Identification of SLC3A2 as a Potential Therapeutic Target of Osteoarthritis Involved in Ferroptosis by Integrating Bioinformatics, Clinical Factors and Experiments
Abstract
1. Background
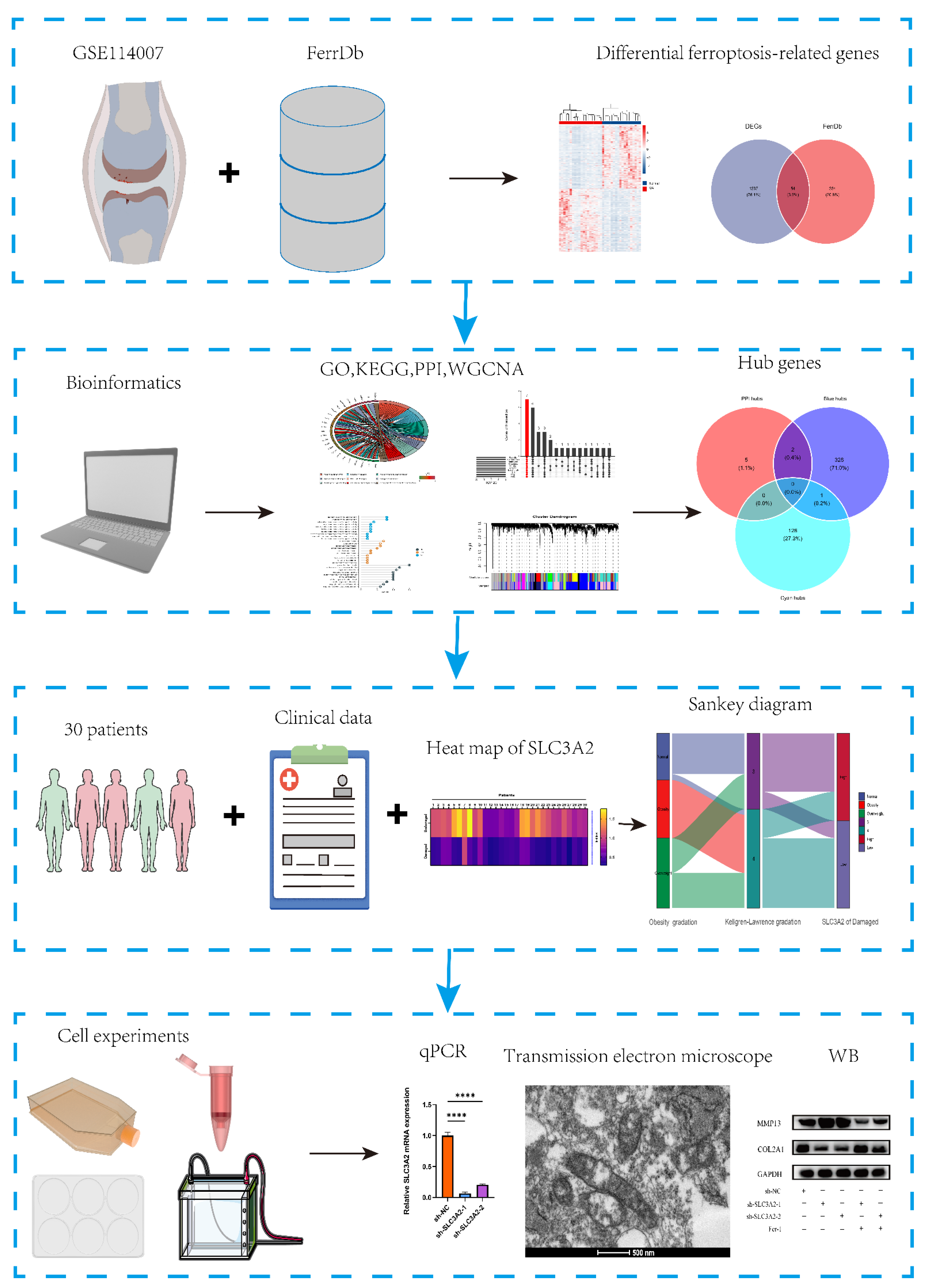
2. Methods
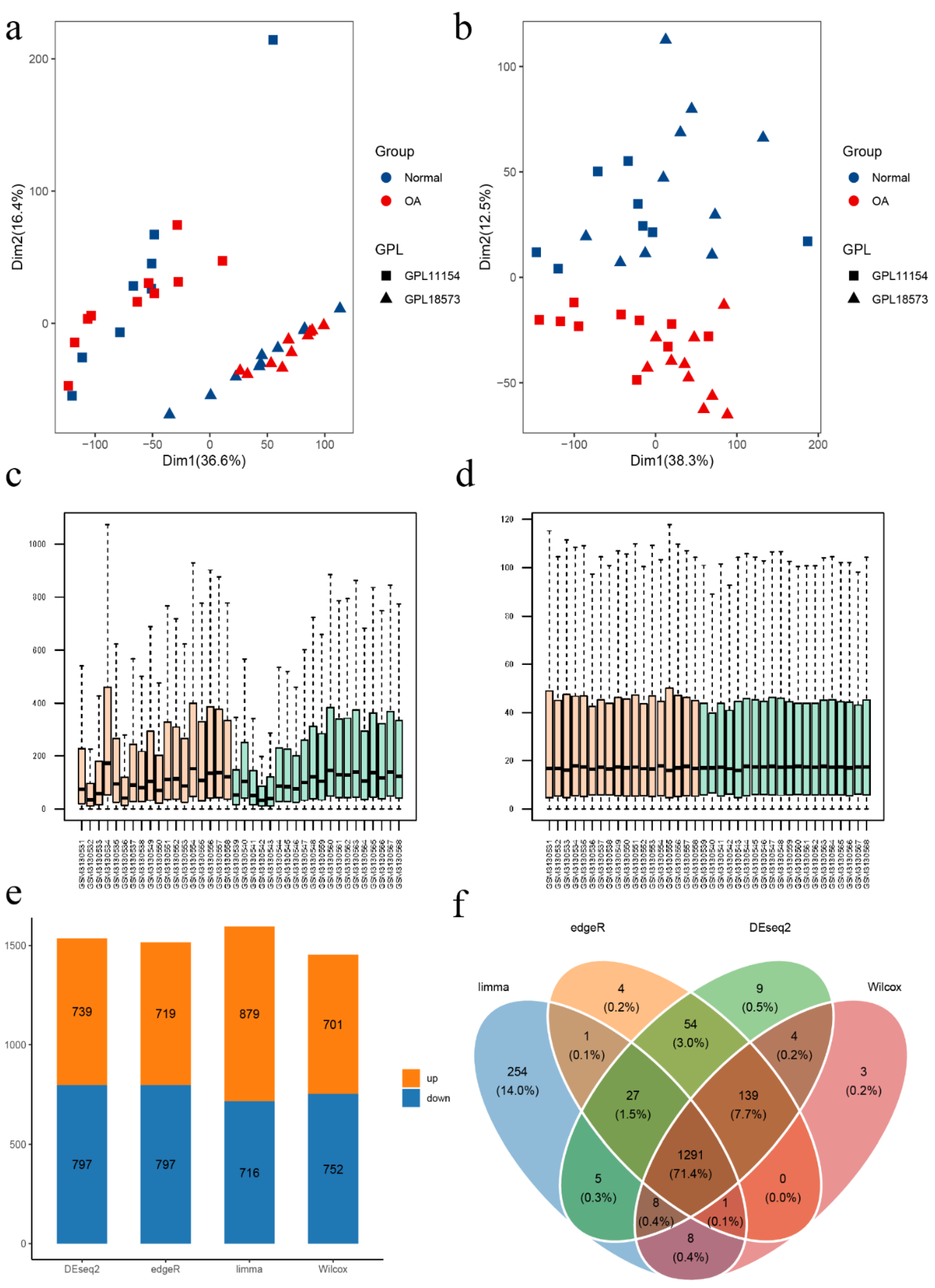
2.1. GEO Datasets Acquisition and Preprocessing
2.2. Ferroptosis-Related Genes Collection
2.3. Screening of Differential Expressed Ferroptosis-Related Genes (DEFRGS)
2.4. Functional Enrichment Analysis and PPI Network Analysis of DEFRGs
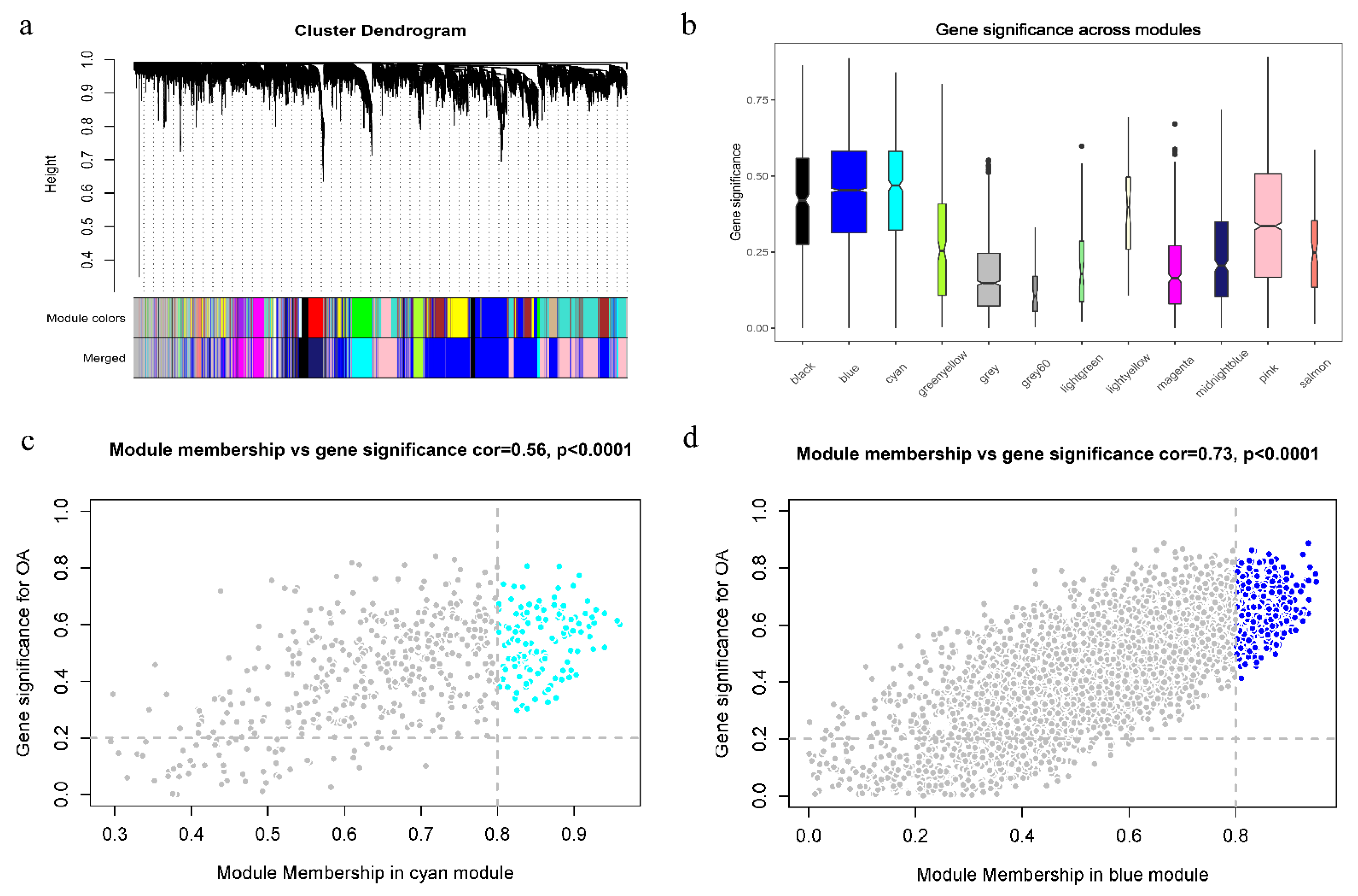
2.5. Weighted Correlation Network Analysis
2.6. Analysis of the Diagnostic Performance of Marker Genes
2.7. Cartilage Samples Collection
2.8. Isolation and Culture of Primary Chondrocytes
2.9. Histological Staining
2.10. Transfection Using SLC3A2 Knockdown (KD) shRNAs
2.11. Transmission Electron Microscopy (TEM)
2.12. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.13. Western Blotting
2.14. Statistical Analysis
3. Results
3.1. Data Processing and Identification of DEFRGs
3.2. GO and KEGG Enrichment Analysis of DEFRGs
3.3. PPI Analysis and Hub Genes of DEFRGs
3.4. WGCNA Analysis
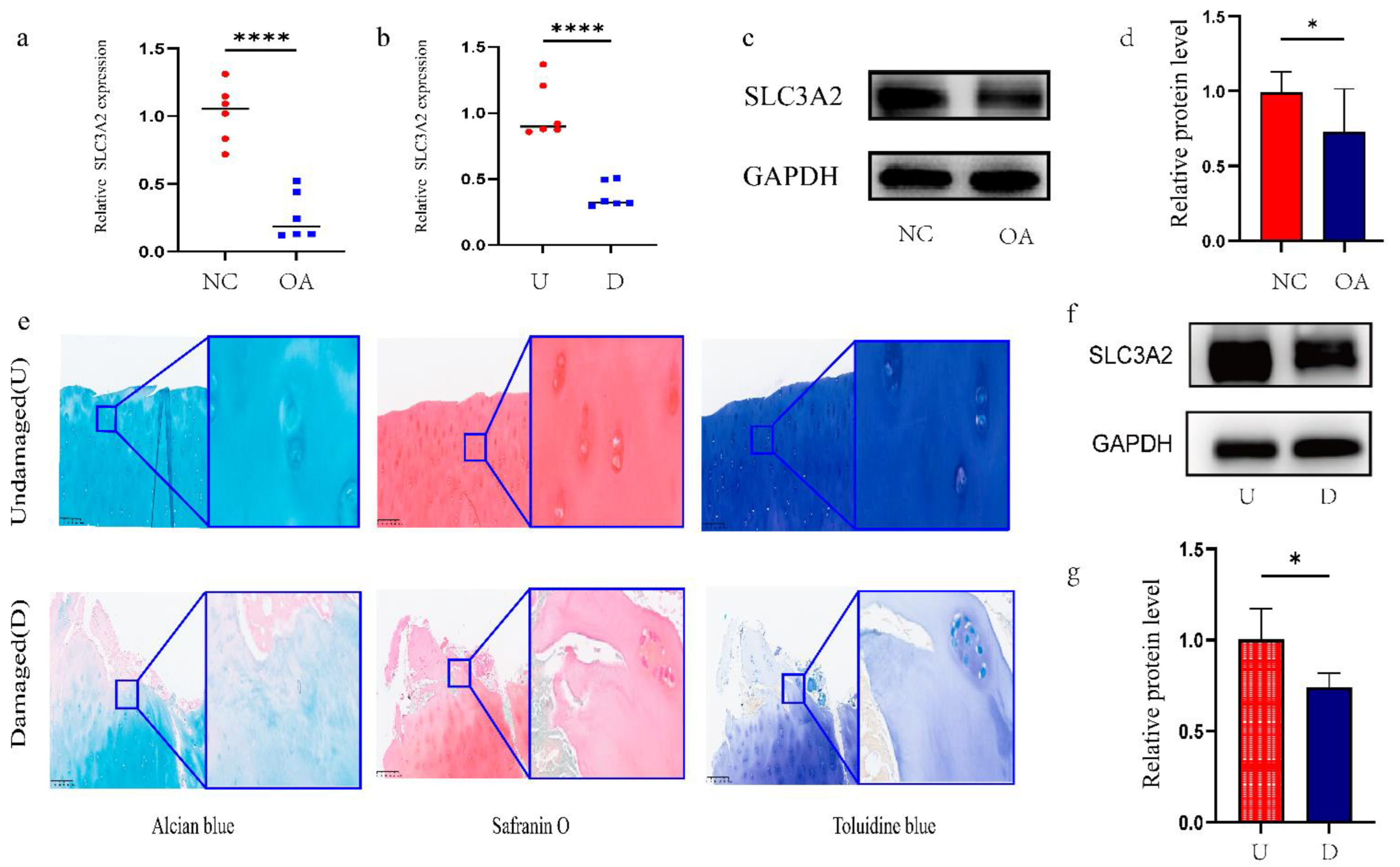
3.5. Identification and Validation of Final Hub Genes
3.6. Associations between Clinicopathological Features and SLC3A2
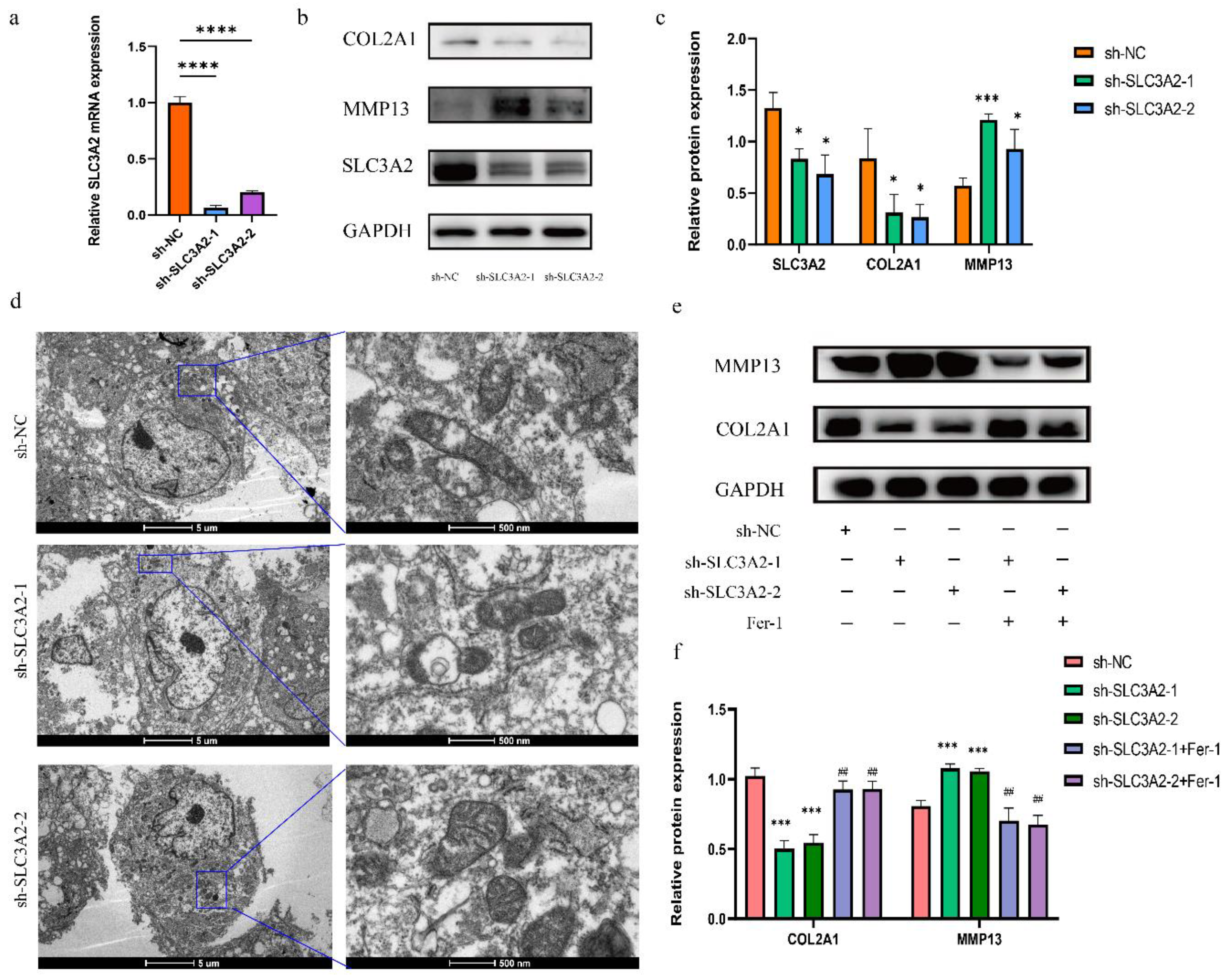
3.7. Effects of SLC3A2 Knockdown on OA Cartilage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Yue, L.; Berman, J. What Is Osteoarthritis? JAMA 2022, 327, 1300. [Google Scholar] [CrossRef]
