1. Introduction
A significant portion, 10–30%, of all metastatic breast cancer (BC) patients develop brain metastases (BM) [
1,
2]. In contrast with other distant metastatic lesions, BM dramatically reduce the quality of life and prognosis of BC patients [
3]. Here, neurological deficits, including both cognitive and sensory dysfunction [
4], are often observed, and the average life expectancy after breast cancer brain metastases (BCBM) diagnosis is 2 to 25 months [
5,
6,
7]. In most cases, BM formation is preceded by metastases in other organs, such as the lung, liver, or bones [
1]. Therapy options in the metastatic setting include surgical resection of metastases, whole-brain radiotherapy, stereotactic radiation, and targeted therapies. However, the chances of complete tumor reduction are frequently limited [
5,
8,
9], in part due to the limited penetration of therapeutic compounds beyond the blood-brain barrier (BBB) [
10]. The BBB represents a highly organized physiological barrier, composed of endothelial cells, pericytes, and astrocytic endfeet, and is characterized by a selective permeability, with the aim to protect the central nervous system (CNS) and to maintain brain homeostasis. In this context, tumor cells require specific molecular characteristics to overcome the BBB and establish brain metastatic lesions [
11,
12,
13]. In order to develop successful therapy options for BCBM patients, but also in order to predict a predisposition of primary BC patients to develop BM, we must improve our knowledge of the molecular characteristics required for tumor cells to interact and successfully pass the BBB.
In BC, the molecular subtype of the primary tumor is strongly associated with BM prevalence. Previous studies have shown that patients of the triple-negative breast cancer (TNBC) and HER2+ subtype have a higher risk of developing BM [
14,
15,
16,
17]. Further, the molecular subtype is not only decisive for the incidence, but also impacts how fast BMs progress, and predicts the survival rates. Patients from the luminal A group showed the highest median survival, whereas patients from the TNBC group showed the lowest median survival (luminal A: 23.1, luminal B: 15.0, HER2: 12.5, and TNBC: 6.4 months, respectively) [
18,
19].
Several studies have shown an association between the BC molecular subtype and the hyaluronan (HA) metabolism rate. HA, a linear glycosaminoglycan (GAG) consisting of repeating disaccharides, is synthesized by HA synthases (HAS), a group of transmembrane proteins directly located at the plasma membrane and cleaved into smaller fragments by different hyaluronidases (HYAL) [
20]. In this context, HA fragments of different molecular weights can be found: high-molecular-weight HA (HMW-HA), which is mainly present under physiological conditions, and low-molecular-weight HA (LMW-HA), which is increasingly secreted under pathological conditions [
21,
22]. While HMW-HA exerts anti-angiogenic, anti-inflammatory, and anti-proliferative functions, LMW-HA activates signaling cascades that promote angiogenesis, immune cell influx proliferation, and migration [
22]. HA molecules can be found as a component of the extracellular matrix, bound to the cell surface, or freely available after being released by many cell types. In recent years, the function of membrane-bound HA has been increasingly studied. Here, HA forms a glycocalyx together with other GAGs, which supports the cell–cell binding. This outer HA layer, which is regarded as a pericellular HA-coat, can be generated via binding to the HA synthases during synthesis and extrusion, or through the interaction to its cognate receptor CD44 or the receptor for hyaluronan-mediated motility (RHAMM), potentially resulting in the activation of a variety of intracellular signaling pathways [
23].
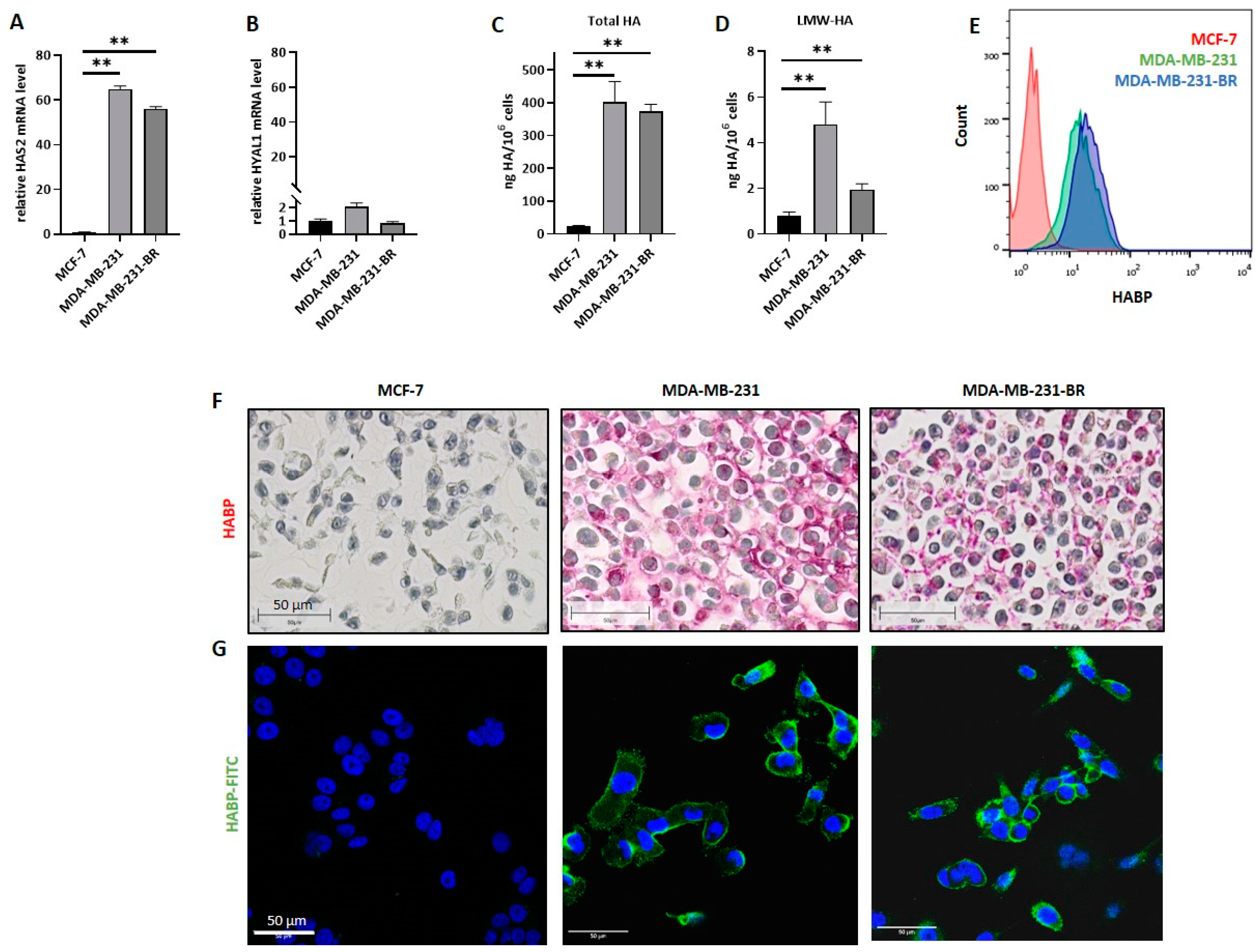
Interestingly, TNBC cells display endogenously higher HA levels compared to hormone-receptor-positive ones [
24], thereby attributing a role to HA during tumor formation and progression. Further, the expression level of enzymes associated with HA metabolism is often increased in tumor cells compared to healthy tissue. Li et al. have shown an association between an increased Has-2 level and BC tumor progression [
25]. We have previously reported the impact of HA metabolism and BM development in BC. Here, an increased Has-2 and especially Hyal-1 level in BC patients is associated with a significantly higher risk of BM formation [
26]. In the present study, we have analyzed the impact of (1) the HA-tumor coat in BC cells and (2) the HYAL1 depletion in TNBC cells on the tumor cell interaction with the BBB in vitro, and on the development of BM in vivo.
