Developmental Potency and Metabolic Traits of Extended Pluripotency Are Faithfully Transferred to Somatic Cells via Cell Fusion-Induced Reprogramming
Abstract
1. Introduction
2. Materials and Methods
2.1. EPSC Cell Line Establishment and Culture
2.2. EGFP Transfection
2.3. Cell Fusion and Hybrid Cell Culture
2.4. Cell Culture
2.5. Karyotype Analysis
2.6. In Vitro Random Differentiation
2.7. In Vitro Extraembryonic Lineage Differentiation
2.8. Chimeric Embryo Generation
2.9. X-Gal Staining
2.10. Electron Microscopy
2.11. Mitochondrial Length Measurement
2.12. Oxygen Consumption Rate Analysis
2.13. Extracellular Acidification Rate Analysis
2.14. RNA Isolation and qRT-PCR
2.15. Immunocytochemistry
2.16. Bulk RNA Sequencing
2.17. Post Sequencing Data Analysis
2.18. Statistical Analysis
2.19. Animal Use Ethical Statement
3. Results
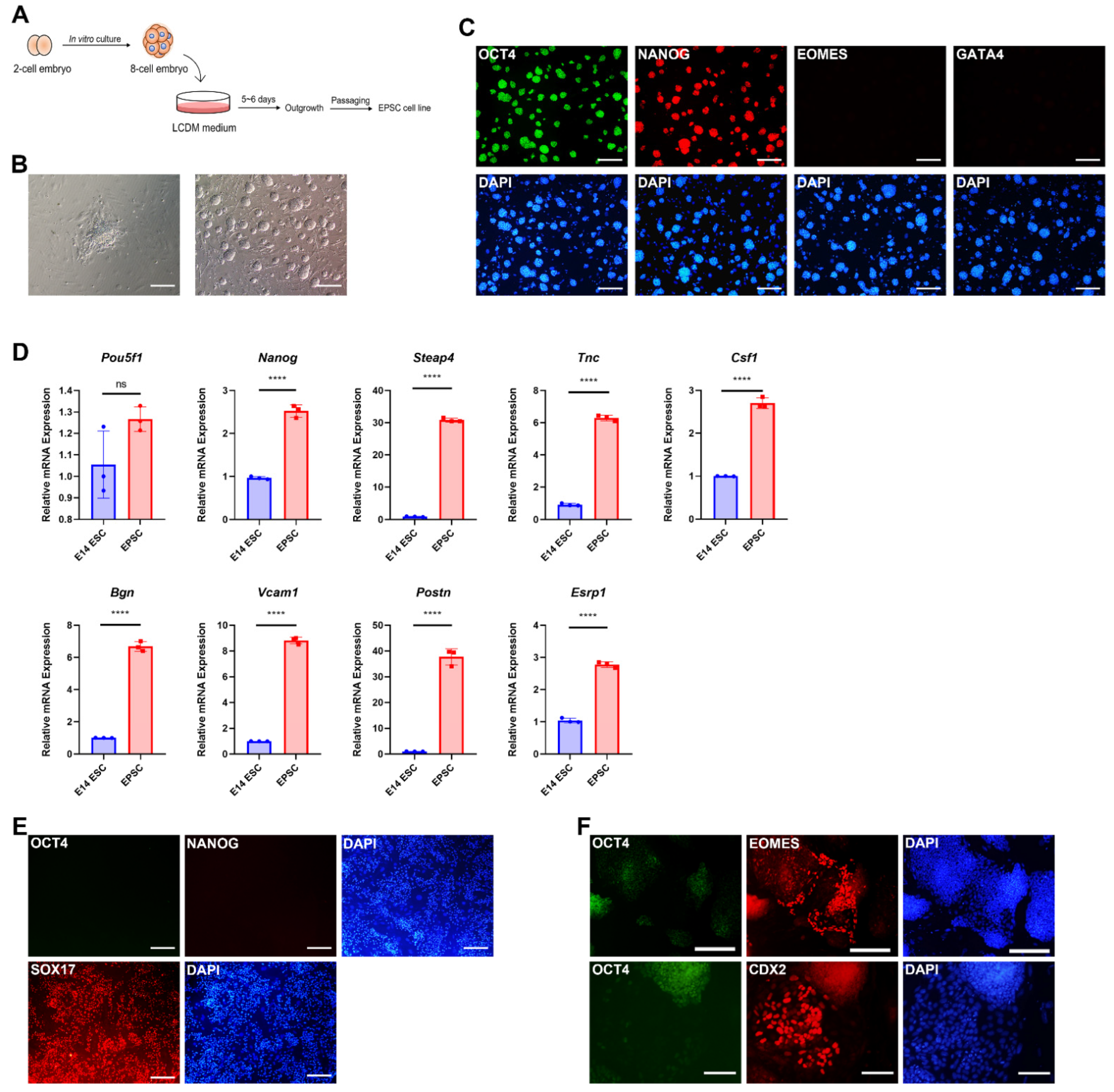
3.1. Derivation and Characterization of Extended Pluripotent Stem Cells
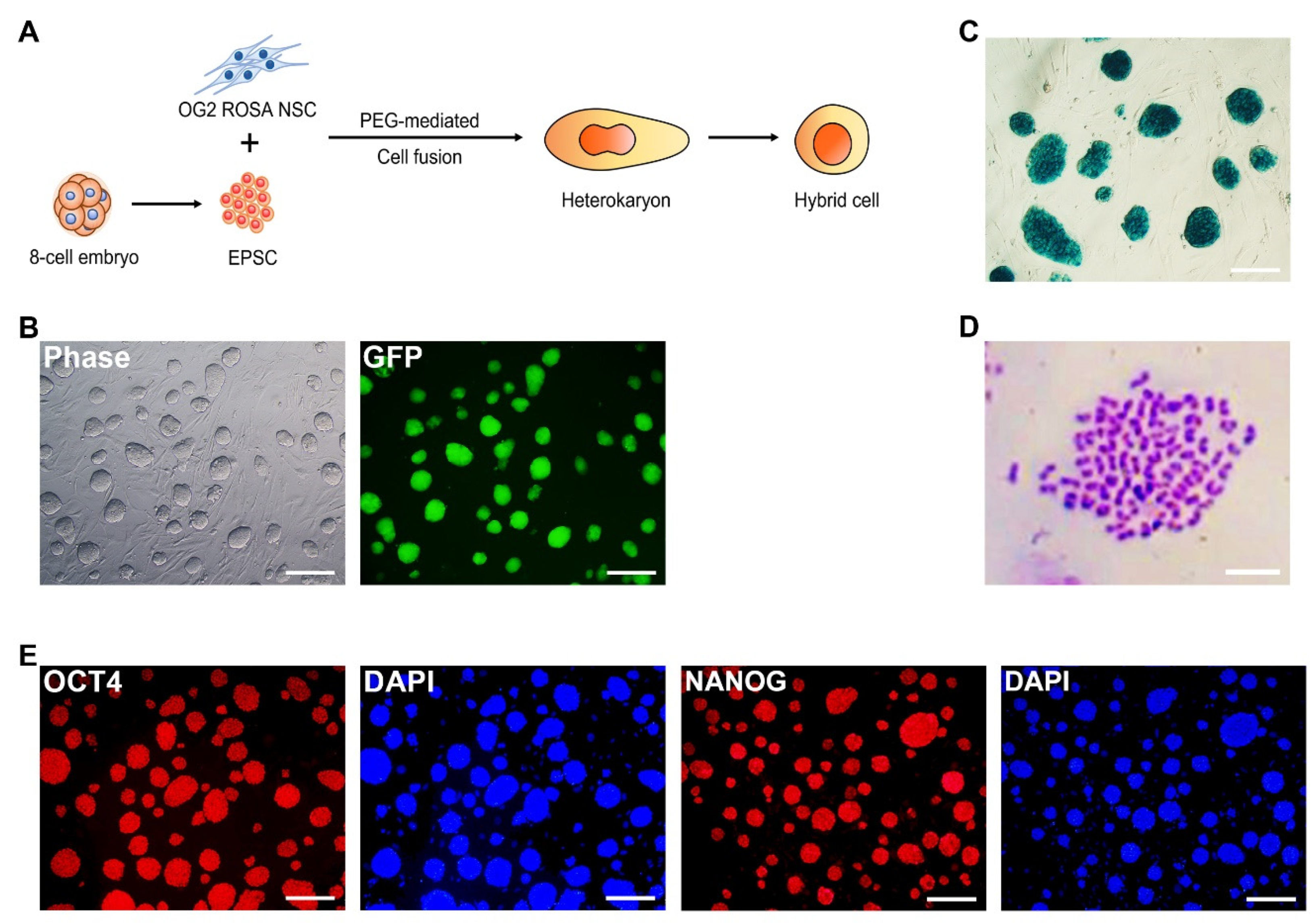
3.2. Extended Potential Reprogramming of Somatic Cells by Cell Fusion with EPSCs
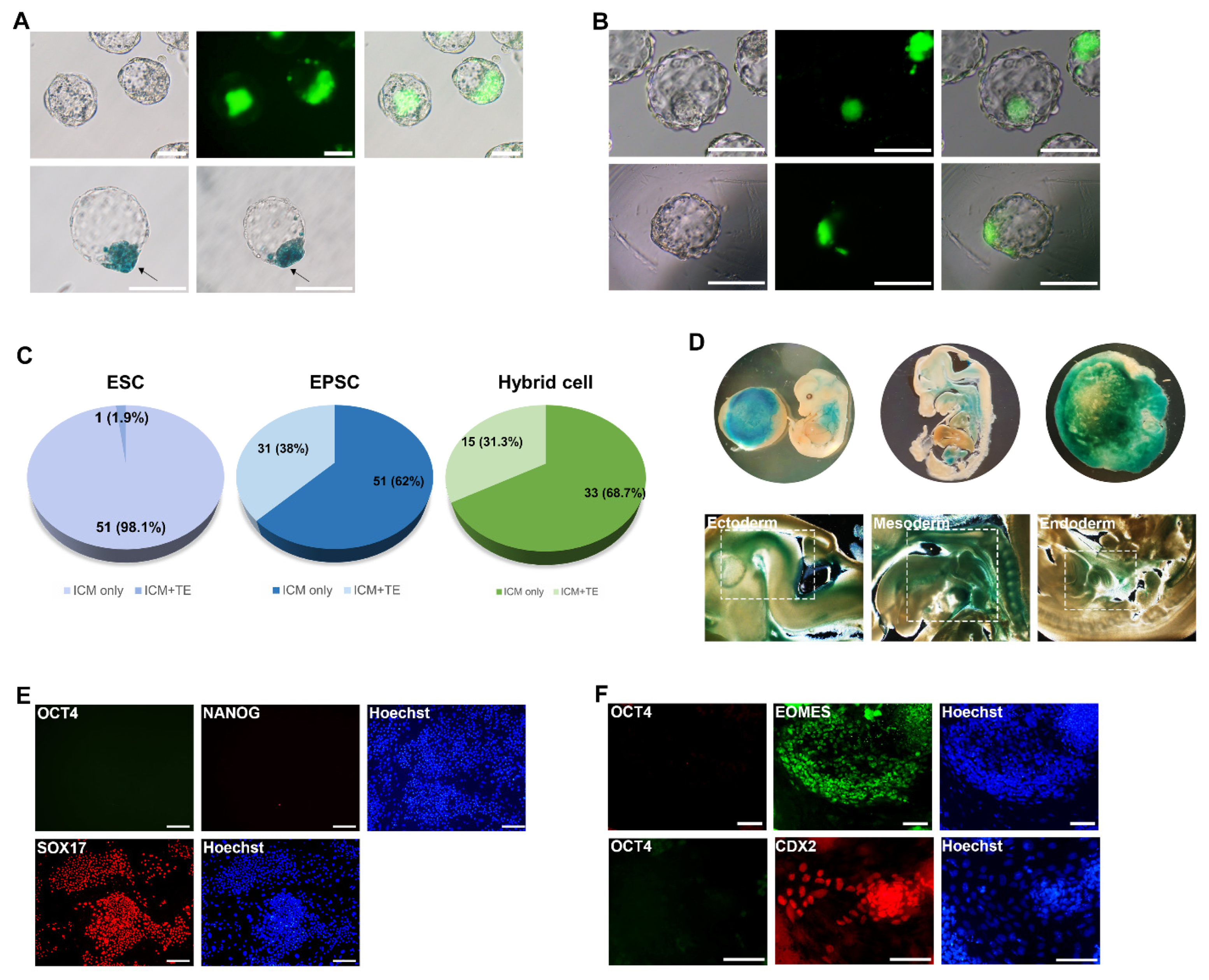
3.3. Developmental Potency of Hybrid Cells into Both Embryonic and Extraembryonic Lineages
