A Novel P53 Nanomedicine Reduces Immunosuppression and Augments Anti-PD-1 Therapy for Non-Small Cell Lung Cancer in Syngeneic Mouse Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Nanocomplex Preparation
2.3. Transfection
2.4. Animal Studies
2.5. ATP Assay
2.6. HMGB1 Assay
2.7. Gal-1 Immunoassay
2.8. Immunohistochemistry
2.9. Flow Cytometry
2.10. RT-qPCR
2.11. Detection of miR-22
2.12. Bioluminescence Imaging
2.13. Transcriptome Analysis
2.14. Patient Survival Data Analysis
2.15. Statistical Analysis
3. Results
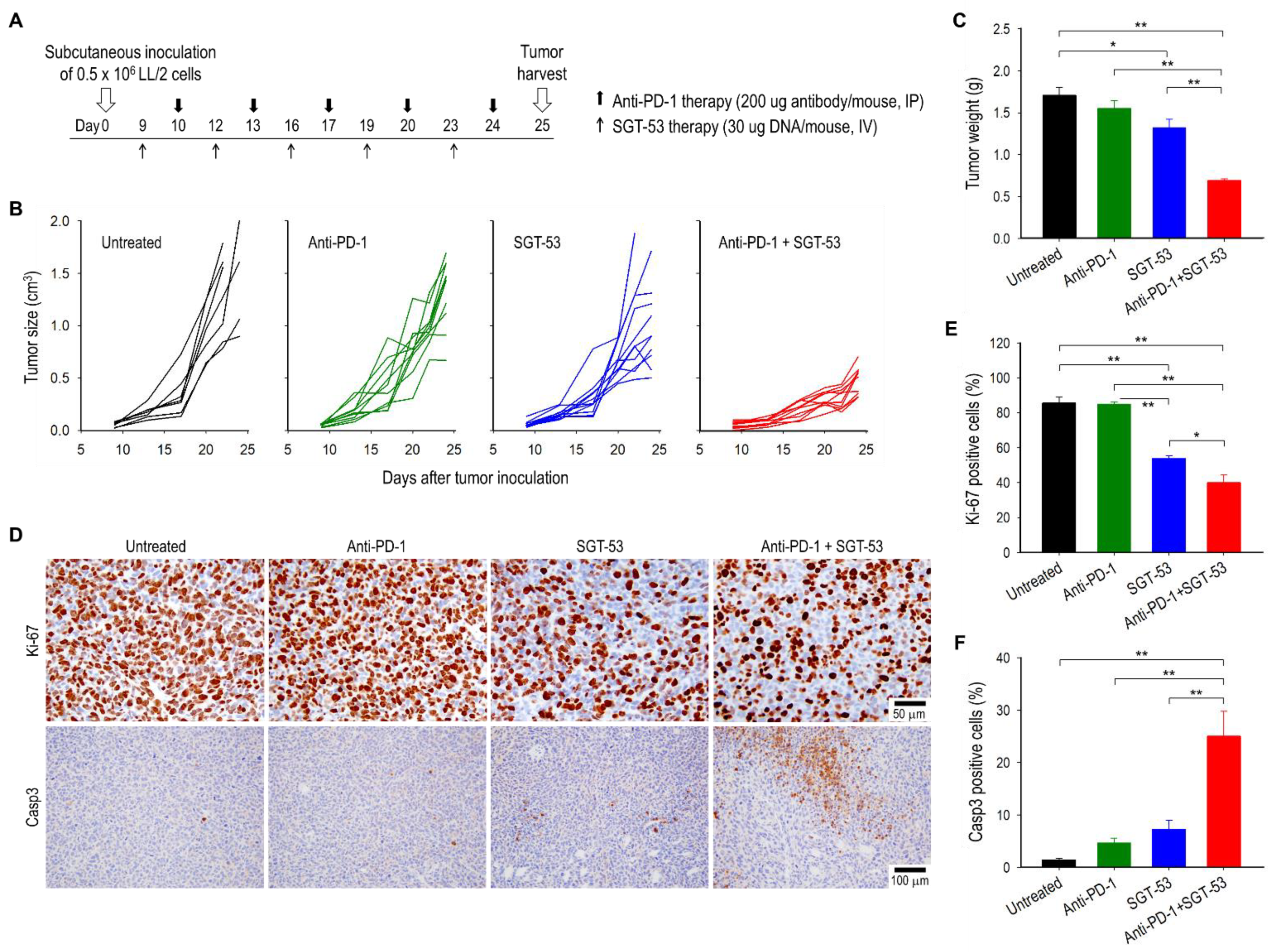
3.1. Combining Anti-PD-1 and SGT-53 Inhibits LL/2 Tumor Growth
3.2. Combining Anti-PD-1 and SGT-53 Improves Survival in a Metastatic LL/2 Tumor Model
3.3. Combining Anti-PD-1 and SGT-53 Enhances Host Immune Responses
3.4. SGT-53 Increases Immunogenicity of Tumor Cells
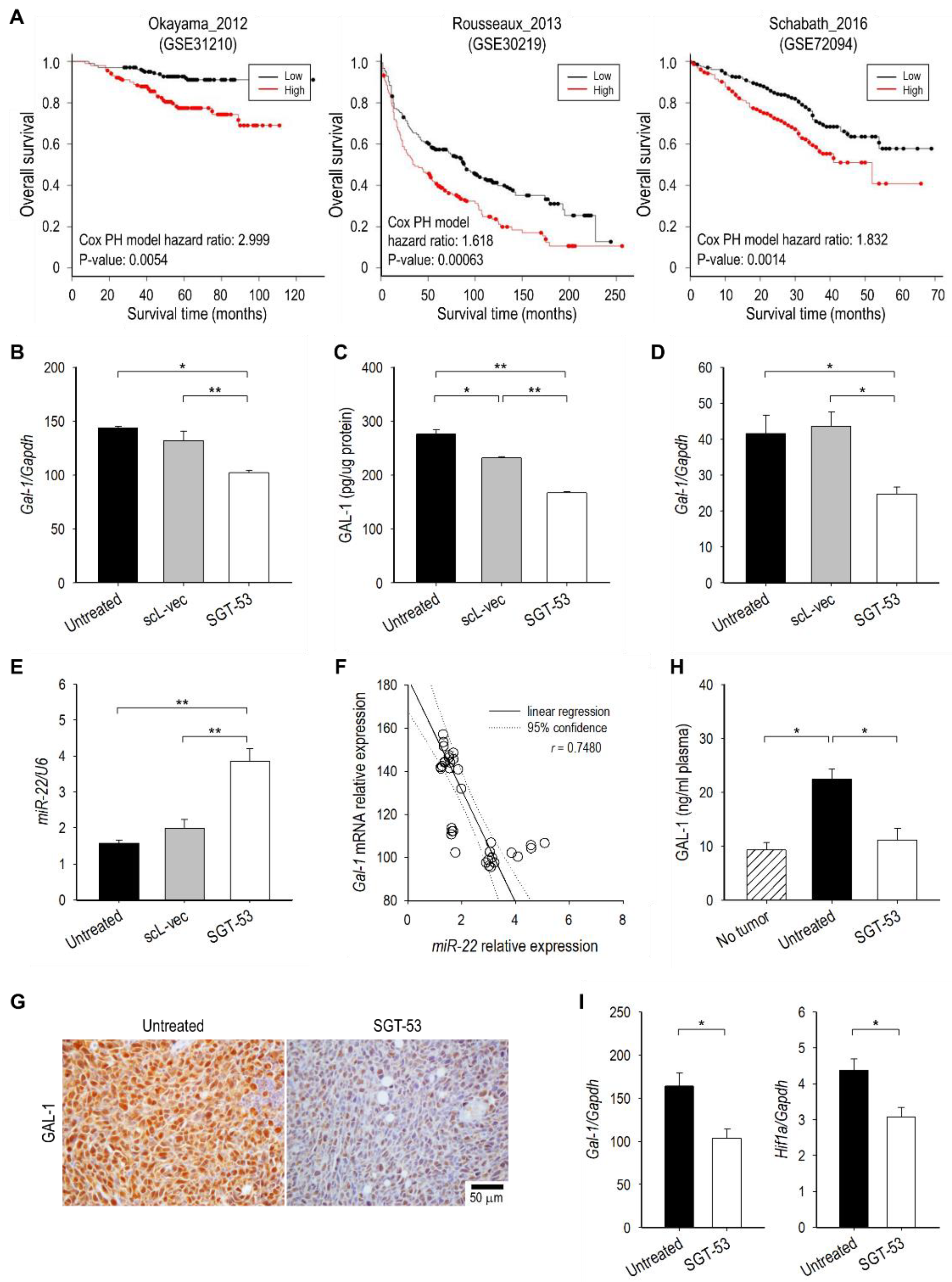
3.5. SGT-53 Represses Immunosuppressive Gal-1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Califano, R.; Gomes, F.; Ackermann, C.J.; Rafee, S.; Tsakonas, G.; Ekman, S. Immune checkpoint blockade for non-small cell lung cancer: What is the role in the special populations? Eur. J. Cancer 2019, 125, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.G.; Shih, J.Y. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol. Cancer 2018, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Dempke, W.C.M.; Fenchel, K.; Dale, S.P. Programmed cell death ligand-1 (PD-L1) as a biomarker for non-small cell lung cancer (NSCLC) treatment-are we barking up the wrong tree? Transl. Lung Cancer Res. 2018, 7, S275–S279. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Smyth, M.J.; Teng, M.W. Acquired resistance to anti-PD1 therapy: Checkmate to checkpoint blockade? Genome Med. 2016, 8, 111. [Google Scholar] [CrossRef]
- Ott, P.A.; Hodi, F.S.; Kaufman, H.L.; Wigginton, J.M.; Wolchok, J.D. Combination immunotherapy: A road map. J. Immunother. Cancer 2017, 5, 16. [Google Scholar] [CrossRef]
- Kim, S.S.; Rait, A.; Kim, E.; Pirollo, K.F.; Nishida, M.; Farkas, N.; Dagata, J.A.; Chang, E.H. A nanoparticle carrying the p53 gene targets tumors including cancer stem cells, sensitizes glioblastoma to chemotherapy and improves survival. ACS Nano 2014, 8, 5494–5514. [Google Scholar] [CrossRef]
