Comprehensive Review of Biomarkers for the Treatment of Locally Advanced Colon Cancer
Abstract
1. Introduction
2. Materials and Methods
3. Role of Biomarkers in LACC Treatment
3.1. Prognostic Biomarkers for LACC
3.2. Predictive Biomarkers for LACC Undergoing Neoadjuvant Chemotherapy
3.2.1. Mismatch Repair Deficiency
3.2.2. Excision Repair Cross-Complementing 1, Thymidylate Synthase, and Glutathione S-Transferase pi
3.3. Predictive Biomarkers for Patients with LACC Undergoing Neoadjuvant Chemoradiotherapy
3.4. Prognostic/Predictive Biomarkers of Postoperative Adjuvant Chemotherapy for LACC
3.4.1. MMR
3.4.2. MSI-High
3.4.3. Epidermal Growth Factor Receptor Expression
3.4.4. KRAS and BRAF
3.4.5. Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha
3.4.6. CpG Island Methylator Phenotype
3.4.7. Deleted in Colorectal Cancer Protein
3.4.8. ERCC1
3.4.9. Circulating Tumor Cells or Circulating Tumor DNA
3.4.10. Blood Sugar Level
4. Advances in Molecular and Epigenetic Biomarkers with Potential Applications in LACC Therapy
4.1. Vascular Endothelial Growth Factor (VEGF), Human Epidermal Growth Factor Receptor 2 (HER2), Hepatocyte Growth Factor (HGF), Tyrosine-Protein Kinase Met (c-Met)
4.2. MicroRNAs and Long Noncoding RNAs
4.3. Genomic and Metabolomic Biomarkers
4.4. Methylation Levels of Long Interspersed Nucleotide Elements
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rosander, E.; Nordenvall, C.; Sjovall, A.; Hjern, F.; Holm, T. Management and Outcome After Multivisceral Resections in Patients with Locally Advanced Primary Colon Cancer. Dis. Colon Rectum 2018, 61, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Tsai, H.L.; Li, C.C.; Huang, C.W.; Chang, T.K.; Su, W.C.; Chen, P.J.; Yin, T.C.; Huang, C.M.; Wang, J.Y. Critical reappraisal of neoadjuvant concurrent chemoradiotherapy for treatment of locally advanced colon cancer. PLoS ONE 2021, 16, e0259460. [Google Scholar] [CrossRef] [PubMed]
- De Nes, L.C.F.; van der Heijden, J.A.G.; Verstegen, M.G.; Drager, L.; Tanis, P.J.; Verhoeven, R.H.A.; de Wilt, J.H.W. Predictors of undergoing multivisceral resection, margin status and survival in Dutch patients with locally advanced colorectal cancer. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2022, 48, 1144–1152. [Google Scholar] [CrossRef] [PubMed]
- Klaver, C.E.; Gietelink, L.; Bemelman, W.A.; Wouters, M.W.; Wiggers, T.; Tollenaar, R.A.; Tanis, P.J. Locally advanced colon cancer: Evaluation of current clinical practice and treatment outcomes at the population level. J. Natl. Compr. Cancer Netw. 2017, 15, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Weiser, M.R. AJCC 8th Edition: Colorectal Cancer. Ann. Surg. Oncol. 2018, 25, 1454–1455. [Google Scholar] [CrossRef]
- Byrd, D.R.; Carducci, M.A.; Compton, C.C.; Fritz, A.G.; Greene, F.L. AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010; Volume 7, pp. 97–100. [Google Scholar]
- Edition, S.; Edge, S.; Byrd, D. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Gunderson, L.L.; Jessup, J.M.; Sargent, D.J.; Greene, F.L.; Stewart, A.K. Revised TN categorization for colon cancer based on national survival outcomes data. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 264–271. [Google Scholar] [CrossRef]
- Gao, P.; Song, Y.-X.; Wang, Z.-N.; Xu, Y.-Y.; Tong, L.-l.; Sun, J.-X.; Yu, M.; Xu, H.-M. Is the prediction of prognosis not improved by the seventh edition of the TNM classification for colorectal cancer? Analysis of the surveilla006Ece, epidemiology, and end results (SEER) database. BMC Cancer 2013, 13, 123. [Google Scholar] [CrossRef]
- Hermanek, P., Jr.; Wiebelt, H.; Riedl, S.; Staimmer, D.; Hermanek, P. Long-term results of surgical therapy of colon cancer. Results of the Colorectal Cancer Study Group. Chir. Z. Alle Geb. Oper. Med. 1994, 65, 287–297. [Google Scholar]
- Oh, S.Y.; Kim, D.Y.; Kim, Y.B.; Suh, K.W. Oncologic outcomes after adjuvant chemotherapy using FOLFOX in MSI-H sporadic stage III colon cancer. World J. Surg. 2013, 37, 2497–2503. [Google Scholar] [CrossRef]
- De Gooyer, J.M.; Verstegen, M.G.; ’t Lam-Boer, J.; Radema, S.A.; Verhoeven, R.H.A.; Verhoef, C.; Schreinemakers, J.M.J.; de Wilt, J.H.W. Neoadjuvant Chemotherapy for Locally Advanced T4 Colon Cancer: A Nationwide Propensity-Score Matched Cohort Analysis. Dig. Surg. 2020, 37, 292–301. [Google Scholar] [CrossRef]
- Zhou, J.; Guo, Z.; Yu, W.; Li, S.; Qiao, W. Clinical Evaluation of Preoperative Radiotherapy Combined with FOLFOX Chemotherapy on Patients with Locally Advanced Colon Cancer. Am. Surg. 2019, 85, 313–320. [Google Scholar] [CrossRef]
- Izbicki, J.R.; Hosch, S.B.; Knoefel, W.T.; Passlick, B.; Bloechle, C.; Broelsch, C.E. Extended resections are beneficial for patients with locally advanced colorectal cancer. Dis. Colon Rectum 1995, 38, 1251–1256. [Google Scholar] [CrossRef]
- Sahakyan, A.M.; Aleksanyan, A.; Batikyan, H.; Petrosyan, H.; Sahakyan, M.A. Standard and multivisceral colectomy in locally advanced colon cancer. Radiol. Oncol. 2020, 54, 341–346. [Google Scholar] [CrossRef]