3.4. Global Gene Expression Patterns of EPSCs, ESCs, NSCs, and EPSC-NSC Hybrid Cells
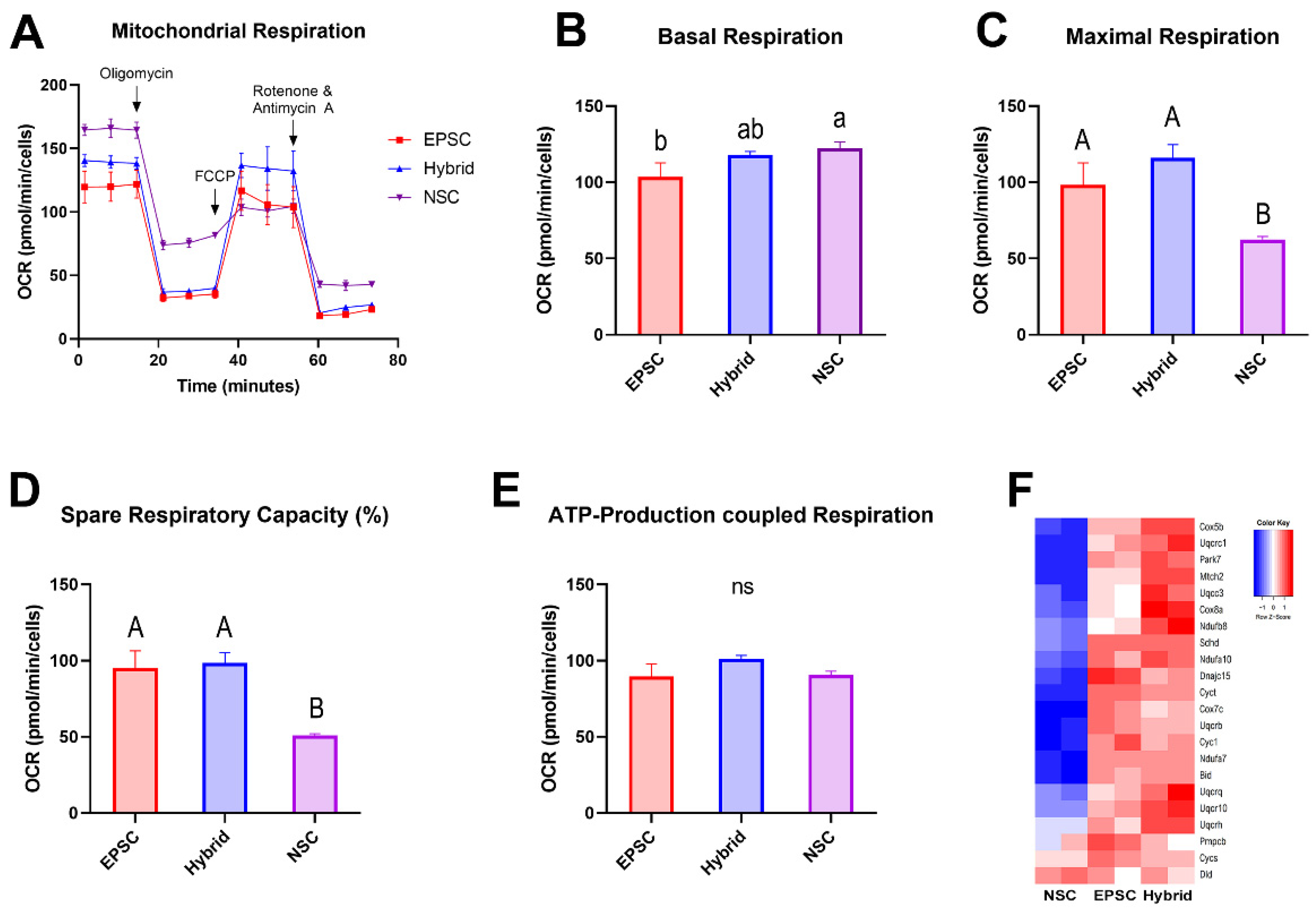
3.5. Mitochondrial Dynamics of EPSC-NSC Hybrid Cells Recapitulate That of EPSCs
3.6. Bioenergetic Metabolism Profiles of NSCs Were Remodeled to the State of EPSCs after Reprogramming by Cell Fusion with EPSCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baker, C.L.; Pera, M.F. Capturing Totipotent Stem Cells. Cell Stem Cell 2018, 22, 25–34. [Google Scholar] [CrossRef]
- Maemura, M.; Taketsuru, H.; Nakajima, Y.; Shao, R.; Kakihara, A.; Nogami, J.; Ohkawa, Y.; Tsukada, Y.I. Totipotency of mouse zygotes extends to single blastomeres of embryos at the four-cell stage. Sci. Rep. 2021, 11, 11167. [Google Scholar] [CrossRef]
- Guo, G.; Huss, M.; Tong, G.Q.; Wang, C.; Li Sun, L.; Clarke, N.D.; Robson, P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 2010, 18, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Boroviak, T.; Loos, R.; Bertone, P.; Smith, A.; Nichols, J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014, 16, 516–528. [Google Scholar] [CrossRef]
