onco. Targets Ther. 2020, 13, 12723–12735. [Google Scholar] [CrossRef]
- Silver, D.L.; Leeds, K.E.; Hwang, H.W.; Miller, E.E.; Pavan, W.J. The EJC component Magoh regulates proliferation and expansion of neural crest-derived melanocytes. Dev. Biol. 2013, 375, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, G.; Mao, P.; Zhang, J.; Zhang, L.; Liu, L.; Wang, J.; Owusu, L.; Ren, B.; Tang, Y.; et al. Down-regulation of GADD45A enhances chemosensitivity in melanoma. Sci. Rep. 2018, 8, 4111. [Google Scholar] [CrossRef] [PubMed]
- Linck-Paulus, L.; Meißgeier, T.; Pieger, K.; Horn, A.H.C.; Matthies, A.; Fischer, S.; Meister, G.; Sticht, H.; Kappelmann-Fenzl, M.; Bosserhoff, A.K. A previously unknown Argonaute 2 variant positively modulates the viability of melanoma cells. Cell. Mol. Life Sci. 2022, 79, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lämmerhirt, L.; Kappelmann-fenzl, M.; Fischer, S.; Pommer, M.; Zimmermann, T.; Kluge, V.; Matthies, A.; Kuphal, S.; Bosserhoff, A.K. Knockdown of Lamin B1 and the Corresponding Lamin B Receptor Leads to Changes in Heterochromatin State and Senescence Induction in Malignant Melanoma. Cells 2022, 11, 2154. [Google Scholar] [CrossRef]
- Fox, M.H. A model for the computer analysis of synchronous DNA distributions obtained by flow cytometry. Cytometry 1980, 1, 71–77. [Google Scholar] [CrossRef]
- Orengo, J.P.; Bundman, D.; Cooper, T.A. A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic Acids Res. 2006, 34, e148. [Google Scholar] [CrossRef]
- Baird, T.D.; Cheng, K.C.C.; Chen, Y.C.; Buehler, E.; Martin, S.E.; Inglese, J.; Hogg, J.R. ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. Elife 2018, 7, e33178. [Google Scholar] [CrossRef]
- Schagat, T.; Paguio, A.; Kopish, K. Normalizing Genetic Reporter Assays: Approaches and Considerations for Increasing Consistency and Statistical Significance. Available online: https://www.promega.de/-/media/files/resources/cell-notes/cn017/normalizing-genetic-reporter-assays.pdf?rev=56fc83d2f445401ea1d003cce3f51be7&sc_lang=en (accessed on 13 October 2022).
- Seefried, F.; Haller, L.; Fukuda, S.; Thongmao, A.; Schneider, N.; Utikal, J.; Higashiyama, S.; Bosserhoff, A.K.; Kuphal, S. Nuclear AREG affects a low-proliferative phenotype and contributes to drug resistance of melanoma. Int. J. Cancer 2022, 151, 2244–2264. [Google Scholar] [CrossRef]
- Expression of MAGOH in Melanoma—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000162385-MAGOH/pathology/melanoma (accessed on 5 October 2021).
- Expression of MAGOHB in Melanoma—The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000111196-MAGOHB/pathology/melanoma (accessed on 5 October 2021).