- Kong, N.; Tao, W.; Ling, X.; Wang, J.; Xiao, Y.; Shi, S.; Ji, X.; Shajii, A.; Gan, S.T.; Kim, N.Y.; et al. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci. Transl. Med. 2019, 11, eaaw1565. [Google Scholar] [CrossRef]
- Cui, Y.; Guo, G. Immunomodulatory Function of the Tumor Suppressor p53 in Host Immune Response and the Tumor Microenvironment. Int. J. Mol. Sci. 2016, 17, 1942. [Google Scholar] [CrossRef]
- Blagih, J.; Zani, F.; Chakravarty, P.; Hennequart, M.; Pilley, S.; Hobor, S.; Hock, A.K.; Walton, J.B.; Morton, J.P.; Gronroos, E.; et al. Cancer-Specific Loss of p53 Leads to a Modulation of Myeloid and T Cell Responses. Cell Rep. 2020, 30, 481–496.e6. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.Y.; Tang, S.J.; Sun, G.H.; Chou, T.Y.; Yeh, T.S.; Yu, S.L.; Sun, K.H. Galectin-1 promotes lung cancer progression and chemoresistance by upregulating p38 MAPK, ERK, and cyclooxygenase-2. Clin. Cancer Res. 2012, 18, 4037–4047. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, C.C.; Huang, W.; Tang, W.H.; Rait, A.; Yin, Y.Z.; Cruz, I.; Xiang, L.M.; Pirollo, K.F.; Chang, E.H. Systemic tumor-targeted gene delivery by anti-transferrin receptor scFv-immunoliposomes. Mol. Cancer Ther. 2002, 1, 337–346. [Google Scholar] [PubMed]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2013. [Google Scholar]
- Cai, L.; Lin, S.; Girard, L.; Zhou, Y.; Yang, L.; Ci, B.; Zhou, Q.; Luo, D.; Yao, B.; Tang, H.; et al. LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2019, 38, 2551–2564. [Google Scholar] [CrossRef] [PubMed]
- Kohli, K.; Pillarisetty, V.G.; Kim, T.S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2022, 29, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Chow, M.T.; Ozga, A.J.; Servis, R.L.; Frederick, D.T.; Lo, J.A.; Fisher, D.E.; Freeman, G.J.; Boland, G.M.; Luster, A.D. Intratumoral Activity of the CXCR3 Chemokine System Is Required for the Efficacy of Anti-PD-1 Therapy. Immunity 2019, 50, 1498–1512.e5. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Tsai, Y.T.; Liang, C.H.; Yu, J.H.; Huang, K.C.; Tung, C.H.; Wu, J.E.; Wu, Y.Y.; Chang, C.H.; Hong, T.M.; Chen, Y.L. A DNA Aptamer Targeting Galectin-1 as a Novel Immunotherapeutic Strategy for Lung Cancer. Mol. Nucleic Acids 2019, 18, 991–998. [Google Scholar] [CrossRef]