- Yamanaka, S.; Blau, H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010, 465, 704–712. [Google Scholar] [CrossRef]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Hackett, J.A.; Surani, M.A. Regulatory principles of pluripotency: From the ground state up. Cell Stem Cell 2014, 15, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Barber, M.; Mansfield, W.; Cui, Y.; Spindlow, D.; Stirparo, G.G.; Dietmann, S.; Nichols, J.; Smith, A. Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency. Cell Stem Cell 2021, 28, 453–471.e8. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.; Wu, J.; Shi, C.; Xu, Y.; Dong, J.; Wang, C.; Lai, W.; et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 2017, 169, 243–257.e25. [Google Scholar] [CrossRef] [PubMed]
- Sozen, B.; Cox, A.L.; De Jonghe, J.; Bao, M.; Hollfelder, F.; Glover, D.M.; Zernicka-Goetz, M. Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid. Dev. Cell 2019, 51, 698–712.e8. [Google Scholar] [CrossRef] [PubMed]
- Sozen, B.; Jorgensen, V.; Weatherbee, B.A.T.; Chen, S.; Zhu, M.; Zernicka-Goetz, M. Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat. Commun. 2021, 12, 5550. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Min, Z.; Alsolami, S.; Ma, Z.; Zhang, E.; Chen, W.; Zhong, K.; Pei, W.; Kang, X.; Zhang, P.; et al. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov. 2021, 7, 81. [Google Scholar] [CrossRef]