- Huang, C.W.; Chen, Y.T.; Tsai, H.L.; Yeh, Y.S.; Su, W.C.; Ma, C.J.; Tsai, T.N.; Wang, J.Y. EGFR expression in patients with stage III colorectal cancer after adjuvant chemotherapy and on cancer cell function. Oncotarget 2017, 8, 114663–114676. [Google Scholar] [CrossRef]
- Kim, J.E.; Hong, Y.S.; Kim, H.J.; Kim, K.P.; Kim, S.Y.; Lim, S.B.; Park, I.J.; Kim, C.W.; Yoon, Y.S.; Yu, C.S.; et al. Microsatellite Instability was not Associated with Survival in Stage III Colon Cancer Treated with Adjuvant Chemotherapy of Oxaliplatin and Infusional 5-Fluorouracil and Leucovorin (FOLFOX). Ann. Surg. Oncol. 2017, 24, 1289–1294. [Google Scholar] [CrossRef]
- Alwers, E.; Jansen, L.; Blaker, H.; Kloor, M.; Tagscherer, K.E.; Roth, W.; Boakye, D.; Herpel, E.; Grullich, C.; Chang-Claude, J.; et al. Microsatellite instability and survival after adjuvant chemotherapy among stage II and III colon cancer patients: Results from a population-based study. Mol. Oncol. 2020, 14, 363–372. [Google Scholar] [CrossRef]
- Andre, T.; Boni, C.; Mounedji-Boudiaf, L.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Zaninelli, M.; Clingan, P.; Bridgewater, J.; et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N. Engl. J. Med. 2004, 350, 2343–2351. [Google Scholar] [CrossRef]
- Taieb, J.; Gallois, C. Adjuvant Chemotherapy for Stage III Colon Cancer. Cancers 2020, 12, 2679. [Google Scholar] [CrossRef]
- Sargent, D.; Sobrero, A.; Grothey, A.; O’Connell, M.J.; Buyse, M.; Andre, T.; Zheng, Y.; Green, E.; Labianca, R.; O’Callaghan, C.; et al. Evidence for cure by adjuvant therapy in colon cancer: Observations based on individual patient data from 20,898 patients on 18 randomized trials. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Ludmir, E.B.; Arya, R.; Wu, Y.; Palta, M.; Willett, C.G.; Czito, B.G. Role of Adjuvant Radiotherapy in Locally Advanced Colonic Carcinoma in the Modern Chemotherapy Era. Ann. Surg. Oncol. 2016, 23, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Leijssen, L.G.J.; Dinaux, A.M.; Amri, R.; Kunitake, H.; Bordeianou, L.G.; Berger, D.L. The Impact of a Multivisceral Resection and Adjuvant Therapy in Locally Advanced Colon Cancer. J. Gastrointest. Surg. Off. J. Soc. Surg. Aliment. Tract 2019, 23, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Karoui, M.; Rullier, A.; Piessen, G.; Legoux, J.; Barbier, E.; De Chaisemartin, C.; Lecaille, C.; Bouche, O.; Ammarguellat, H.; Brunetti, F. Perioperative FOLFOX 4 versus FOLFOX 4 plus cetuximab versus immediate surgery for high-risk stage II and III colon cancers: A phase II multicenter randomized controlled trial (PRODIGE 22). Ann. Surg. 2020, 271, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Karoui, M.; Rullier, A.; Luciani, A.; Bonnetain, F.; Auriault, M.-L.; Sarran, A.; Monges, G.; Trillaud, H.; Le Malicot, K.; Leroy, K. Neoadjuvant FOLFOX 4 versus FOLFOX 4 with Cetuximab versus immediate surgery for high-risk stage II and III colon cancers: A multicentre randomised controlled phase II trial–the PRODIGE 22-ECKINOXE trial. BMC Cancer 2015, 15, 511. [Google Scholar] [CrossRef]
- Zhou, Z.; Nimeiri, H.S.; Benson III, A.B. Preoperative chemotherapy for locally advanced resectable colon cancer-a new treatment paradigm in colon cancer? Ann. Transl. Med. 2013, 1, 11. [Google Scholar]
- Seligmann, J.F.; Group, F.C. FOxTROT: Neoadjuvant FOLFOX chemotherapy with or without panitumumab (Pan) for patients (pts) with locally advanced colon cancer (CC). J. Clin. Oncol. 2020, 38, 4013. [Google Scholar] [CrossRef]
- Jakobsen, A.; Andersen, F.; Fischer, A.; Jensen, L.H.; Jorgensen, J.C.; Larsen, O.; Lindebjerg, J.; Ploen, J.; Rafaelsen, S.R.; Vilandt, J. Neoadjuvant chemotherapy in locally advanced colon cancer. A phase II trial. Acta Oncol. 2015, 54, 1747–1753. [Google Scholar] [CrossRef]
- Group, F.C. Feasibility of preoperative chemotherapy for locally advanced, operable colon cancer: The pilot phase of a randomised controlled trial. Lancet Oncol. 2012, 13, 1152–1160. [Google Scholar]
- Cheong, C.K.; Nistala, K.R.Y.; Ng, C.H.; Syn, N.; Chang, H.S.Y.; Sundar, R.; Yang, S.Y.; Chong, C.S. Neoadjuvant therapy in locally advanced colon cancer: A meta-analysis and systematic review. J. Gastrointest. Oncol. 2020, 11, 847–857. [Google Scholar] [CrossRef]
- Benson, A.B., 3rd; Venook, A.P.; Cederquist, L.; Chan, E.; Chen, Y.J.; Cooper, H.S.; Deming, D.; Engstrom, P.F.; Enzinger, P.C.; Fichera, A.; et al. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 2017, 15, 370–398. [Google Scholar] [CrossRef]
- Mathis, K.L.; Nelson, H.; Pemberton, J.H.; Haddock, M.G.; Gunderson, L.L. Unresectable colorectal cancer can be cured with multimodality therapy. Ann. Surg. 2008, 248, 592–598. [Google Scholar] [CrossRef]
- Sauer, R.; Liersch, T.; Merkel, S.; Fietkau, R.; Hohenberger, W.; Hess, C.; Becker, H.; Raab, H.-R.; Villanueva, M.-T.; Witzigmann, H. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: Results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 1926–1933. [Google Scholar] [CrossRef]