2. Materials and Methods
2.1. Cell Lines
All used cell lines were purchased and cultured as previously described by Hamester et al. [
27].
2.2. CRISPR/Cas9-HYAL1 Knockout in MDA-MB-231-BR
CRISPR/Cas9-HYAL1 knockout (KO) in MDA-MB-231-BR cells was performed according to the plasmid-based procedure described by Ran et al. [
28]. Two guide sequences, both located on exon 2 of the HYAL1 gene, were selected with the provided CRISPR guide RNA design tool guide scan. Guide sequences and primers used for sgRNA oligo insert construction can be found in the
supplementary materials and methods file. The sgRNA oligos were cloned into pSpCas9(BB)-2A-GFP plasmid (Addgene (Watertown, MA, USA) plasmid ID: 48138). MDA-MB-231-BR cells were transfected with HYAL1-specific CRISPR/Cas9 plasmid, as well as empty vector (Ctrl), by using Lipofectamine
® LTX Reagent (#15338100, Thermo Fisher Scientific, Waltham, MA, USA). Single-cell-colonies were assessed using the FACS Sorting method, and HYAL1 KO was checked via sequencing of genomic DNA. The primer for sequencing can also be found in the
supplementary materials and methods file.
2.3. HYAL1 and HAS2 Overexpression in MCF-7
The full HAS2 and HYAL1 cDNA sequences were obtained from a commercially available vector (pCR-Blunt II-TOPO-HAS2 Plasmid, Source Bioscience, Nottingham, UK; IRCMp5012E0230D, and pCMV-Sport6-HYAL1 Plasmid; Source Bioscience, Nottingham, UK; IRATp970C1157D) and cloned into LeGO-iC2-Puro+ Plasmid and LeGO-iG2-Neo+ Plasmid, respectively (a kind gift from AG Fehse, Center for Oncology, Department of Stem Cell Transplantation, UKE, Hamburg, Germany). After lentiviral production in HEK293T cells, MCF-7 cells were transduced. The corresponding empty vector was taken as a negative control.
2.4. CD44 Knockdown (KD) in MDA-MB-231-BR
CD44 knockdown in MDA-MB-231-BR cell line was generated by lentiviral transduction (lentiviral vectors were a kind gift from Daniel Wicklein and Kristoffer Riecken from the Department of Anatomy and Experimental Morphology and the Department of Stem Cell Transplantation, at the UKE, Hamburg, Germany, respectively) using vectors containing shRNA-sequences which targeted specific regions of the CD44 mRNA sequence (pLVX-shCD44 (1 CD44 IV)). Similarly, a control cell line was established using a scramble shRNA sequence (pLVX-shScrambled).
2.5. Treatment with Hyaluronidase
All cell lines were incubated with 10 U/mL Hyaluronidase from bovine testes (#H3506-100MG, Sigma Aldrich, St. Louis, MI, USA) in serum-reduced media (3% (v/v) FBS (#F9665, Thermo Fisher Scientific Waltham, MA, USA)) overnight, and functional assays were subsequently performed.
2.6. HA-Binding Protein (HABP) Staining and Immunohistochemistry
For HABP histochemistry, cells were first embedded in paraffin (
supplementary materials and methods), and a staining procedure was performed as previously described [
29]. Briefly, antigen retrieval was performed using the 1× Target Retrieval Buffer (#S1699, Dako, Agilent Technologies Inc., Santa Clara, CA, USA) in a water bath at 60–65 °C overnight, and, subsequently, slides were blocked with a 1% (
w/
v) BSA/TBS (bovine serum albumin (BSA, #8076.1, Carl Roth, Karlsruhe, Germany)/Tris-Buffered Saline (TBS)) solution for 30 min at room temperature (RT), and incubated with an HABP (#385911, Merck Millipore, Burlington, MA, USA) solution (1:75, c = 6.6 g/mL) for 90 min at room temperature. The HA-binding protein was visualized using the ABC-AP and the permanent AP Red Kit (#AK-5000, Vector Laboratories). For the luciferase staining, antigen retrieval was performed using 1× Citrate-buffer (pH 6) in a steamer (100 °C, 20 min), and subsequent incubation with Luciferase primary antibody (#ab181640, Abcam, Cambridge, UK) was performed (1:500 in Antibody Diluent (#S0809, Dako, Agilent Technologies Inc., Santa Clara, CA, USA, overnight at 4 °C), followed by incubation with a biotinylated anti-Goat IgG secondary antibody (#BA-9500, Vector Laboratories, Burlingame, CA, USA; 10 mL TBS, 50 µL Normal Horse Serum (#S-2000-20, Vector Laboratories) and 100 µL anti-Goat IgG), for 30 min at RT). Antibody visualization and nuclei counterstaining were performed as described above.
The HABP fluorescence staining was performed on cells previously seeded on coverslips. After cell fixation (20 min) with a 3.7% formaldehyde solution (#7398.1, Carl Roth), a blocking step was performed (1 h with a 1% BSA/PBS solution) followed by incubation with the biotinylated hyaluronic acid-binding protein solution (HABP, #385911, Merck Millipore, Burlington, MA, USA; 1:75 in 1% BSA/PBS, 45 min, RT). Cells were then incubated with a secondary antibody solution (Streptavidin-FITC, #405202, BioLegend, San Diego, CA, USA; 1:500 in 1% BSA/PBS, 60 min, RT) and subsequently with DAPI (#9542, Sigma Aldrich, St. Louis, MI, USA; 1:1000 in PBS [#D8537, Merck Millipore, Burlington, MA, USA], 10 min, RT, dark). After washing, coverslips were transferred to glass slides and mounted with Fluoromount-G™ (#00-4958-02, Invitrogen, Waltham, MA, USA).