- Global Burden of Disease Study 2013 Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet Lond. Engl. 2015, 386, 743–800. [CrossRef]
- Xue, J.-F.; Shi, Z.-M.; Zou, J.; Li, X.-L. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomed. Pharmacother. 2017, 89, 1252–1261. [Google Scholar] [CrossRef]
- Yang, H.; Wen, Y.; Zhang, M.; Liu, Q.; Zhang, H.; Zhang, J.; Lu, L.; Ye, T.; Bai, X.; Xiao, G.; et al. MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint. Autophagy 2019, 16, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Noh, H.-J.; Lee, H.; Park, H.-H.; Ha, Y.-J.; Park, S.H.; Lee, H.; Kim, S.-J.; Kang, H.C.; Eyun, S.; et al. TRIM24-RIP3 axis perturbation accelerates osteoarthritis pathogenesis. Ann. Rheum. Dis. 2020, 79, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Duan, X.; Fu, X.; Jiang, Y.; Yang, P.; Cao, C.; Li, Q.; Zhang, J.; Hu, X.; Zhang, X.; et al. RIP1 Perturbation Induces Chondrocyte Necroptosis and Promotes Osteoarthritis Pathogenesis via Targeting BMP7. Front. Cell Dev. Biol. 2021, 9, 638382. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Novak, K.; Haqqi, T.M. ERK1/2-mediated activation of DRP1 regulates mitochondrial dynamics and apoptosis in chondrocytes. Osteoarthr. Cartil. 2022, 30, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Park, D.R.; Kim, J.; Kim, G.M.; Lee, H.; Kim, M.; Hwang, D.; Lee, H.; Kim, H.-S.; Kim, W.; Park, M.C.; et al. Osteoclast-associated receptor blockade prevents articular cartilage destruction via chondrocyte apoptosis regulation. Nat. Commun. 2020, 11, 4343. [Google Scholar] [CrossRef]