- Rubinstein, N.; Alvarez, M.; Zwirner, N.W.; Toscano, M.A.; Ilarregui, J.M.; Bravo, A.; Mordoh, J.; Fainboim, L.; Podhajcer, O.L.; Rabinovich, G.A. Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection; A potential mechanism of tumor-immune privilege. Cancer Cell 2004, 5, 241–251. [Google Scholar] [CrossRef]
- Nambiar, D.K.; Aguilera, T.; Cao, H.; Kwok, S.; Kong, C.; Bloomstein, J.; Wang, Z.; Rangan, V.S.; Jiang, D.; von Eyben, R.; et al. Galectin-1-driven T cell exclusion in the tumor endothelium promotes immunotherapy resistance. J. Clin. Investig. 2019, 129, 5553–5567. [Google Scholar] [CrossRef]
- You, Y.; Tan, J.X.; Dai, H.S.; Chen, H.W.; Xu, X.J.; Yang, A.G.; Zhang, Y.J.; Bai, L.H.; Bie, P. MiRNA-22 inhibits oncogene galectin-1 in hepatocellular carcinoma. Oncotarget 2016, 7, 57099–57116. [Google Scholar] [CrossRef] [PubMed]
- White, N.M.; Masui, O.; Newsted, D.; Scorilas, A.; Romaschin, A.D.; Bjarnason, G.A.; Siu, K.W.; Yousef, G.M. Galectin-1 has potential prognostic significance and is implicated in clear cell renal cell carcinoma progression through the HIF/mTOR signaling axis. Br. J. Cancer 2014, 110, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Chen, T.T.; Xia, L.; Guo, M.; Xu, Y.; Yue, F.; Jiang, Y.; Chen, G.Q.; Zhao, K.W. Hypoxia inducible factor-1 mediates expression of galectin-1: The potential role in migration/invasion of colorectal cancer cells. Carcinogenesis 2010, 31, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Ajina, R.; Zahavi, D.J.; Zhang, Y.W.; Weiner, L.M. Overcoming malignant cell-based mechanisms of resistance to immune checkpoint blockade antibodies. Semin. Cancer Biol. 2020, 65, 28–37. [Google Scholar] [CrossRef]
- Malhotra, J.; Jabbour, S.K.; Aisner, J. Current state of immunotherapy for non-small cell lung cancer. Transl. Lung Cancer Res. 2017, 6, 196–211. [Google Scholar] [CrossRef]
- Moya-Horno, I.; Viteri, S.; Karachaliou, N.; Rosell, R. Combination of immunotherapy with targeted therapies in advanced non-small cell lung cancer (NSCLC). Ther. Adv. Med. Oncol. 2018, 10, 1758834017745012. [Google Scholar] [CrossRef]
- Kim, S.S.; Rait, A.; Rubab, F.; Rao, A.K.; Kiritsy, M.C.; Pirollo, K.F.; Wang, S.; Weiner, L.M.; Chang, E.H. The clinical potential of targeted nanomedicine: Delivering to cancer stem-like cells. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 278–291. [Google Scholar] [CrossRef]