2.7. F-Actin Fluorescence Staining
For staining of cytoskeletal F-actin, 5 × 104 cells were seeded on sterile coverslips into 24-well plates and cultured for 72 h. Cells were washed and fixed as described above. After blocking for 1 h at room temperature with 1% BSA/PBS, cells were stained with phalloidin (#ab235138, Abcam, 1:2000 in PBS (#D8537, Merck)) for 30 min at RT, and finally cover-slipped with Mounting Medium for Fluorescence with DAPI (#H-1200-10, Vector Laboratories). Slides were stored at −20 °C in the dark and imaged with a fluorescence microscope (Olympus, Tokyo, Japan; IXplore Live-System).
2.8. Western Blot
Western blot was performed as previously described [
29]. The following antibodies were used: β-Actin (#sc-47778, Santa Cruz Biotechnology (SCB)), β-Catenin (#8480, Cell Signaling Technology (CST)), CD44 (#sc-7297, SCB), HSC-70 (#sc-7298, SCB), RHAMM (#sc-515221, SCB), Slug (#5741, CST), Snail (#3879, CST), TCF8/ZEB1 (#3396, CST), Vimentin (#5741, CST), ZO-1 (#8193, CST), Anti-mouse (#sc-2055, SCB), and Anti-rabbit (#sc-2357, SCB). Detection was performed using the Westar Nova 2.0 Chemiluminescent Substrate for Western blotting Kit (#XLS071,0250, Cyanagen Srl., Bologna, Italy). Densitometric evaluation of the protein bands was performed using the GS-800 Calibrated Densitometer and Quantity-One-4.6.3 software from Bio-Rad, Hercules, CA, USA. Two independent western blot analyses, as well as subsequent quantification and statistical analysis, have been performed for all experiments shown in this study. Figures display one representative Western blot image.
2.9. Enzyme-Linked Immunosorbent Assay (ELISA)
In order to analyze the amount of HA in cell culture supernatants, 1 × 106 cells were seeded into T75-flasks and cultured to a confluence of 80%. Cells were washed 2 times with PBS (#D8537, Merck Millipore, Burlington, MA, USA), and cultured with serum-reduced media (3% FBS) for another 48 h. Cell culture media were collected and centrifuged (10 min, 12,000 rpm, RT). The resulting supernatant was used for HA-ELISA (ELISA-like Assay for HA/Hyaluronan, #AMS.CSR-HA-96KIT, Amsbio, Abingdon, UK). ELISA was performed according to the manufacturer’s protocol. The amount of HA was normalized to the cell count at the time of taking supernatants.
2.10. HYAL1-Activity Gel-Assay
Tumor cells were seeded in 6-well-plates and cultivated until they reached 80% confluency. Isolated protein lysates were incubated with HMW-HA in a Hyal-1 working buffer (1% Triton X-100 (#9036-19-5, Merck Millipore, Burlington, MA, USA), 150 mM NaCl (#7647-14-5, Merck Millipore, Burlington, MA, USA), 100 mM formic acid (#,64-18-6, Merck Millipore, Burlington, MA, USA, pH 3.7) for 14 h at 37 °C in a heating block. A 0.5 cm thick 1% agarose (#15581044, Thermo Fisher Scientific, Waltham, MA, USA) gel in 1× TAE buffer (2 M Trizma® base (#77-86-1, Merck Millipore, Burlington, MA, USA), 1 M pure acetic acid (#64-19-7, Merck Millipore, Burlington, MA, USA), 0.05 M EDTA (#60-00-4, Merck Millipore, Burlington, MA, USA, pH 8.3), was prepared and pre-runed for 6 h at 80 V. All samples were subsequently electrophoresed for 2 h at 100 V (10 µL sample + 2 µL 6× loading Dye (#R0611, Thermo Fisher Scientific, Waltham, MA, USA)).The gel was equilibrated in a box with 30% ethanol in water by shaking for 1 h at RT. Ethanol was replaced, and the gel was incubated in Stains-All working solution (500 µL 1× Stains-All (#7423-31-6, Merck Millipore, Burlington, MA, USA, 200 mL 30% ethanol, overnight, dark, RT). The gel was de-stained with 30% ethanol in water (overnight, dark, RT).
2.11. Quantitative Real-Time PCR
2.12. Flow Cytometry
Next, 2 × 105 cells were stained with biotinylated hyaluronic acid binding protein (HABP, #385911, Merck Millipore, Burlington, MA, USA; 1:75 in 1% BSA/PBS, 30 min, 4 °C) or CD44-AF488 (#338829, clone BJ18, BioLegend, San Diego, CA, USA; 1:200 in 1% BSA/PBS, for 20 min at 4 °C), and, after a washing step, were incubated with a secondary antibody (Streptavidin-FITC, #405202, BioLegend, San Diego, CA, USA; 1:500 in 1% BSA/PBS) for 20 min at 4 °C. Labeled cells were measured with FACSCalibur (Becton Dickinson, Franklin Lakes, NJ, USA). The analysis was performed using FlowJoV10 Software (Version v10.8.1). At least three independent flow cytometry analyses, as well as subsequent quantification and statistical analysis, have been performed for all experiments shown in this study. If this was not the case for individual experiments, it was mentioned separately in the figure legends. Figures display one image from a representative flow cytometry experiment.
2.13. Static Cell Adhesion Assay
Static cell adhesion assay to analyze the adhesion ability of different tumor cells to human brain endothelial cells was performed as previously described [
27].
2.14. Electrical Cell-Substrate Impedance Sensing (ECIS)
The effect of different tumor cells on the BBB integrity was examined by an electrical cell-substrate impedance sensing system (ECIS), as previously described [
27]. Results represent three to five replicates from each cell type or donor.
2.15. Transwell Invasion Assay
Transwell invasion assay was used to analyze the potential of different tumor cells to invade through a brain endothelial cell monolayer. The assay was performed as previously described by Hamester et al. [
27].
2.16. Migration Assay
Cells (2 × 105 per well) were seeded in six-well plates and cultured until confluence. An artificial wound was created with the help of a pipette tip in each well. The migration potential of the cells was determined by analyzing the wound area at different time points (0, 10, and 24 h) with the ImageJ Wound Healing Tool (Version 1.52t, Wayne Rasband, National Institute of Health, Bethesda, MA, USA). We have assessed the change in the wound width over time for each individual well. This width is defined as the average distance between the two margins of the scratch. Precisely, for each individual well wound, widths at the same position were measured at different time points and calculated as a percentage of the value at time point 0 h (100%).