- Lee, Y.C.; Hsieh, C.C.; Chuang, J.P. Prognostic significance of partial tumor regression after preoperative chemoradiotherapy for rectal cancer: A meta-analysis. Dis. Colon Rectum 2013, 56, 1093–1101. [Google Scholar] [CrossRef]
- Huang, C.-M.; Huang, M.-Y.; Ma, C.-J.; Yeh, Y.S.; Tsai, H.-L.; Huang, C.-W.; Huang, C.-J.; Wang, J.-Y. Neoadjuvant FOLFOX chemotherapy combined with radiotherapy followed by radical resection in patients with locally advanced colon cancer. Radiat. Oncol. 2017, 12, 48. [Google Scholar] [CrossRef]
- Huang, C.M.; Huang, C.W.; Ma, C.J.; Tsai, H.L.; Su, W.C.; Chang, T.K.; Huang, M.Y.; Wang, J.Y. Outcomes of neoadjuvant chemoradiotherapy followed by radical resection for T4 colorectal cancer. World J. Gastrointest. Oncol. 2020, 12, 1428–1442. [Google Scholar] [CrossRef]
- Cukier, M.; Smith, A.; Milot, L.; Chu, W.; Chung, H.; Fenech, D.; Herschorn, S.; Ko, Y.; Rowsell, C.; Soliman, H. Neoadjuvant chemoradiotherapy and multivisceral resection for primary locally advanced adherent colon cancer: A single institution experience. Eur. J. Surg. Oncol. 2012, 38, 677–682. [Google Scholar] [CrossRef]
- Yuan, Y.; Xiao, W.-W.; Xie, W.-H.; Cai, P.-Q.; Wang, Q.-X.; Chang, H.; Chen, B.-Q.; Zhou, W.-H.; Zeng, Z.-F.; Wu, X.-J.; et al. Neoadjuvant chemoradiotherapy for patients with unresectable radically locally advanced colon cancer: A potential improvement to overall survival and decrease to multivisceral resection. BMC Cancer 2021, 21, 179. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Cho, K.R.; Vogelstein, B. Genetic alterations in the adenoma–carcinoma sequence. Cancer 1992, 70, 1727–1731. [Google Scholar] [CrossRef]
- Antelo, M.; Balaguer, F.; Shia, J.; Shen, Y.; Hur, K.; Moreira, L.; Cuatrecasas, M.; Bujanda, L.; Giraldez, M.D.; Takahashi, M.; et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS ONE 2012, 7, e45357. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Worthley, D.L.; Leggett, B.A. Colorectal cancer: Molecular features and clinical opportunities. Clin. Biochem. Rev. 2010, 31, 31. [Google Scholar] [PubMed]
- Aghagolzadeh, P.; Radpour, R. New trends in molecular and cellular biomarker discovery for colorectal cancer. World J. Gastroenterol. 2016, 22, 5678–5693. [Google Scholar] [CrossRef] [PubMed]
- Chand, M.; Keller, D.S.; Mirnezami, R.; Bullock, M.; Bhangu, A.; Moran, B.; Tekkis, P.P.; Brown, G.; Mirnezami, A.; Berho, M. Novel biomarkers for patient stratification in colorectal cancer: A review of definitions, emerging concepts, and data. World J. Gastrointest. Oncol. 2018, 10, 145–158. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Mahmood, F.; Akingboye, A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int. J. Mol. Sci. 2020, 21, 5311. [Google Scholar] [CrossRef]
- Klingbiel, D.; Saridaki, Z.; Roth, A.D.; Bosman, F.T.; Delorenzi, M.; Tejpar, S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: Results of the PETACC-3 trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 126–132. [Google Scholar] [CrossRef]
- Taieb, J.; Le Malicot, K.; Shi, Q.; Penault-Llorca, F.; Bouche, O.; Tabernero, J.; Mini, E.; Goldberg, R.M.; Folprecht, G.; Luc Van Laethem, J.; et al. Prognostic Value of BRAF and KRAS Mutations in MSI and MSS Stage III Colon Cancer. J. Natl. Cancer Inst. 2017, 109, djw272. [Google Scholar] [CrossRef]
- Zaanan, A.; Shi, Q.; Taieb, J.; Alberts, S.R.; Meyers, J.P.; Smyrk, T.C.; Julie, C.; Zawadi, A.; Tabernero, J.; Mini, E.; et al. Role of Deficient DNA Mismatch Repair Status in Patients With Stage III Colon Cancer Treated With FOLFOX Adjuvant Chemotherapy: A Pooled Analysis From 2 Randomized Clinical Trials. JAMA Oncol. 2018, 4, 379–383. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef]
- Yang, I.P.; Miao, Z.F.; Huang, C.W.; Tsai, H.L.; Yeh, Y.S.; Su, W.C.; Chang, T.K.; Chang, S.F.; Wang, J.Y. High blood sugar levels but not diabetes mellitus significantly enhance oxaliplatin chemoresistance in patients with stage III colorectal cancer receiving adjuvant FOLFOX6 chemotherapy. Ther. Adv. Med. Oncol. 2019, 11, 1758835919866964. [Google Scholar] [CrossRef]
- Sobrero, A.F.; Puccini, A.; Shi, Q.; Grothey, A.; Andrè, T.; Shields, A.F.; Souglakos, I.; Yoshino, T.; Iveson, T.; Ceppi, M.; et al. A new prognostic and predictive tool for shared decision making in stage III colon cancer. Eur. J. Cancer 2020, 138, 182–188. [Google Scholar] [CrossRef]
- Chu, Q.D.; Zhou, M.; Medeiros, K.L.; Peddi, P.; Kavanaugh, M.; Wu, X.C. Poor survival in stage IIB/C (T4N0) compared to stage IIIA (T1-2 N1, T1N2a) colon cancer persists even after adjusting for adequate lymph nodes retrieved and receipt of adjuvant chemotherapy. BMC Cancer 2016, 16, 460. [Google Scholar] [CrossRef]