- Senzer, N.; Nemunaitis, J.; Nemunaitis, D.; Bedell, C.; Edelman, G.; Barve, M.; Nunan, R.; Pirollo, K.F.; Rait, A.; Chang, E.H. Phase I Study of a Systemically Delivered p53 Nanoparticle in Advanced Solid Tumors. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 1096–1103. [Google Scholar] [CrossRef]
- Pirollo, K.F.; Nemunaitis, J.; Leung, P.K.; Nunan, R.; Adams, J.; Chang, E.H. Safety and Efficacy in Advanced Solid Tumors of a Targeted Nanocomplex Carrying the p53 Gene Used in Combination with Docetaxel: A Phase 1b Study. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 1697–1706. [Google Scholar] [CrossRef]
- Leung, C.P.; Barve, M.A.; Wu, M.-S.; Pirollo, K.F.; Strauss, J.F.; Liao, W.-C.; Yang, S.-H.; Nunan, R.A.; Adams, J.; Harford, J.B.; et al. A phase II trial combining tumor-targeting TP53 gene therapy with gemcitabine/nab-paclitaxel as a second-line treatment for metastatic pancreatic cancer. J. Clin. Oncol. 2021, 39, 4139. [Google Scholar] [CrossRef]
- Gudkov, A.V.; Komarova, E.A. Pathologies associated with the p53 response. Cold Spring Harb. Perspect. Biol. 2010, 2, a001180. [Google Scholar] [CrossRef]
- Wellenstein, M.D.; Coffelt, S.B.; Duits, D.E.M.; van Miltenburg, M.H.; Slagter, M.; de Rink, I.; Henneman, L.; Kas, S.M.; Prekovic, S.; Hau, C.S.; et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 2019, 572, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.C.; Sun, L.; Clavijo, P.E.; Friedman, J.; Harford, J.B.; Saleh, A.D.; Van Waes, C.; Chang, E.H.; Allen, C.T. Nanocomplex-based TP53 gene therapy promotes anti-tumor immunity through TP53- and STING-dependent mechanisms. Oncoimmunology 2018, 7, e1404216. [Google Scholar] [CrossRef]
- Kim, S.S.; Harford, J.B.; Moghe, M.; Rait, A.; Chang, E.H. Combination with SGT-53 overcomes tumor resistance to a checkpoint inhibitor. Oncoimmunology 2018, 7, e1484982. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Harford, J.B.; Moghe, M.; Slaughter, T.; Doherty, C.; Chang, E.H. A tumor-targeting nanomedicine carrying the p53 gene crosses the blood-brain barrier and enhances anti-PD-1 immunotherapy in mouse models of glioblastoma. Int. J. Cancer 2019, 145, 2535–2546. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, R.; Cadieux, E.L.; Salgado, R.; Bakir, M.A.; Moore, D.A.; Hiley, C.T.; Lund, T.; Tanic, M.; Reading, J.L.; Joshi, K.; et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 2019, 567, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Niu, D.; Lai, L.; Ren, E.C. p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat. Commun. 2013, 4, 2359. [Google Scholar] [CrossRef]