2.17. Intracardiac Metastasis Mouse Model
An intracardiac mouse model to analyze development of brain metastases was conducted as previously described [
27]. In contrast to the method described so far, anesthetized mice were imaged from both lateral and ventral views approximately 10–15 min after intraperitoneal injection of D-luciferin, using the IVIS
® Spectrum in vivo imaging system (PerkinElmer, Waltham, MA, USA). The animal experiments were approved by the Authority for Social Affairs, Family, Health, and Consumer Protection of the Free and Hanseatic City of Hamburg, through application N005/2020.
2.18. Statistics
Statistical analysis was conducted using SPSS software Version 25 (IBM SPSS Statistics, Armonk, NY, USA). For all functional assays, cells were plated in triplicates and each experiment was performed three times (n = 3). Statistical significance was determined using unpaired two-tailed Student’s t-tests. The assumption of homogeneity of variance was tested using Levene’s Test of Equality of Variances (p > 0.05). Results are given as mean +/− s.d. All p-values less than 0.05 were considered statistically significant.
4. Discussion
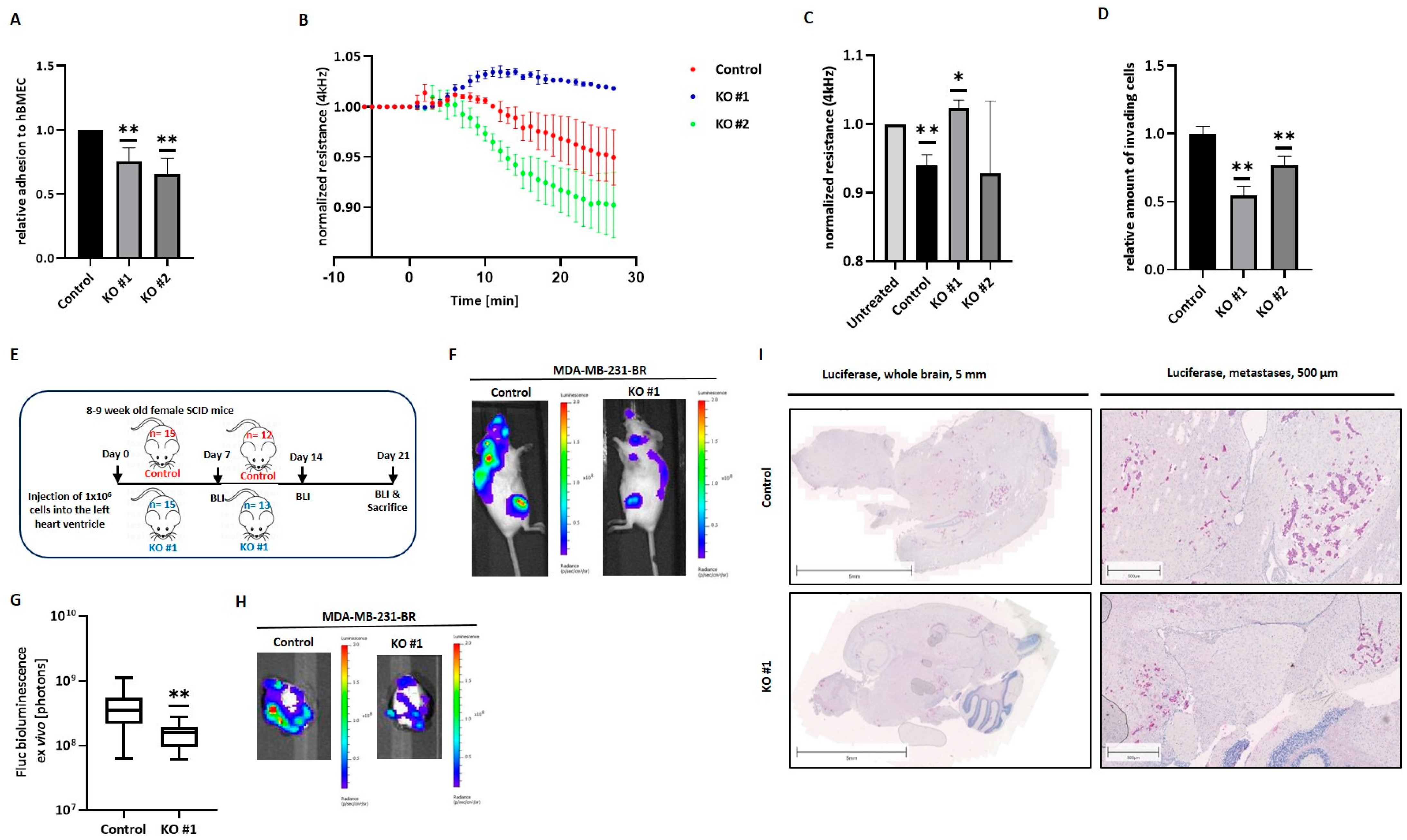
BC patients with BM suffer from reduced life quality and expectancy. In this context, many efforts have been made to identify primary BC patients with a high risk of developing BM. Screening options should be examined in future studies, as these patients represent an important group for preventive strategies. Although certain BC molecular subtypes exhibit a higher predisposition to development of BM, little is known about the specific molecular players required for a tumor cell to successfully overcome the BBB and colonize in the brain. We could previously show a significant association between the expression of the synthase HAS2 and the hyaluronidase HYAL1 in the primary tumor of BC patients and the development of BM [
26]. While Has-2 is responsible for the synthesis of HA polymers, Hyal-1 cleaves these into low molecular weight fragments, which have been described to exert alternative functions. In this study, we have explored the relevance of the HA metabolism and, more precisely, of the LMW-HA fragments, on the ability of BC cells to pass the BBB and develop cerebral metastasis. We found that Hyal-1 and, specifically, the Hyal-1-generated LMW-HA fragments, are essential for the formation of a functional pericellular HA-coat, which in turn promotes the tumor cell interaction with the brain endothelium and supports BCBM development.