- Huang, C.-W.; Tsai, H.-L.; Huang, M.-Y.; Huang, C.-M.; Yeh, Y.-S.; Ma, C.-J.; Wang, J.-Y. Different clinicopathologic features and favorable outcomes of patients with stage III left-sided colon cancer. World J. Surg. Oncol. 2015, 13, 257. [Google Scholar] [CrossRef]
- Ulanja, M.B.; Rishi, M.; Beutler, B.D.; Sharma, M.; Patterson, D.R.; Gullapalli, N.; Ambika, S. Colon Cancer Sidedness, Presentation, and Survival at Different Stages. J. Oncol. 2019, 2019, 4315032. [Google Scholar] [CrossRef]
- Su, M.W.; Chang, C.K.; Lin, C.W.; Chu, H.W.; Tsai, T.N.; Su, W.C.; Chen, Y.C.; Chang, T.K.; Huang, C.W.; Tsai, H.L.; et al. Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers. Cells 2022, 11, 527. [Google Scholar] [CrossRef]
- Petrelli, F.; Tomasello, G.; Borgonovo, K.; Ghidini, M.; Turati, L.; Dallera, P.; Passalacqua, R.; Sgroi, G.; Barni, S. Prognostic Survival Associated With Left-Sided vs. Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol. 2017, 3, 211–219. [Google Scholar] [CrossRef]
- Dehal, A.; Graff-Baker, A.N.; Vuong, B.; Fischer, T.; Klempner, S.J.; Chang, S.-C.; Grunkemeier, G.L.; Bilchik, A.J.; Goldfarb, M. Neoadjuvant chemotherapy improves survival in patients with clinical T4b colon cancer. J. Gastrointest. Surg. 2018, 22, 242–249. [Google Scholar] [CrossRef]
- Lindor, N.M.; Burgart, L.J.; Leontovich, O.; Goldberg, R.M.; Cunningham, J.M.; Sargent, D.J.; Walsh-Vockley, C.; Petersen, G.M.; Walsh, M.D.; Leggett, B.A.; et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2002, 20, 1043–1048. [Google Scholar] [CrossRef]
- Kawakami, H.; Zaanan, A.; Sinicrope, F.A. Implications of mismatch repair-deficient status on management of early stage colorectal cancer. J. Gastrointest. Oncol. 2015, 6, 676–684. [Google Scholar] [CrossRef]
- Yunlong, W.; Tongtong, L.; Hua, Z. The efficiency of neoadjuvant chemotherapy in colon cancer with mismatch repair deficiency. Cancer Med. 2022, 00, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Chalabi, M.; Fanchi, L.F.; Dijkstra, K.K.; Van den Berg, J.G.; Aalbers, A.G.; Sikorska, K.; Lopez-Yurda, M.; Grootscholten, C.; Beets, G.L.; Snaebjornsson, P.; et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 2020, 26, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Chalabi, M.; Verschoor, Y.L.; van den Berg, J.; Sikorska, K.; Beets, G.; Lent, A.V.; Grootscholten, M.C.; Aalbers, A.; Buller, N.; Marsman, H.; et al. Neoadjuvant immune checkpoint inhibition in locally advanced MMR-deficient colon cancer: The NICHE-2 study. In Proceedings of the 2022 ESMO Congress, Paris, France, 11 September 2022. [Google Scholar]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): Final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022, 23, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kwon, H.C.; Oh, S.Y.; Lee, D.M.; Lee, S.; Lee, J.H.; Roh, M.S.; Kim, D.C.; Park, K.J.; Choi, H.J.; et al. Prognostic value of ERCC1, thymidylate synthase, and glutathione S-transferase pi for 5-FU/oxaliplatin chemotherapy in advanced colorectal cancer. Am. J. Clin. Oncol. 2009, 32, 38–43. [Google Scholar] [CrossRef]
- Noda, E.; Maeda, K.; Inoue, T.; Fukunaga, S.; Nagahara, H.; Shibutani, M.; Amano, R.; Nakata, B.; Tanaka, H.; Muguruma, K.; et al. Predictive value of expression of ERCC 1 and GST-p for 5-fluorouracil/oxaliplatin chemotherapy in advanced colorectal cancer. Hepato-Gastroenterol. 2012, 59, 130–133. [Google Scholar] [CrossRef]
- Huang, M.Y.; Lee, H.H.; Huang, C.W.; Huang, C.M.; Ma, C.J.; Yin, T.C.; Tsai, H.L.; Chai, C.Y.; Chen, Y.T.; Wang, J.Y. ERCC overexpression associated with a poor response of cT4b colorectal cancer with FOLFOX-based neoadjuvant concurrent chemoradiation. Oncol. Lett. 2020, 20, 212. [Google Scholar] [CrossRef]
- Chan, A.K.; Wong, A.; Jenken, D.; Heine, J.; Buie, D.; Johnson, D. Posttreatment TNM staging is a prognostic indicator of survival and recurrence in tethered or fixed rectal carcinoma after preoperative chemotherapy and radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2005, 61, 665–677. [Google Scholar] [CrossRef]
- Qin, Q.; Zhou, A.P.; Yang, L.; Xu, C.; Sun, Y.K.; Zhang, W.; Wang, J.W.; Zhong, D.S. Prognostic and predictive roles of DNA mismatch repair status in colon cancer patients treated with oxaliplatin-based chemotherapy: A retrospective study. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2020, 71, 573–580. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Ou, F.-S.; Zemla, T.; Nixon, A.B.; Mody, K.; Levasseur, A.; Dueck, A.C.; Dhanarajan, A.R.; Lieu, C.H.; Cohen, D.J. Randomized trial of standard chemotherapy alone or combined with atezolizumab as adjuvant therapy for patients with stage III colon cancer and deficient mismatch repair (ATOMIC, Alliance A021502). J. Clin. Oncol. 2019, 37, e15169. [Google Scholar] [CrossRef]
- Lau, D.; Kalaitzaki, E.; Church, D.N.; Pandha, H.; Tomlinson, I.; Annels, N.; Gerlinger, M.; Sclafani, F.; Smith, G.; Begum, R.; et al. Rationale and design of the POLEM trial: Avelumab plus fluoropyrimidine-based chemotherapy as adjuvant treatment for stage III mismatch repair deficient or POLE exonuclease domain mutant colon cancer: A phase III randomised study. ESMO Open 2020, 5, E000638. [Google Scholar] [CrossRef]