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Michelle, L.; Cloutier, A.; Toutant, J.; Shkreta, L.; Thibault, P.; Durand, M.; Garneau, D.; Gendron, D.; Lapointe, E.; Couture, S.; et al. Proteins Associated with the Exon Junction Complex Also Control the Alternative Splicing of Apoptotic Regulators. Mol. Cell. Biol. 2012, 32, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.O.; Moore, K.A.; Chapin, A.; Hollien, J.; Metzstein, M.M. Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. Elife 2016, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, G.; Fernandes, R.; García-Moreno, J.F.; Romão, L. Nonsense-mediated RNA decay and its bipolar function in cancer. Mol. Cancer 2021, 20. [Google Scholar] [CrossRef]
- Gao, M.; Guo, N.; Huang, C.; Song, L. Diverse roles of GADD45alpha in stress signaling. Curr. Protein Pept. Sci. 2009, 10, 388–394. [Google Scholar] [CrossRef]
- Salvador, J.M.; Brown-Clay, J.D.; Fornace, A.J. Gadd45 in Stress Signaling, Cell Cycle Control, and Apoptosis. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2013; Volume 793, pp. 1–19. ISBN 9781461482888. [Google Scholar]
- Litchfield, K.; Reading, J.L.; Lim, E.L.; Xu, H.; Liu, P.; Al-Bakir, M.; Wong, Y.N.S.; Rowan, A.; Funt, S.A.; Merghoub, T.; et al. Escape from nonsense-mediated decay associates with anti-tumor immunogenicity. Nat. Commun. 2020, 11, 3800. [Google Scholar] [CrossRef]


| Cell Line | Origin | Medium | CO2 | ID |
|---|---|---|---|---|
| Sbcl2 | PT 1 | TU | 5% | RRID:CVCL_D732 |
| Mel Ho | PT | RPM1-1640 | 8% | RRID:CVCL_1402 |
| WM1366 | PT | TU | 5% | RRID:CVCL_6789 |
| WM3211 | PT | TU | 5% | RRID:CVCL_6797 |
| 501Mel | MET 2 | RPMI-1640 | 8% | RRID:CVCL_4633 |
| SKMel28 | MET | DMEM | 8% | RRID:CVCL_0526 |
| WM1158 | MET | TU | 5% | RRID:CVCL_6785 |
| siRNA | Sequence (5′-3′) | Manufacturer |
|---|---|---|
| siMAGOH | GCGUGAUGGAGGAACUGAATT 1 | Sigma-Aldrich Chemie GmbH, Steinheim, Germany |
| siMAGOH2 | “Hs_MAGOH_8 FlexiTube siRNA” GeneGlobe ID: SI05064101 | QIAGEN GmbH, Hilden, Germany |
| siMAGOHB | GGCUGUUUGUAUAUUUAAUTT 1 | Sigma-Aldrich Chemie GmbH, Steinheim, Germany |
| siCtrl | “AllStars Negative Control siRNA” | QIAGEN GmbH, Hilden, Germany |
| siGADD45A | GGAGGAAGUGCUAGCAAATT 2 | Sigma-Aldrich Chemie GmbH, Steinheim, Germany |
| siMAGOH/B Pool | Mixture of 30 different siRNAs targeting MAGOH and MAGOHB | siTools Biotech, Planegg, Germany |
| siCtrl Pool | Mixture of 30 non-targeting siRNAs | siTools Biotech, Planegg, Germany |
| Primer Name | Sequence (5′-3′) | Measurement (°C) |
|---|---|---|
| hBcl-x 1095 rev | GGGAGGGTAGAGTGGATGGT | ≤85 |
| hBcl-XL 887 fwd | TGACCACCTAGAGCCTTGGA | ≤85 |
| hBcl-Xs 749 fwd | TGAACAGGATACTTTTGTGGAACT | ≤85 |
| Beta actin_cyto fwd | CTACGTCGCCCTGGACTTCGAGC | ≤85 |
| Beta-Actin_cyto rev | GATGGAGCCGCCGATCCACACGG | ≤85 |
| hGADD45A 399 fwd | ATCACTGTCGGGGTGTACGA | ≤86 |
| hGADD45A 619 rev | CAGCGTCGGTCTCCAAGA | ≤86 |
| hGADD45B 443 fwd | TGCAAATCCACTTCACGCTC | ≤88 |
| hGADD45B 613 rev | CGTGTGAGGGTTCGTGACC | ≤88 |
| hGADD56G 235 fwd | CGTCTACGAGTCAGCCAAAGT | ≤88 |
| hGADD45G 394 rev | ACGCGCACTATGTCGATGT | ≤88 |
| hMAGOH 150 fwd | GACCGGACGGGAAGTTAAGA | ≤75 |
| hMAGOH 251 rev | TCAGTTCCTCCATCACGCTTT | ≤75 |
| hMAGOHB 106 fwd | GCCGGACGGAAAGCTTAGAT | ≤75 |
| hMAGOHB 341 rev | GGCCTTCAGGATCCTTTGACT | ≤75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soederberg, A.; Meißgeier, T.; Bosserhoff, A.K.; Linck-Paulus, L. MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A. Cells 2022, 11, 3859. https://doi.org/10.3390/cells11233859
Soederberg A, Meißgeier T, Bosserhoff AK, Linck-Paulus L. MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A. Cells. 2022; 11(23):3859. https://doi.org/10.3390/cells11233859
Chicago/Turabian StyleSoederberg, Agnes, Tina Meißgeier, Anja Katrin Bosserhoff, and Lisa Linck-Paulus. 2022. "MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A" Cells 11, no. 23: 3859. https://doi.org/10.3390/cells11233859
APA StyleSoederberg, A., Meißgeier, T., Bosserhoff, A. K., & Linck-Paulus, L. (2022). MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A. Cells, 11(23), 3859. https://doi.org/10.3390/cells11233859