- Ran, Q.; Liang, H.; Gu, M.; Qi, W.; Walter, C.A.; Roberts, L.J.; Herman, B.; Richardson, A.; Van Remmen, H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004, 279, 55137–55146. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Z.; Zhang, H.; Lu, J.; Tian, Y.; Wei, Y.; Yang, Y.; Bai, L. P2X7 Receptor Induces Pyroptotic Inflammation and Cartilage Degradation in Osteoarthritis via NF-κB/NLRP3 Crosstalk. Oxid. Med. Cell. Longev. 2021, 2021, 8868361. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Friedmann Angeli, J.P.; Schneider, M.; Proneth, B.; Tyurina, Y.Y.; Tyurin, V.A.; Hammond, V.J.; Herbach, N.; Aichler, M.; Walch, A.; Eggenhofer, E.; et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014, 16, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Guo, Z.; Lin, J.; Sun, K.; Guo, J.; Yao, X.; Wang, G.; Hou, L.; Xu, J.; Guo, J.; Guo, F. Deferoxamine Alleviates Osteoarthritis by Inhibiting Chondrocyte Ferroptosis and Activating the Nrf2 Pathway. Front. Pharmacol. 2022, 13, 791376. [Google Scholar] [CrossRef] [PubMed]
- Zilka, O.; Shah, R.; Li, B.; Friedmann Angeli, J.P.; Griesser, M.; Conrad, M.; Pratt, D.A. On the Mechanism of Cytoprotection by Ferrostatin-1 and Liproxstatin-1 and the Role of Lipid Peroxidation in Ferroptotic Cell Death. ACS Cent. Sci. 2017, 3, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Badgley, M.A.; Kremer, D.M.; Maurer, H.C.; DelGiorno, K.E.; Lee, H.-J.; Purohit, V.; Sagalovskiy, I.R.; Ma, A.; Kapilian, J.; Firl, C.E.M.; et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 2020, 368, 85–89. [Google Scholar] [CrossRef]