- Cohen, R.; Hain, E.; Buhard, O.; Guilloux, A.; Bardier, A.; Kaci, R.; Bertheau, P.; Renaud, F.; Bibeau, F.; Fléjou, J.-F. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019, 5, 551–555. [Google Scholar] [CrossRef]
- Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [CrossRef]
- Roth, A.D.; Tejpar, S.; Delorenzi, M.; Yan, P.; Fiocca, R.; Klingbiel, D.; Dietrich, D.; Biesmans, B.; Bodoky, G.; Barone, C. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010, 28, 466–474. [Google Scholar] [CrossRef]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Foster, N.R.; Thibodeau, S.N.; Marsoni, S.; Monges, G.; Labianca, R.; Kim, G.P.; Yothers, G.; Allegra, C.; Moore, M.J.; et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J. Natl. Cancer Inst. 2011, 103, 863–875. [Google Scholar] [CrossRef]
- Ciardiello, F.; Tortora, G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef]
- Moroni, M.; Veronese, S.; Benvenuti, S.; Marrapese, G.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Gambacorta, M.; Siena, S.; Bardelli, A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 2005, 6, 279–286. [Google Scholar] [CrossRef]
- Bertotti, A.; Papp, E.; Jones, S.; Adleff, V.; Anagnostou, V.; Lupo, B.; Sausen, M.; Phallen, J.; Hruban, C.A.; Tokheim, C.; et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015, 526, 263–267. [Google Scholar] [CrossRef]
- Huang, C.W.; Ma, C.J.; Su, W.C.; Chen, Y.T.; Tsai, H.L.; Yeh, Y.S.; Chang, T.K.; Hsu, W.H.; Yu, F.J.; Wang, J.Y. Prognostic Value of EGFR Expression for Patients With Stage III Colorectal Cancer Receiving Fluoropyrimidine Metronomic Maintenance Therapy After Radical Resection and Adjuvant Oxaliplatin-Based Chemotherapy. Oncol. Res. 2021, 28, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Mireia, G.R.; Gallach, S.; Jantus-Lewintre, E.; Safont, M.J.; Maria Luisa, G.M.; Garde-Noguera, J.; Maria Dolores, R.C.; Blasco, S.; Giner-Bosch, V.; Camps, C. Molecular subtyping of colon cancer (CC) based on mutational status of RAS, BRAF, and DNA mismatch repair (MMR) proteins. Prognostic value. J. Clin. Oncol. 2016, 34, e15094. [Google Scholar] [CrossRef]
- André, T.; De Gramont, A.; Vernerey, D.; Chibaudel, B.; Bonnetain, F.; Tijeras-Raballand, A.; Scriva, A.; Hickish, T.; Tabernero, J.; Van Laethem, J.L.; et al. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: Updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 4176–4187. [Google Scholar]
- Sinicrope, F.A.; Shi, Q.; Allegra, C.J.; Smyrk, T.C.; Thibodeau, S.N.; Goldberg, R.M.; Meyers, J.P.; Pogue-Geile, K.L.; Yothers, G.; Sargent, D.J.; et al. Association of DNA Mismatch Repair and Mutations in BRAF and KRAS With Survival After Recurrence in Stage III Colon Cancers: A Secondary Analysis of 2 Randomized Clinical Trials. JAMA Oncol. 2017, 3, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Taieb, J.; Shi, Q.; Pederson, L.; Alberts, S.; Wolmark, N.; Van Cutsem, E.; de Gramont, A.; Kerr, R.; Grothey, A.; Lonardi, S.; et al. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: Results of an ACCENT pooled analysis of seven studies. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1466–1471. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Church, D.N.; Sieber, O.; Ramamoorthy, R.; Yanagisawa, Y.; Johnstone, E.; Davidson, B.; Kerr, D.J.; Tomlinson, I.P.; Midgley, R. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 4297–4305. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Lochhead, P.; Nishihara, R.; Morikawa, T.; Kuchiba, A.; Yamauchi, M.; Imamura, Y.; Qian, Z.R.; Baba, Y.; Shima, K.; et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N. Engl. J. Med. 2012, 367, 1596–1606. [Google Scholar] [CrossRef]
- Michel, P.; Boige, V.; Andre, T.; Aparicio, T.; Bachet, J.B.; Dahan, L.; Guimbaud, R.; Lepage, C.; Manfredi, S.; Tougeron, D.; et al. Aspirin versus placebo in stage III or high-risk stage II colon cancer with PIK3CA mutation: A French randomised double-blind phase III trial (PRODIGE 50-ASPIK). Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2018, 50, 305–307. [Google Scholar] [CrossRef]
- Gallois, C.; Taieb, J.; Le Corre, D.; Le Malicot, K.; Tabernero, J.; Mulot, C.; Seitz, J.F.; Aparicio, T.; Folprecht, G.; Lepage, C.; et al. Prognostic Value of Methylator Phenotype in Stage III Colon Cancer Treated with Oxaliplatin-based Adjuvant Chemotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 4745–4753. [Google Scholar] [CrossRef]
- Shiovitz, S.; Bertagnolli, M.M.; Renfro, L.A.; Nam, E.; Foster, N.R.; Dzieciatkowski, S.; Luo, Y.; Lao, V.V.; Monnat, R.J., Jr.; Emond, M.J.; et al. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology 2014, 147, 637–645. [Google Scholar] [CrossRef]