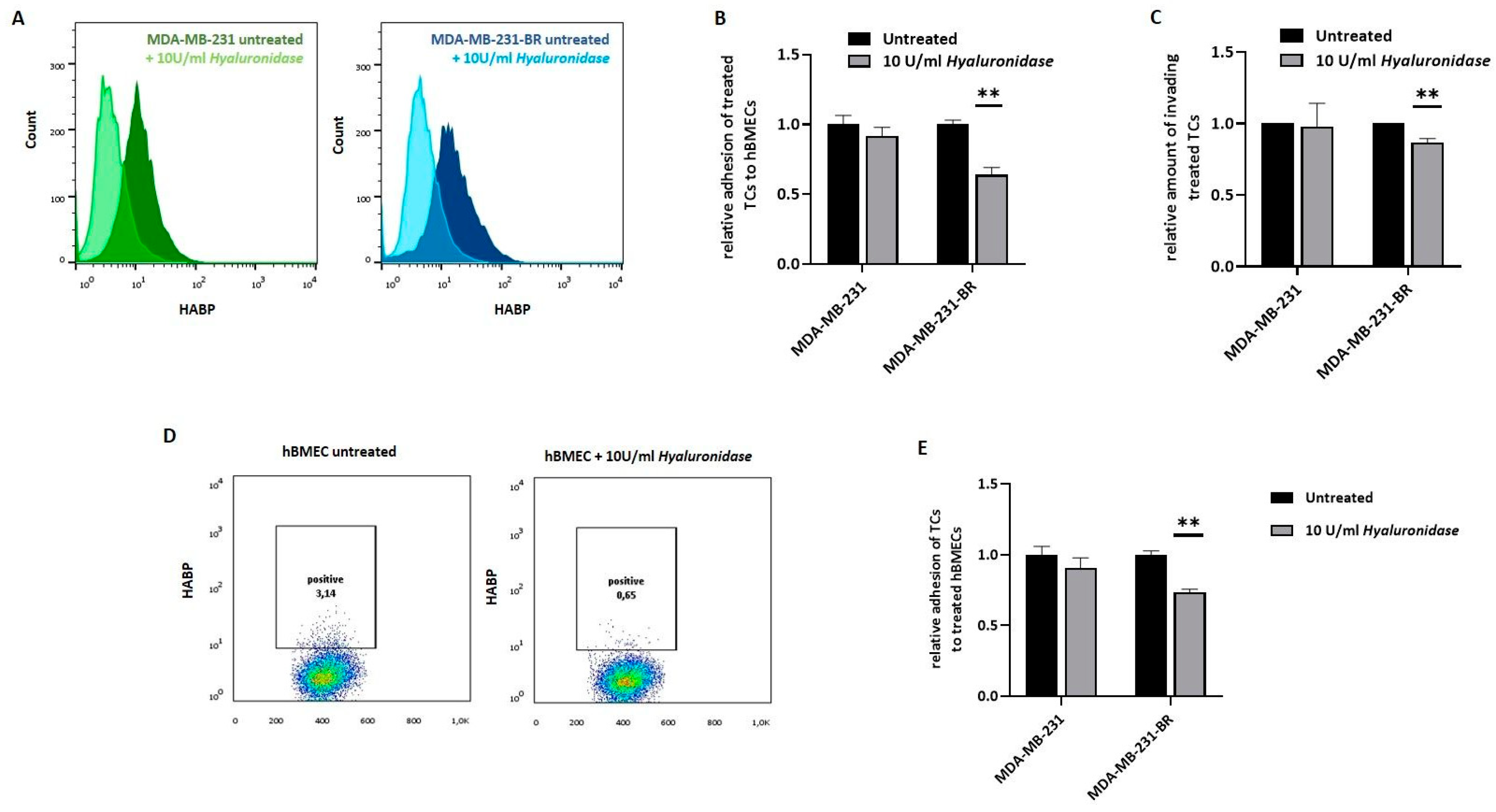
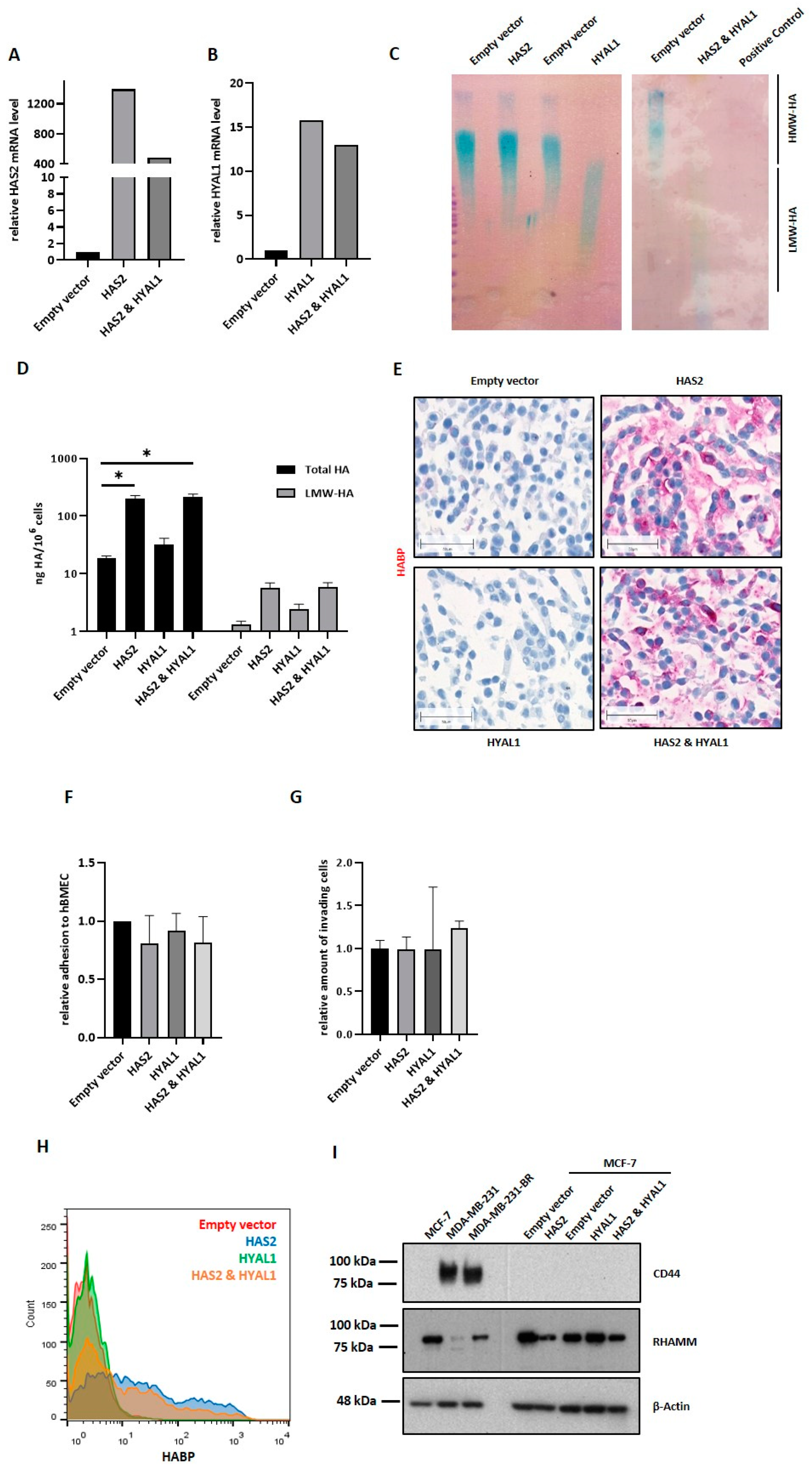
Several studies have previously demonstrated the association of hyaluronan with more aggressive phenotypes in cancer, including BC, being recently included in the “hallmarks of cancer” [
33]. In line with these findings, our present results demonstrate a higher HA metabolism rate in the more aggressive TNBC cell line, MDA-MB-231, and its brain-seeking subline, MDA-MB-231-BR, compared to the luminal BC cell line MCF-7. Here, a robust pericellular HA-coat was detected in the TNBC cell lines, with the brain-seeking subline displaying a more expressed surface HA deposit. Remarkably, although both TNBC cell lines displayed a strong ability to adhere to the brain endothelial monolayer and invade in vitro, only the brain-seeking BC cell line was dependent on its membrane-associated HA, as shown after pericellular HA-removal with exogenous hyaluronidases. Similarly, the reduction in the surface-associated endothelial HA via hyaluronidase treatment resulted in decreased adhesion and invasion ability of brain-metastatic BC cells, but not of the parental cell line, underlining the key role of hyaluronan for the development of BM. To our knowledge, this is the first report on the role of pericellular HA for adhesion and the invasion of BC cells through the BBB in vitro. An association between the presence of a cellular HA-coat and increased cell adhesion, motility, and invasive potential has been previously described in several tumor entities, including BC [
23]. A study by Brett et al. showed that the enzymatic removal of HA significantly inhibits cancer cell extravasation and invasion through human umbilical vein endothelial cells (HUVECs) [
34]. These data and our own suggest that membrane-bound HA represents a docking structure that mediates tumor cell adhesion to the endothelial layer and, additionally, triggers pro-invasive signaling. Although the parental and the brain-seeking subline synthesize approximately the same amount of HA, the flow cytometry and HA-fluorescence staining analyses showed higher levels of HA and a more compact staining pattern on the surface of MDA-MB-231-BR cells in comparison to the parental cell line. This result indicates a more pronounced and functional HA-coat in the brain-seeking cells. One explanation might be the presence of