- Dierge, E.; Debock, E.; Guilbaud, C.; Corbet, C.; Mignolet, E.; Mignard, L.; Bastien, E.; Dessy, C.; Larondelle, Y.; Feron, O. Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell Metab. 2021, 33, 1701–1715.e5. [Google Scholar] [CrossRef]
- Derry, P.J.; Hegde, M.L.; Jackson, G.R.; Kayed, R.; Tour, J.M.; Tsai, A.-L.; Kent, T.A. Revisiting the intersection of amyloid, pathologically modified tau and iron in Alzheimer’s disease from a ferroptosis perspective. Prog. Neurobiol. 2020, 184, 101716. [Google Scholar] [CrossRef]
- Mahoney-Sánchez, L.; Bouchaoui, H.; Ayton, S.; Devos, D.; Duce, J.A.; Devedjian, J.-C. Ferroptosis and its potential role in the physiopathology of Parkinson’s Disease. Prog. Neurobiol. 2021, 196, 101890. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Feng, Y.; Li, H.; Chen, X.; Wang, G.; Xu, S.; Li, Y.; Zhao, L. Ferrostatin-1 alleviates lipopolysaccharide-induced acute lung injury via inhibiting ferroptosis. Cell. Mol. Biol. Lett. 2020, 25, 10. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Sun, K.; Yu, S.; Luo, J.; Guo, J.; Lin, J.; Wang, G.; Guo, Z.; Ye, Y.; Guo, F. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J. Orthop. Transl. 2021, 27, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Chen, Y.; Xue, F.; Liu, K.; Zhu, B.; Gao, J.; Yin, J.; Zhang, C.; Li, G. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine 2022, 76, 103847. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, A.; Bhattacharyya, B. Common Subcluster Mining in Microarray Data for Molecular Biomarker Discovery. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- van den Borne, M.P.J.; Raijmakers, N.J.H.; Vanlauwe, J.; Victor, J.; de Jong, S.N.; Bellemans, J.; Saris, D.B.F. International Cartilage Repair Society International Cartilage Repair Society (ICRS) and Oswestry macroscopic cartilage evaluation scores validated for use in Autologous Chondrocyte Implantation (ACI) and microfracture. Osteoarthr. Cartil. 2007, 15, 1397–1402. [Google Scholar] [CrossRef]
- Raymond, L.; Eck, S.; Hays, E.; Tomek, I.; Kantor, S.; Vincenti, M. RelA is required for IL-1beta stimulation of Matrix Metalloproteinase-1 expression in chondrocytes. Osteoarthr. Cartil. 2007, 15, 431–441. [Google Scholar] [CrossRef][Green Version]
- Ushita, M.; Saito, T.; Ikeda, T.; Yano, F.; Higashikawa, A.; Ogata, N.; Chung, U.; Nakamura, K.; Kawaguchi, H. Transcriptional induction of SOX9 by NF-kappaB family member RelA in chondrogenic cells. Osteoarthr. Cartil. 2009, 17, 1065–1075. [Google Scholar] [CrossRef]
- Kobayashi, H.; Chang, S.H.; Mori, D.; Itoh, S.; Hirata, M.; Hosaka, Y.; Taniguchi, Y.; Okada, K.; Mori, Y.; Yano, F.; et al. Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat. Commun. 2016, 7, 13336. [Google Scholar] [CrossRef]