- Shibata, D.; Reale, M.A.; Lavin, P.; Silverman, M.; Fearon, E.R.; Steele, G., Jr.; Jessup, J.M.; Loda, M.; Summerhayes, I.C. The DCC protein and prognosis in colorectal cancer. N. Engl. J. Med. 1996, 335, 1727–1732. [Google Scholar] [CrossRef] [PubMed]
- Gal, R.; Sadikov, E.; Sulkes, J.; Klein, B.; Koren, R. Deleted in colorectal cancer protein expression as a possible predictor of response to adjuvant chemotherapy in colorectal cancer patients. Dis. Colon Rectum 2004, 47, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.Y.; Tsai, H.L.; Lin, C.H.; Huang, C.W.; Ma, C.J.; Huang, C.M.; Chai, C.Y.; Wang, J.Y. Predictive value of ERCC1, ERCC2, and XRCC1 overexpression for stage III colorectal cancer patients receiving FOLFOX-4 adjuvant chemotherapy. J. Surg. Oncol. 2013, 108, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.Y.; Tsai, H.L.; Uen, Y.H.; Hu, H.M.; Chen, C.W.; Cheng, T.L.; Lin, S.R.; Wang, J.Y. Circulating tumor cells as a surrogate marker for determining clinical outcome to mFOLFOX chemotherapy in patients with stage III colon cancer. Br. J. Cancer 2013, 108, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.Y.; Tsai, H.L.; Huang, J.J.; Wang, J.Y. Clinical Implications and Future Perspectives of Circulating Tumor Cells and Biomarkers in Clinical Outcomes of Colorectal Cancer. Transl. Oncol. 2016, 9, 340–347. [Google Scholar] [CrossRef]
- Wang, J.Y.; Hsieh, J.S.; Chang, M.Y.; Huang, T.J.; Chen, F.M.; Cheng, T.L.; Alexandersen, K.; Huang, Y.S.; Tzou, W.S.; Lin, S.R. Molecular detection of APC, K- ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J. Surg. 2004, 28, 721–726. [Google Scholar] [CrossRef]
- Larsson, S.C.; Orsini, N.; Wolk, A. Diabetes mellitus and risk of colorectal cancer: A meta-analysis. J. Natl. Cancer Inst. 2005, 97, 1679–1687. [Google Scholar] [CrossRef]
- Huang, C.W.; Sun, L.C.; Shih, Y.L.; Tsai, H.L.; Chen, C.W.; Yeh, Y.S.; Ma, C.J.; Huang, C.J.; Wang, J.Y. The impact on clinical outcome of high prevalence of diabetes mellitus in Taiwanese patients with colorectal cancer. World J. Surg. Oncol. 2012, 10, 76. [Google Scholar] [CrossRef]
- Chuang, J.P.; Lee, J.C.; Leu, T.H.; Hidajah, A.C.; Chang, Y.H.; Li, C.Y. Association of gout and colorectal cancer in Taiwan: A nationwide population-based cohort study. BMJ Open 2019, 9, e028892. [Google Scholar] [CrossRef]
- Ikemura, M.; Hashida, T. Effect of Hyperglycemia on Antitumor Activity and Survival in Tumor-bearing Mice Receiving Oxaliplatin and Fluorouracil. Anticancer Res. 2017, 37, 5463–5468. [Google Scholar] [CrossRef]
- Ahluwalia, A.; Jones, M.K.; Szabo, S.; Tarnawski, A.S. Aberrant, ectopic expression of VEGF and VEGF receptors 1 and 2 in malignant colonic epithelial cells. Implications for these cells growth via an autocrine mechanism. Biochem. Biophys. Res. Commun. 2013, 437, 515–520. [Google Scholar] [CrossRef]
- Lee, J.C.; Chow, N.H.; Wang, S.T.; Huang, S.M. Prognostic value of vascular endothelial growth factor expression in colorectal cancer patients. Eur. J. Cancer 2000, 36, 748–753. [Google Scholar] [CrossRef]
- Ahluwalia, A.; Jones, M.K.; Matysiak-Budnik, T.; Tarnawski, A.S. VEGF and colon cancer growth beyond angiogenesis: Does VEGF directly mediate colon cancer growth via a non-angiogenic mechanism? Curr. Pharm. Des. 2014, 20, 1041–1044. [Google Scholar] [CrossRef]
- Moasser, M.M. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007, 26, 6469–6487. [Google Scholar] [CrossRef]
- Laurent-Puig, P.; Balogoun, R.; Cayre, A.; Le Malicot, K.; Tabernero, J.; Mini, E.; Folprecht, G.; Van Laethem, J.-L.; Thaler, J.; Petersen, L.N. ERBB2 alterations a new prognostic biomarker in stage III colon cancer from a FOLFOX based adjuvant trial (PETACC8). Ann. Oncol. 2016, 27, vi151. [Google Scholar] [CrossRef]
- Richman, S.D.; Southward, K.; Chambers, P.; Cross, D.; Barrett, J.; Hemmings, G.; Taylor, M.; Wood, H.; Hutchins, G.; Foster, J.M. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: Analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J. Pathol. 2016, 238, 562–570. [Google Scholar] [CrossRef]
- Kermorgant, S.; Zicha, D.; Parker, P.J. PKC controls HGF-dependent c-Met traffic, signalling and cell migration. EMBO J. 2004, 23, 3721–3734. [Google Scholar] [CrossRef]
- Li, F.; Zhu, Y.T. HGF-activated colonic fibroblasts mediates carcinogenesis of colonic epithelial cancer cells via PKC-cMET-ERK1/2-COX-2 signaling. Cell. Signal. 2015, 27, 860–866. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, J.; Park, S.H.; Park, J.O.; Lim, H.Y.; Kang, W.K.; Park, Y.S.; Kim, S.T. c-MET Overexpression in Colorectal Cancer: A Poor Prognostic Factor for Survival. Clin. Color. Cancer 2018, 17, 165–169. [Google Scholar] [CrossRef]
- Gao, H.; Guan, M.; Sun, Z.; Bai, C. High c-Met expression is a negative prognostic marker for colorectal cancer: A meta-analysis. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 515–520. [Google Scholar] [CrossRef]