HA fragments of different sizes in the two TNBC cell lines. It is known that large HA fragments, so-called HMW-HA, which are mainly present under physiological conditions, exert anti-angiogenic, anti-inflammatory, and anti-proliferative functions, whereas smaller fragments or LMW-HA, which are, rather, found under pathological conditions, activate signaling cascades that promote angiogenesis, immune cell influx proliferation, and migration [
21,
22]. As mentioned before, we have previously demonstrated that high tumoral Hyal-1 expression levels in the primary tumor significantly correlate with BM development in BC patients [
26]. Taking these data into consideration, we hypothesized that the brain-seeking cell line preferentially synthetizes LMW-HA, which might be responsible for the formation of a compact and functional HA-coat, thereby enhancing tumor cell adhesion to the endothelium and invasion through the BBB. However, there are no methods currently available to discriminate between the HMW-HA and LMW-HA structures at the cell membrane. The HA-binding protein used to visualize and quantify HA via fluorescence staining, which is also used in flow cytometry analysis, recognizes and binds both structures and provides information about the total HA amount. Therefore, in order to explore the specific role of LMW-HA, we generated an HYAL1 knockout in the TNBC brain seeking subline. As expected, both HYAL1 KO clones were confirmed to secrete less LMW-HA, while the total amount of secreted HA was not altered. Indeed, at the cell surface, the amount of total bound HA measured by flow cytometry was similar for all KO clones and the corresponding control cells. This means that, despite HYAL1 KO, these cells were able to generate a pericellular HA-coat. HA fluorescence staining, however, revealed a remarkably reduced and thinner HA coat in the HYAL1 KO clones, indicating that LMW-HA is essential, but is not the only component of the pericellular HA-coat. A study conducted by Reiprich et al. recently showed the formation of a thicker pericellular HA coat after HAS3 overexpression when compared with HAS2 [
35]. Thus, the smaller HA polymers synthesized by Has-3 appear to form a thicker coat compared to the larger Has-2-generated HA polymers.
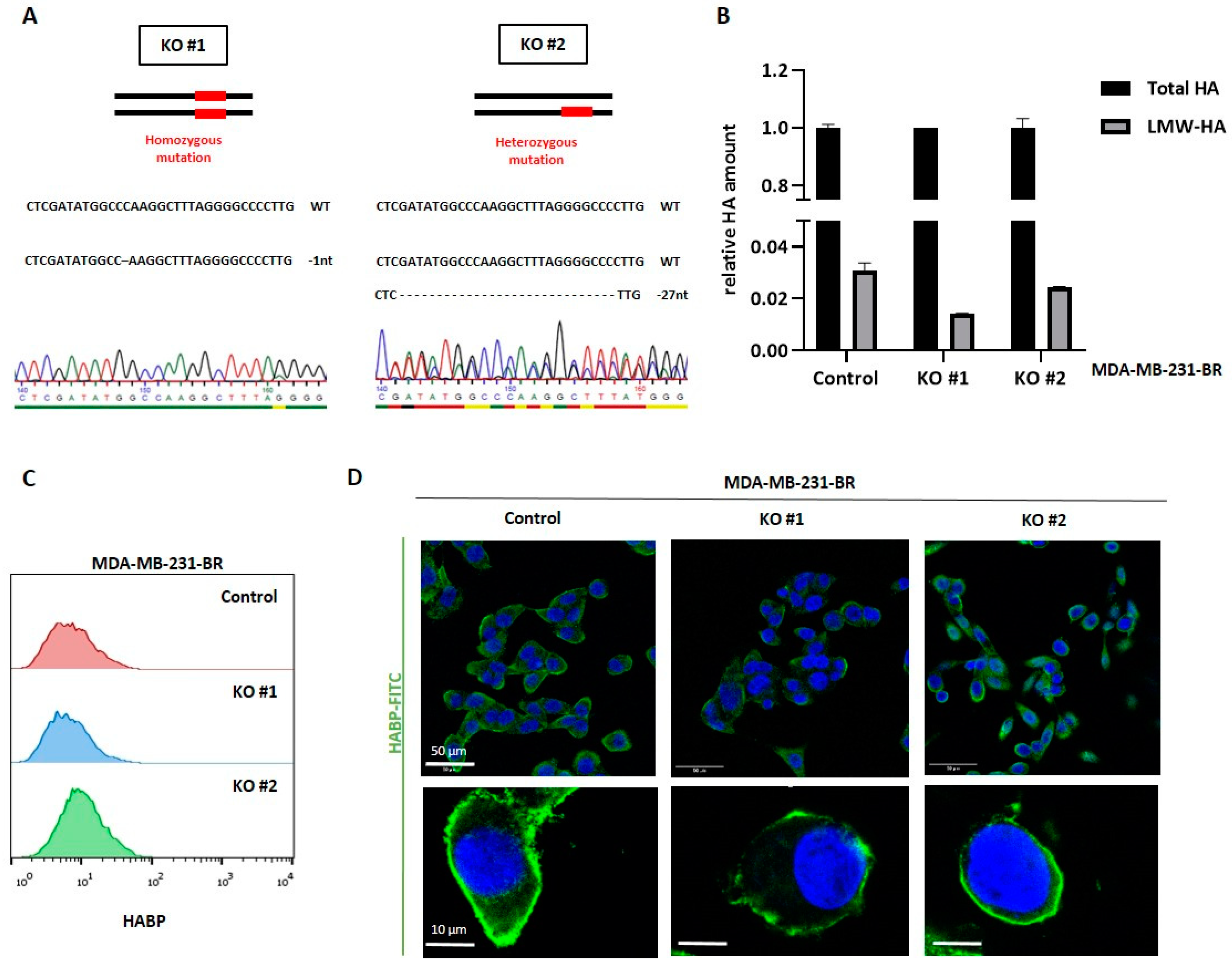