- Xia, L.; Gong, N. Identification and verification of ferroptosis-related genes in the synovial tissue of osteoarthritis using bioinformatics analysis. Front. Mol. Biosci. 2022, 9, 992044. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Y.; Gan, F.; Jiang, M.; Chen, D.; Xie, M.; Jike, Y.; Bo, Z. Identification of Potential Therapeutic Target Genes in Osteoarthritis. Evid.-Based Complement. Altern. Med. ECAM 2022, 2022, 8027987. [Google Scholar] [CrossRef]
- Duan, Z.-X.; Li, Y.-S.; Tu, C.; Xie, P.; Li, Y.-H.; Qi, L.; Li, Z.-H. Identification of a potential gene target for osteoarthritis based on bioinformatics analyses. J. Orthop. Surg. 2020, 15, 228. [Google Scholar] [CrossRef]
- Li, Y.; Ge, X.; Peng, F.; Li, W.; Li, J.J. Exaggerated false positives by popular differential expression methods when analyzing human population samples. Genome Biol. 2022, 23, 79. [Google Scholar] [CrossRef] [PubMed]
- Fort, J.; de la Ballina, L.R.; Burghardt, H.E.; Ferrer-Costa, C.; Turnay, J.; Ferrer-Orta, C.; Usón, I.; Zorzano, A.; Fernández-Recio, J.; Orozco, M.; et al. The Structure of Human 4F2hc Ectodomain Provides a Model for Homodimerization and Electrostatic Interaction with Plasma Membrane. J. Biol. Chem. 2007, 282, 31444–31452. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, D.; Kanai, Y.; Palacín, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 2013, 34, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Patel, D.N.; Welsch, M.; Skouta, R.; Lee, E.D.; Hayano, M.; Thomas, A.G.; Gleason, C.E.; Tatonetti, N.P.; Slusher, B.S.; et al. Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 2014, 3, e02523. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Green, M.; Choi, J.E.; Gijón, M.; Kennedy, P.D.; Johnson, J.K.; Liao, P.; Lang, X.; Kryczek, I.; Sell, A.; et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019, 569, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Yang, S.; Li, L.; Dai, Q.; Borcherding, N.; Wagner, B.A.; Buettner, G.R.; Spitz, D.R.; Leslie, K.K.; Zhang, J.; et al. Metadherin enhances vulnerability of cancer cells to ferroptosis. Cell Death Dis. 2019, 10, 682. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, X.; Yu, K.; Xu, X.; Chen, T.; Shi, Y.; Wang, Y.; Qiu, S.; Guo, S.; Cui, J.; et al. Targeting SLC3A2 subunit of system XC- is essential for m6A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic. Biol. Med. 2021, 168, 25–43. [Google Scholar] [CrossRef]


| Characteristics | Number of Cases (%) |
|---|---|
| Age (years) | |
| <65 | 9 (30) |
| ≥65 | 21 (70) |
| Gender | |
| Male | 8 (26.7) |
| Female | 22 (73.3) |
| Obesity gradation 1 | |
| Underweight | 0 (0) |
| Normal | 8 (26.7) |
| Overweight | 12 (40) |
| Obesity | 10 (33.3) |
| Kellgren-Lawrence gradation | |
| Ⅲ | 12 (40) |
| Ⅳ | 18 (60) |
| Expression of SLC3A2 2 | |
| Low expression | 15 (50) |
| High expression | 15 (50) |
| Characteristics | SLC3A2 Expression | p Value | |
|---|---|---|---|
| Low | High | ||
| Age (years) | |||
| <65 | 4 | 5 | >0.999 |
| ≥65 | 11 | 10 | |
| Gender | |||
| Male | 3 | 5 | 0.682 |
| Female | 12 | 10 | |
| Obesity gradation | |||
| Underweight | 0 | 0 | 0.601 |
| Normal | 3 | 5 | |
| Overweight | 6 | 6 | |
| Obesity | 6 | 4 | |
| Kellgren-Lawrence gradation | |||
| Ⅲ | 2 | 10 | 0.008 |
| Ⅳ | 13 | 5 | |