- Li, L.; Ng, D.S.; Mah, W.C.; Almeida, F.F.; Rahmat, S.A.; Rao, V.K.; Leow, S.C.; Laudisi, F.; Peh, M.T.; Goh, A.M.; et al. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ. 2015, 22, 1081–1093. [Google Scholar] [CrossRef]
- Kawashima, H.; Takatori, H.; Suzuki, K.; Iwata, A.; Yokota, M.; Suto, A.; Minamino, T.; Hirose, K.; Nakajima, H. Tumor suppressor p53 inhibits systemic autoimmune diseases by inducing regulatory T cells. J. Immunol. 2013, 191, 3614–3623. [Google Scholar] [CrossRef]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar] [CrossRef]
- Fu, S.; Zhang, N.; Yopp, A.C.; Chen, D.; Mao, M.; Zhang, H.; Ding, Y.; Bromberg, J.S. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25—Precursors. Am. J. Transplant. 2004, 4, 1614–1627. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Suano, A.; Hamilton, A.B.; Betz, A.G. Gimme shelter: The immune system during pregnancy. Immunol. Rev. 2011, 241, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Camby, I.; Le Mercier, M.; Lefranc, F.; Kiss, R. Galectin-1: A small protein with major functions. Glycobiology 2006, 16, 137R–157R. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Wu, C.Y.; Hung, J.Y.; Lin, Y.S.; Huang, M.S.; Kuo, P.L. Galectin-1 promotes lung cancer tumor metastasis by potentiating integrin alpha6beta4 and Notch1/Jagged2 signaling pathway. Carcinogenesis 2013, 34, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Puchades, M.; Nilsson, C.L.; Emmett, M.R.; Aldape, K.D.; Ji, Y.; Lang, F.F.; Liu, T.J.; Conrad, C.A. Proteomic investigation of glioblastoma cell lines treated with wild-type p53 and cytotoxic chemotherapy demonstrates an association between galectin-1 and p53 expression. J. Proteome Res. 2007, 6, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Girotti, M.R.; Salatino, M.; Dalotto-Moreno, T.; Rabinovich, G.A. Sweetening the hallmarks of cancer: Galectins as multifunctional mediators of tumor progression. J. Exp. Med. 2020, 217, e20182041. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Huo, R.; Xiao, L.; Zhu, X.; Xie, J.; Sun, S.; He, Y.; Zhang, J.; Sun, Y.; Zhou, Z.; et al. A novel p53/microRNA-22/Cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritis. Arthritis Rheumatol. 2014, 66, 49–59. [Google Scholar] [CrossRef]