Despite the not yet fully elucidated composition of the pericellular HA coat on tumor cells, and, in particular, the specific role of Hyal-1-synthesized LMW-HA fragments in this coat, we could demonstrate a significant impact of Hyal-1 on the development of BM in vitro as well as in vivo. In all three major steps involved in the brain-metastatic process, namely adhesion, disruption of the BBB, and invasion, HYAL1 KO cells showed a less aggressive phenotype and reduced metastasis-promoting properties. The reduced adhesiveness to the brain endothelium argues for a crucial mechanical role of LMW-HA fragments. On the other hand, aberrant signaling also occurs due to the lower amount of LMW-HA, likely resulting in reduced BBB integrity and invasion through the BBB. These effects were confirmed in vivo using a mouse model with intracardiac injection. For the HYAL1 KO clone, significantly reduced brain metastasis was detected, either by BLI signals within the brains, or macroscopically by histochemical HA-BP staining in mice injected with HYAL1 KO, compared to the control group.
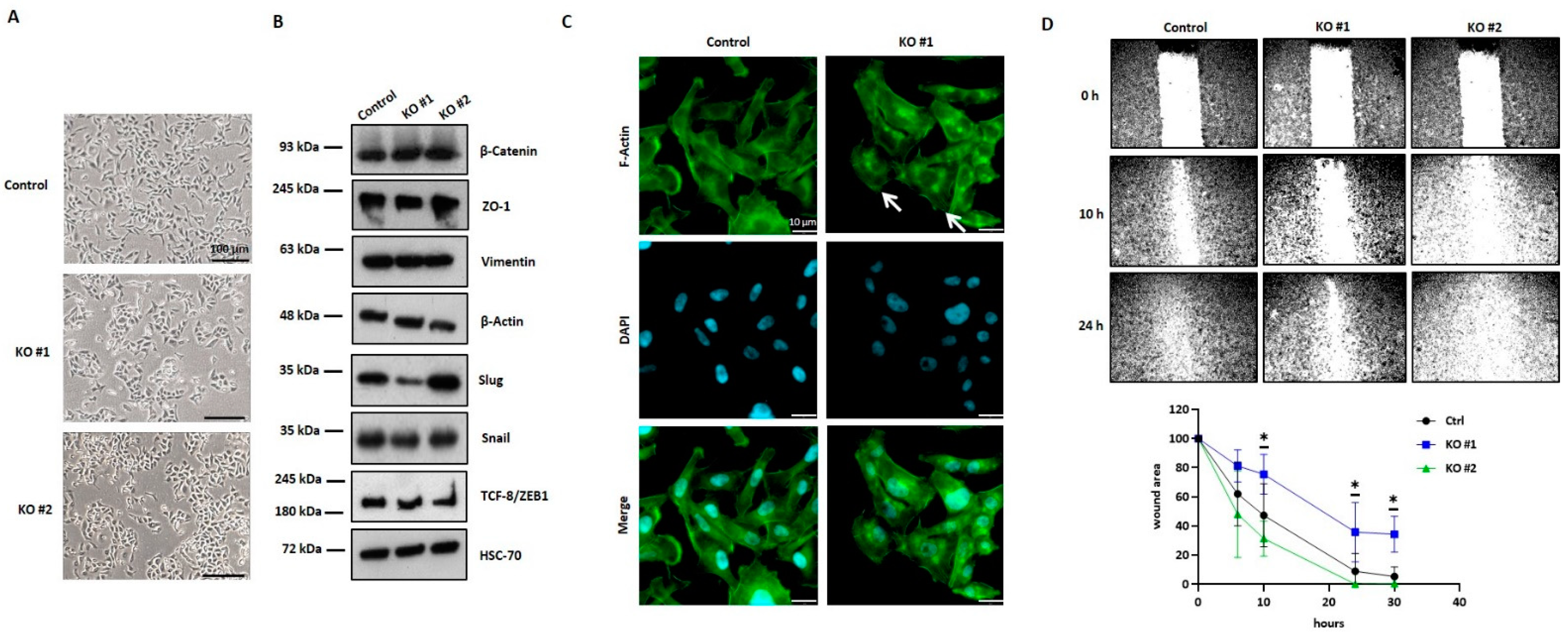
Particularly striking were the morphological alterations observed in MDA-MB-231-BR cells with HYAL1 KO, which displayed a more epithelial phenotype. These newly acquired cellular characteristics might also explain the significantly reduced adhesion and invasion related to the BBB observed in these cells. A previous study by Zoltan-Jones et al. showed that HA can promote cell invasiveness and EMT; more precisely, overexpression of HAS2 in the epithelial BC cell line MCF10A promoted both invasiveness and EMT [
36]. In contrast, in the present work, the HYAL1 depletion and, in turn, the reduction of LMW-HA fragments in the brain-seeking TNBC cell line did not affect MET. This observation allowed us to assume that high expression levels of Has-2 and, therefore, high HMW-HA production promotes a mesenchymal phenotype in tumor cells, and is independent of the amount of LMW-HA. Moreover, we found that HA also affected the cytoskeleton organization, likely through the binding to its cognate receptor CD44. To date, a modulating role of HYAL2, but not HYAL1, within the ERM-related cytoskeletal interactions has been shown only in fibroblasts [
37]. However, future experiments are required to elucidate the mechanism underlying the observed changes in the actin cytoskeleton in HYAL1 depleted cells.