| Variables | SLC3A2 Expression | |
|---|---|---|
| Spearman | p Value | |
| Age | −0.294 | 0.115 |
| Gender | −0.200 | 0.289 |
| Weight | −0.234 | 0.213 |
| Height | 0.074 | 0.697 |
| BMI | −0.462 | 0.010 |
| Obesity gradation | −0.485 | 0.007 |
| Kellgren-Lawrence gradation | −0.742 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Deng, Z.; Yu, B.; Liu, H.; Yang, Z.; Zeng, A.; Fu, M. Identification of SLC3A2 as a Potential Therapeutic Target of Osteoarthritis Involved in Ferroptosis by Integrating Bioinformatics, Clinical Factors and Experiments. Cells 2022, 11, 3430. https://doi.org/10.3390/cells11213430
Liu H, Deng Z, Yu B, Liu H, Yang Z, Zeng A, Fu M. Identification of SLC3A2 as a Potential Therapeutic Target of Osteoarthritis Involved in Ferroptosis by Integrating Bioinformatics, Clinical Factors and Experiments. Cells. 2022; 11(21):3430. https://doi.org/10.3390/cells11213430
Chicago/Turabian StyleLiu, Hailong, Zengfa Deng, Baoxi Yu, Hui Liu, Zhijian Yang, Anyu Zeng, and Ming Fu. 2022. "Identification of SLC3A2 as a Potential Therapeutic Target of Osteoarthritis Involved in Ferroptosis by Integrating Bioinformatics, Clinical Factors and Experiments" Cells 11, no. 21: 3430. https://doi.org/10.3390/cells11213430
APA StyleLiu, H., Deng, Z., Yu, B., Liu, H., Yang, Z., Zeng, A., & Fu, M. (2022). Identification of SLC3A2 as a Potential Therapeutic Target of Osteoarthritis Involved in Ferroptosis by Integrating Bioinformatics, Clinical Factors and Experiments. Cells, 11(21), 3430. https://doi.org/10.3390/cells11213430