- Xue, M.; Zhuo, Y.; Shan, B. MicroRNAs, Long Noncoding RNAs, and Their Functions in Human Disease. Methods Mol. Biol. 2017, 1617, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Z.; Wang, N.; Zhang, M.; Zeng, X.; Zhao, W. MicroRNA-1299 is a negative regulator of STAT3 in colon cancer. Oncol. Rep. 2017, 37, 3227–3234. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Huang, T.; Guo, H.; Yang, J.; Li, M.; Chen, Z.; Zhang, Y. LncRNA CASC15 promotes colon cancer cell proliferation and metastasis by regulating the miR4310/LGR5/Wnt/betacatenin signaling pathway. Mol. Med. Rep. 2018, 18, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bian, Y.; Zhu, Y.; Wan, L.; Kong, L.; Hu, J.; Yang, M.; Li, L.; Liu, B.; Qian, X. Integrative analysis and validation of dysregulated long non-coding RNAs in colon cancer. J. Cell. Mol. Med. 2020, 24, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.M.; Tsai, H.L.; Chen, Y.C.; Huang, C.W.; Li, C.C.; Su, W.C.; Chang, T.K.; Yeh, Y.S.; Chen, P.J.; Huang, M.Y.; et al. Role of non-coding RNAs in radiosensitivity of colorectal cancer: A narrative review. Front. Oncol. 2022, 12, 889658. [Google Scholar] [CrossRef]
- Schetter, A.J.; Okayama, H.; Harris, C.C. The role of microRNAs in colorectal cancer. Cancer J. 2012, 18, 244–252. [Google Scholar] [CrossRef]
- Lopez-Urrutia, E.; Bustamante Montes, L.P.; Ladron de Guevara Cervantes, D.; Perez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Nakajima, G.; Hayashi, K.; Xi, Y.; Kudo, K.; Uchida, K.; Takasaki, K.; Yamamoto, M.; Ju, J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics 2006, 3, 317–324. [Google Scholar]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef]
- Chang, C.M.; Wong, H.S.; Huang, C.Y.; Hsu, W.L.; Maio, Z.F.; Chiu, S.J.; Tsai, Y.T.; Chen, B.K.; Wan, Y.Y.; Wang, J.Y.; et al. Functional Effects of let-7g Expression in Colon Cancer Metastasis. Cancers 2019, 11, 489. [Google Scholar] [CrossRef]
- Kim, J.K.; Qu, X.; Chen, C.T.; Smith, J.J.; Sanchez-Vega, F.; Garcia-Aguilar, J. Identifying Diagnostic MicroRNAs and Investigating Their Biological Implications in Rectal Cancer. JAMA Netw. Open 2021, 4, e2136913. [Google Scholar] [CrossRef]
- Song, B.; Wang, Y.; Xi, Y.; Kudo, K.; Bruheim, S.; Botchkina, G.I.; Gavin, E.; Wan, Y.; Formentini, A.; Kornmann, M.; et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 2009, 28, 4065–4074. [Google Scholar] [CrossRef]
- Song, B.; Wang, Y.; Titmus, M.A.; Botchkina, G.; Formentini, A.; Kornmann, M.; Ju, J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. Cancer 2010, 9, 96. [Google Scholar] [CrossRef]
- Chai, J.; Dong, W.; Xie, C.; Wang, L.; Han, D.L.; Wang, S.; Guo, H.L.; Zhang, Z.L. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life 2015, 67, 191–201. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Takahashi, Y.; Sawada, G.; Kurashige, J.; Uchi, R.; Matsumura, T.; Ueo, H.; Takano, Y.; Eguchi, H.; Sudo, T.; Sugimachi, K.; et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br. J. Cancer 2014, 110, 164–171. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, X.; Pu, C.; Song, X. Upregulated plasmacytoma variant translocation 1 promotes cell proliferation, invasion and metastasis in colorectal cancer. Mol. Med. Rep. 2018, 17, 6598–6604. [Google Scholar] [CrossRef]
- Fan, H.; Zhu, J.H.; Yao, X.Q. Long non-coding RNA PVT1 as a novel potential biomarker for predicting the prognosis of colorectal cancer. Int. J. Biol. Mrk. 2018, 33, 415–422. [Google Scholar] [CrossRef]
- Yang, X.D.; Xu, H.T.; Xu, X.H.; Ru, G.; Liu, W.; Zhu, J.J.; Wu, Y.Y.; Zhao, K.; Wu, Y.; Xing, C.G.; et al. Knockdown of long non-coding RNA HOTAIR inhibits proliferation and invasiveness and improves radiosensitivity in colorectal cancer. Oncol. Rep. 2016, 35, 479–487. [Google Scholar] [CrossRef]
- Wang, G.; Li, Z.; Zhao, Q.; Zhu, Y.; Zhao, C.; Li, X.; Ma, Z.; Zhang, Y. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/beta-catenin signaling pathway. Oncol. Rep. 2014, 31, 1839–1845. [Google Scholar] [CrossRef] [PubMed]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; Orlando, G.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10, 2154. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Sakoda, L.C.; Hoffmeister, M.; Rosenthal, E.A.; Lee, J.K.; van Duijnhoven, F.J.B.; Platz, E.A.; Wu, A.H.; Dampier, C.H.; de la Chapelle, A.; et al. Genome-wide Modeling of Polygenic Risk Score in Colorectal Cancer Risk. Am. J. Hum. Genet. 2020, 107, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Puzio-Kuter, A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef] [PubMed]
- Amir Hashim, N.A.; Ab-Rahim, S.; Wan Ngah, W.Z.; Nathan, S.; Ab Mutalib, N.S.; Sagap, I.; AR, A.J.; Mazlan, M. Global metabolomics profiling of colorectal cancer in Malaysian patients. BioImpacts BI 2021, 11, 33–43. [Google Scholar] [CrossRef]
- Ogino, S.; Nosho, K.; Kirkner, G.J.; Kawasaki, T.; Chan, A.T.; Schernhammer, E.S.; Giovannucci, E.L.; Fuchs, C.S. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl. Cancer Inst. 2008, 100, 1734–1738. [Google Scholar] [CrossRef]