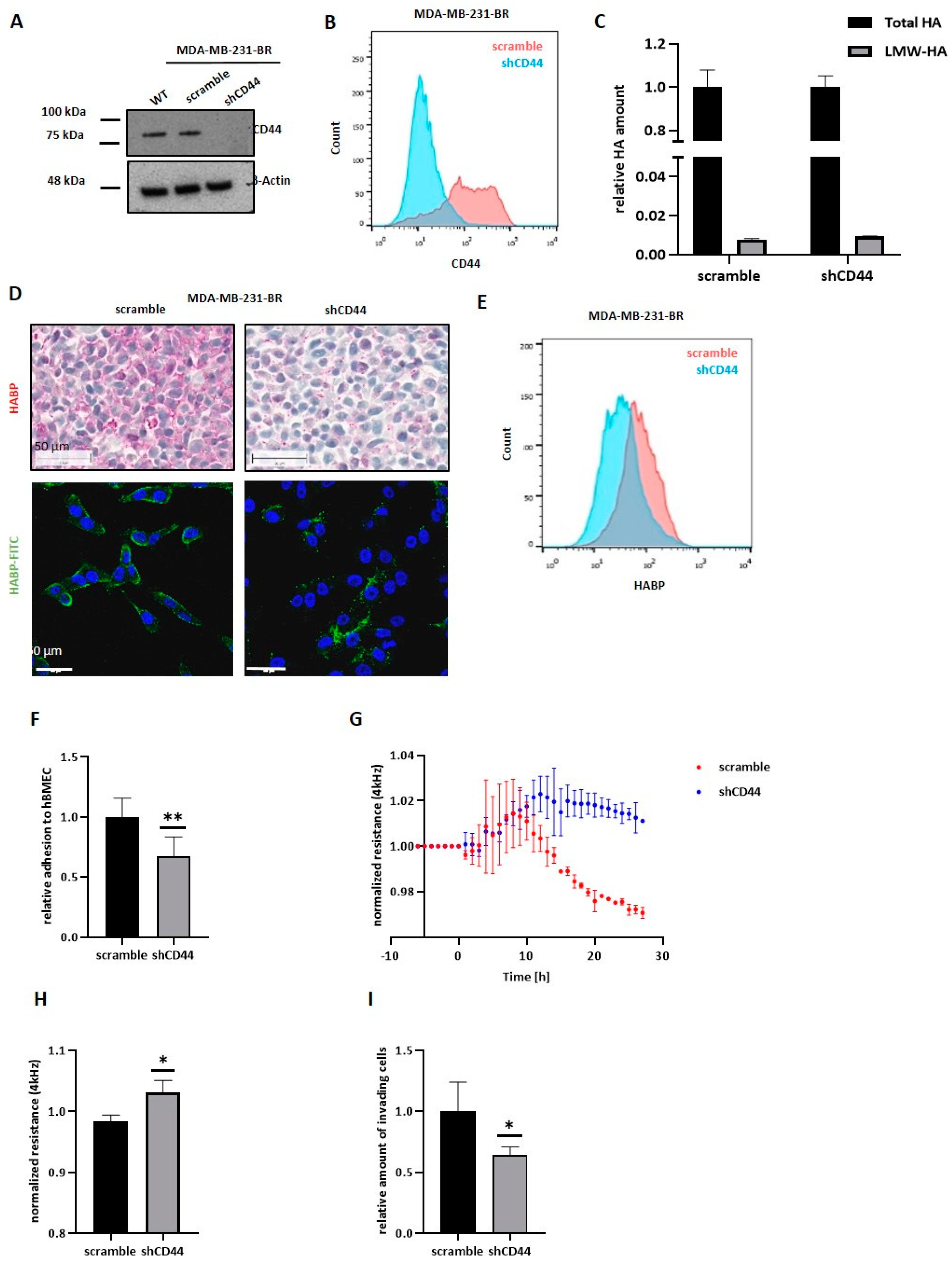
To our surprise, when we overexpressed HAS2 and HYAL1 in the luminal BC cell line MCF7, neither increased tumor cell adhesion, nor migration through the brain endothelium, nor any morphological alteration was observed. MCF-7 overexpressing cells synthesized and secreted a similar amount of LMW-HA compared to MDA-MB-231-BR cells, but their pericellular HA-coat was considerably weaker, suggesting that LMW-HA fragments might only have an impact on the BBB as a part of the pericellular coat, and not in their secreted and soluble form. Here, a possible explanation for the lack of a proper HA-coat, even in the presence of sufficient HA molecules, might be the absence of the HA receptor CD44 in MCF-7 cells. Indeed, we could further confirm the key role of CD44 in the formation of the HA-coat in MDA-MB-231-BR cells. Here, a less pronounced HA-coat was detected in CD44 knockdown cells in comparison to the control cells. Moreover, down-regulation of CD44 in MDA-MB-231-BR resulted in a reduced HA-coat. This points to a key role of CD44 as a mediator for the formation of a functional HA-coat, rather than during HA-internalization and subsequent synthesis of LMW-HA.
Taken together, the HA metabolism plays a key role in the development of BM in BC. Precisely, low molecular weight HA, mainly generated by the hyaluronidase Hyal1, supports the formation of a pericellular HA coat as well as pro-metastatic signaling in BC cells. Both effects are dependent on the binding of LMW-HA to its receptor, CD44, and ultimately promote BM development. In this context, inhibition of the interaction between Hyal-1 generated LMW-HA fragments and CD44 might represent a target for future therapeutic interventions.
5. Conclusions
Due to an existing lack of effective treatment, BCBM is becoming a critical issue. Due to the semi-permeable BBB, the passage of therapeutic agents into the brain is difficult, and effective treatment is generally not possible. In order to enhance options and success of treatment, an improved understanding of the molecular mechanisms of how tumor cells pass the BBB and, thus, develop metastatic lesions is required. In the present work, the precise and outstanding role of Hyal-1 during BCBM development was revealed for the first time. The down-regulation of the hyaluronidase Hyal-1 in the brain-seeking BC cell line MDA-MB-231-BR, and the associated reduced amount of LMW-HA, significantly reduced the essential steps during brain metastasis in vitro, as well as the number of brain metastases in vivo. In this regard, LMW-HA appears to be responsible for the formation of a pericellular HA-coat around tumor cells, and interacts with brain endothelium. A prerequisite for the HA-coat-mediated impact on the metastatic behavior of BC cells is the presence of the HA receptor CD44. Aside from its essential function in the context of the HA metabolism, and, thus, the synthesis of LMW-HA fragments, CD44 plays a key role in the formation of the pericellular HA coat on brain metastatic BC cells. By inhibiting the interaction with the brain endothelium, tumor cells could lose the prerequisite for the formation of brain metastases by inhibiting the “signaling” of the LMW-HA-CD44 axis, and, thus, the formation of a pericellular HA coat.