- Leung, Z.; Ko, F.C.F.; Tey, S.K.; Kwong, E.M.L.; Mao, X.; Liu, B.H.M.; Ma, A.P.Y.; Fung, Y.M.E.; Che, C.M.; Wong, D.K.H.; et al. Galectin-1 promotes hepatocellular carcinoma and the combined therapeutic effect of OTX008 galectin-1 inhibitor and sorafenib in tumor cells. J. Exp. Clin. Cancer Res. CR 2019, 38, 423. [Google Scholar] [CrossRef]
- Qiu, B.Q.; Zhang, P.F.; Xiong, D.; Xu, J.J.; Long, X.; Zhu, S.Q.; Ye, X.D.; Wu, Y.; Pei, X.; Zhang, X.M.; et al. CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non-small cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. J. Cell Physiol. 2019, 234, 11256–11264. [Google Scholar] [CrossRef]
- Ravi, R.; Mookerjee, B.; Bhujwalla, Z.M.; Sutter, C.H.; Artemov, D.; Zeng, Q.; Dillehay, L.E.; Madan, A.; Semenza, G.L.; Bedi, A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000, 14, 34–44. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Yagi, S.; Ito, T.; Lowenstein, C.J. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS ONE 2011, 6, e20291. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-S.; Harford, J.B.; Moghe, M.; Doherty, C.; Chang, E.H. A Novel P53 Nanomedicine Reduces Immunosuppression and Augments Anti-PD-1 Therapy for Non-Small Cell Lung Cancer in Syngeneic Mouse Models. Cells 2022, 11, 3434. https://doi.org/10.3390/cells11213434
Kim S-S, Harford JB, Moghe M, Doherty C, Chang EH. A Novel P53 Nanomedicine Reduces Immunosuppression and Augments Anti-PD-1 Therapy for Non-Small Cell Lung Cancer in Syngeneic Mouse Models. Cells. 2022; 11(21):3434. https://doi.org/10.3390/cells11213434
Chicago/Turabian StyleKim, Sang-Soo, Joe B. Harford, Manish Moghe, Caroline Doherty, and Esther H. Chang. 2022. "A Novel P53 Nanomedicine Reduces Immunosuppression and Augments Anti-PD-1 Therapy for Non-Small Cell Lung Cancer in Syngeneic Mouse Models" Cells 11, no. 21: 3434. https://doi.org/10.3390/cells11213434
APA StyleKim, S.-S., Harford, J. B., Moghe, M., Doherty, C., & Chang, E. H. (2022). A Novel P53 Nanomedicine Reduces Immunosuppression and Augments Anti-PD-1 Therapy for Non-Small Cell Lung Cancer in Syngeneic Mouse Models. Cells, 11(21), 3434. https://doi.org/10.3390/cells11213434