- Kim, M.S.; Lee, J.; Sidransky, D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010, 29, 181–206. [Google Scholar] [CrossRef]
- Lou, Y.T.; Chen, C.W.; Fan, Y.C.; Chang, W.C.; Lu, C.Y.; Wu, I.C.; Hsu, W.H.; Huang, C.W.; Wang, J.Y. LINE-1 Methylation Status Correlates Significantly to Post-Therapeutic Recurrence in Stage III Colon Cancer Patients Receiving FOLFOX-4 Adjuvant Chemotherapy. PLoS ONE 2014, 10, e0123973. [Google Scholar] [CrossRef]
- Grothey, A.; Sobrero, A.F.; Shields, A.F.; Yoshino, T.; Paul, J.; Taieb, J.; Souglakos, J.; Shi, Q.; Kerr, R.; Labianca, R.; et al. Duration of Adjuvant Chemotherapy for Stage III Colon Cancer. N. Engl. J. Med. 2018, 378, 1177–1188. [Google Scholar] [CrossRef]
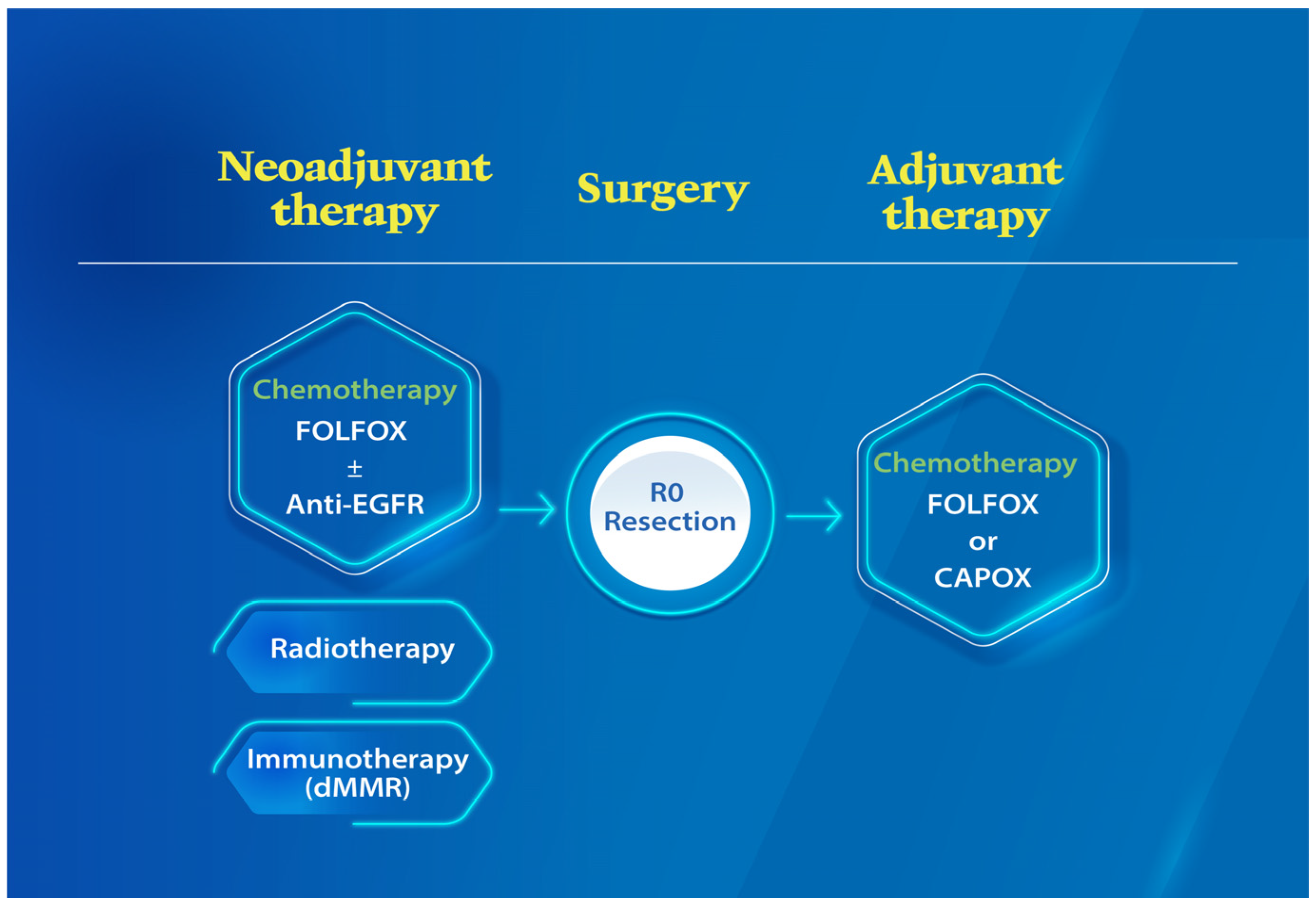
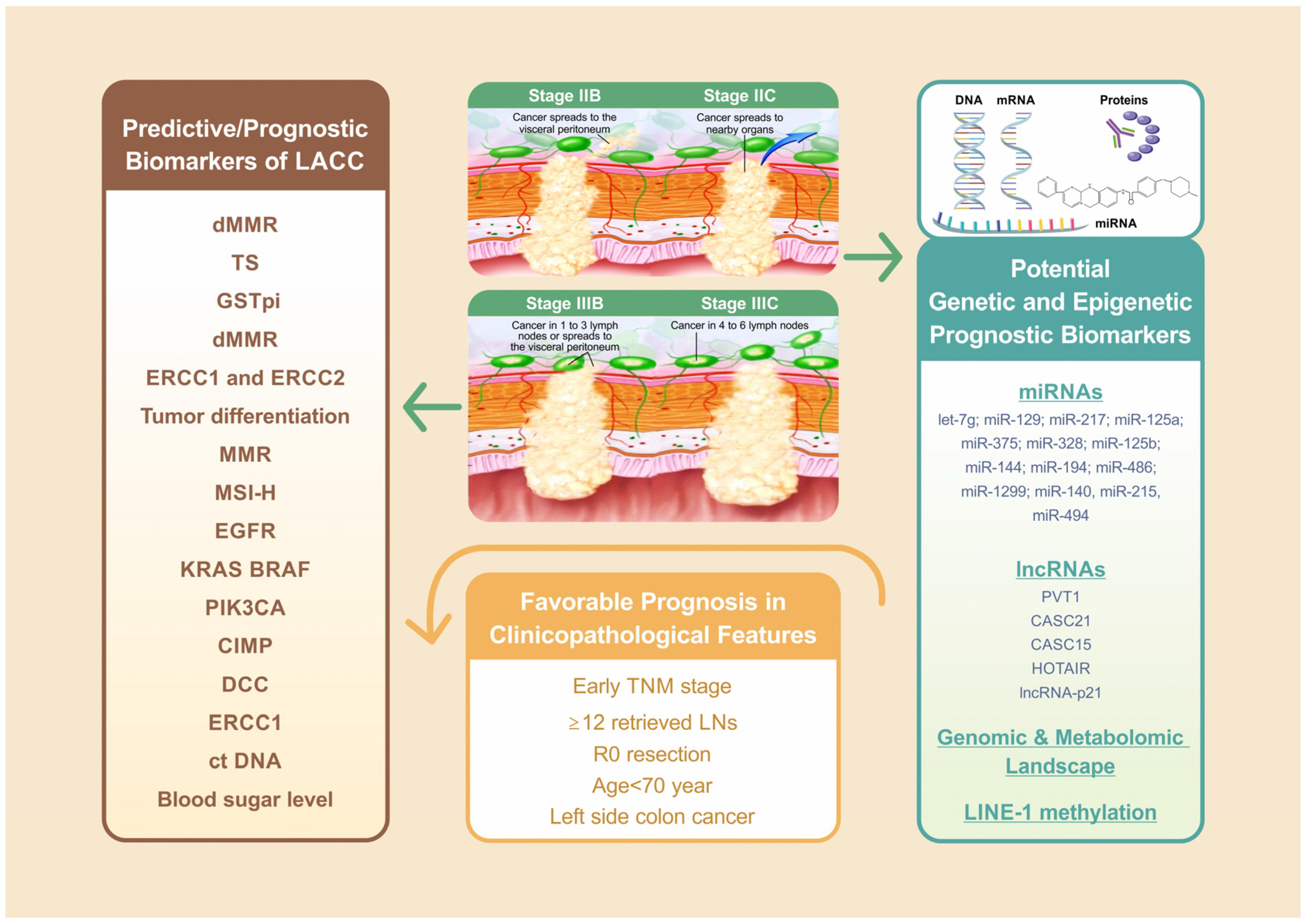
| LACC Treatment Phases | Biomarkers (Predictive/Prognostic) | Prediction Value (Favorable/Worse) | References |
|---|---|---|---|
| Clinicopathological features | TNM staging (Prognostic) | Favorable (early TNM) | [4,9] |
| ≥12 retrieved LNs (Prognostic) | Favorable | [54] | |
| R0 resection (Prognostic) | Favorable | [4,24] | |
| Age ≥ 70 year (Prognostic) | Worse | [4] | |
| Sidedness of colon cancer (Prognostic) | Favorable (Left side colon cancer) | [55] | |
| Neoadjuvant chemotherapy | dMMR (Predictive/Prognostic) | Worse | [28,62] |
| ERCC1 (Predictive/Prognostic), TS (Prognostic), GSTpi (Predictive) | Worse | [68,69] | |
| Neoadjuvant Immunotherapy | dMMR (Predictive) | Favorable | [65] |
| Neoadjuvant Chemoradiotherapy | ERCC1 and ERCC2 (Predictive) | Worse | [70] |
| Tumor differentiation (Prognostic) | Worse (Low differentiation) | [39] | |
| Postoperative adjuvant chemotherapy | MMR (Prognostic) | Controversial | [50,72,73,74,86,87] |
| MSI-H (Prognostic) | Favorable | [19,48,49,78,79,87] | |
| EGFR (Prognostic) | Worse | [17,83] | |
| KRAS and BRAF(Prognostic) | Worse | [49,86,87] | |
| PIK3CA (Prognostic) 1 | Favorable | [90] | |
| CIMP (Prognostic) | Worse | [91,92] | |
| DCC (Prognostic) | Favorable | [93,94] | |
| ERCC1 (Prognostic) | Worse | [95] | |
| ct DNA (Prognostic) | Worse | [51,96,97,98] | |
| Blood sugar level (Prognostic) | Worse (Fasting blood sugar ≥ 126 mg/dL) | [52] |
| Category | Biomarkers | Predictive/Prognostic Value | References |
|---|---|---|---|
| Molecular Factors | VEGF | Associated with advanced lymph node status and distant metastasis | [103,104,105] |
| HER2 | Negative predictive biomarker in metastatic CRC | [106,107,108] | |
| HGF & c-Met | Poor survival and shorter PFS during bevacizumab treatment in patients with stage IV CRC | [110,111,112] | |
| miRNAs | let-7g | Poor chemo-response and disease progression of CRC; migration, invasion in CRC cell lines | [121,123] |
| miR-129; miR-217; miR-125a; miR-375; miR-328; miR-125b; miR-144; miR-194; miR-486 | Overall survival in colon cancer (Prognostic) | [116] | |
| miR-1299 | TNM staging; colon cancer prognosis (Prognostic) | [114] | |
| miR-140, miR-215, miR-494 | Chemo-resistance in colon cell lines | [125,126,127] | |
| lncRNAs | PVT1 | Overall survival of CRC patients; proliferation and invasion capabilities in CRC cell lines; tumor differentiation, TNM staging, lymphatic node metastasis (Prognostic) | [129,130,131] |
| CASC21 | TNM staging (Prognostic) | [116] | |
| CASC15 | TNM staging (Prognostic) | [115] | |
| HOTAIR | Radiosensitivity of CRC cells | [132] | |
| lncRNA-p21 | CRC radiosensitivity | [133] | |
| Genomic& Metabolomic Landscape | PRS from GWASs (KRAS, NRAS, BRAF, MSI, Her-2 etc.) | Early detection for CRC | [134,135] |
| Blood metabolite levels profile | Early detection for CRC; differentiate left-side CRC and right-side CRC | [57,136,137] | |
| LINE-1 | Degree of LINE-1 methylation | OS in early-onset CRC patients; survival in colon cancer; postoperative recurrence and DFS (Prognostic) | [42,138,140] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, J.-P.; Tsai, H.-L.; Chen, P.-J.; Chang, T.-K.; Su, W.-C.; Yeh, Y.-S.; Huang, C.-W.; Wang, J.-Y. Comprehensive Review of Biomarkers for the Treatment of Locally Advanced Colon Cancer. Cells 2022, 11, 3744. https://doi.org/10.3390/cells11233744
Chuang J-P, Tsai H-L, Chen P-J, Chang T-K, Su W-C, Yeh Y-S, Huang C-W, Wang J-Y. Comprehensive Review of Biomarkers for the Treatment of Locally Advanced Colon Cancer. Cells. 2022; 11(23):3744. https://doi.org/10.3390/cells11233744
Chicago/Turabian StyleChuang, Jen-Pin, Hsiang-Lin Tsai, Po-Jung Chen, Tsung-Kun Chang, Wei-Chih Su, Yung-Sung Yeh, Ching-Wen Huang, and Jaw-Yuan Wang. 2022. "Comprehensive Review of Biomarkers for the Treatment of Locally Advanced Colon Cancer" Cells 11, no. 23: 3744. https://doi.org/10.3390/cells11233744
APA StyleChuang, J.-P., Tsai, H.-L., Chen, P.-J., Chang, T.-K., Su, W.-C., Yeh, Y.-S., Huang, C.-W., & Wang, J.-Y. (2022). Comprehensive Review of Biomarkers for the Treatment of Locally Advanced Colon Cancer. Cells, 11(23), 3744. https://doi.org/10.3390/cells11233744