- Min, Z.; Zhong, K.; Luo, Y.; Fan, Y.; Yu, Y. Protein Expression Landscape Defines the Formation Potential of Mouse Blastoids From EPSCs. Front. Cell Dev. Biol. 2022, 10, 840492. [Google Scholar] [CrossRef]
- Tsubouchi, T.; Soza-Ried, J.; Brown, K.; Piccolo, F.M.; Cantone, I.; Landeira, D.; Bagci, H.; Hochegger, H.; Merkenschlager, M.; Fisher, A.G. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell 2013, 152, 873–883. [Google Scholar] [CrossRef]
- Hong, Y.J.; Hong, K.; Byun, S.; Choi, H.W.; Do, J.T. Reprogramming of Extraembryonic Trophoblast Stem Cells into Embryonic Pluripotent State by Fusion with Embryonic Stem Cells. Stem Cells Dev. 2018, 27, 1350–1359. [Google Scholar] [CrossRef]
- Tat, P.A.; Sumer, H.; Pralong, D.; Verma, P.J. The efficiency of cell fusion-based reprogramming is affected by the somatic cell type and the in vitro age of somatic cells. Cell. Reprogram. 2011, 13, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Do, J.T.; Han, D.W.; Scholer, H.R. Reprogramming somatic gene activity by fusion with pluripotent cells. Stem Cell Rev. 2006, 2, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Sumer, H.; Nicholls, C.; Liu, J.; Tat, P.A.; Liu, J.P.; Verma, P.J. Comparison of reprogramming ability of mouse ES and iPS cells measured by somatic cell fusion. Int. J. Dev. Biol. 2010, 54, 1723–1728. [Google Scholar] [CrossRef]
- Szabo, P.E.; Hubner, K.; Scholer, H.; Mann, J.R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 2002, 115, 157–160. [Google Scholar] [CrossRef]
- Do, J.T.; Scholer, H.R. Nuclei of embryonic stem cells reprogram somatic cells. Stem Cells 2004, 22, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Niakan, K.K.; Schrode, N.; Cho, L.T.; Hadjantonakis, A.K. Derivation of extraembryonic endoderm stem (XEN) cells from mouse embryos and embryonic stem cells. Nat. Protoc. 2013, 8, 1028–1041. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Hadley, W. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Taiyun Wei, V.S. R Package ‘Rorrplot’: Visualization of a Correlation Matrix. 2021. Available online: https://github.com/taiyun/corrplot (accessed on 13 May 2022).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. 2022. Available online: https://cran.r-project.org/web/packages/gplots (accessed on 13 May 2022).
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Geng, T.; Wu, D.Y.; Zhang, T.; He, H.N.; Du, H.N.; Zhang, D.; Miao, Y.L.; Jiang, W. Derivation of feeder-free human extended pluripotent stem cells. Stem Cell Rep. 2021, 16, 1686–1696. [Google Scholar] [CrossRef]
- Do, J.T.; Han, D.W.; Gentile, L.; Sobek-Klocke, I.; Stehling, M.; Lee, H.T.; Scholer, H.R. Erasure of cellular memory by fusion with pluripotent cells. Stem Cells 2007, 25, 1013–1020. [Google Scholar] [CrossRef]
- Pesce, M.; Scholer, H.R. Oct-4: Control of totipotency and germline determination. Mol. Reprod. Dev. 2000, 55, 452–457. [Google Scholar] [CrossRef]
- Choi, H.W.; Joo, J.Y.; Hong, Y.J.; Kim, J.S.; Song, H.; Lee, J.W.; Wu, G.; Scholer, H.R.; Do, J.T. Distinct Enhancer Activity of Oct4 in Naive and Primed Mouse Pluripotency. Stem Cell Rep. 2016, 7, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, J.C.; Lan, G.; Masaki, H.; Gao, X.; Antunes, L.; Yu, Y.; Zhu, Z.; et al. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Seo, B.J.; Han, M.J.; Hong, Y.J.; Hong, K.; Song, H.; Lee, J.W.; Do, J.T. Changes in the Expression of Mitochondrial Morphology-Related Genes during the Differentiation of Murine Embryonic Stem Cells. Stem Cells Int. 2020, 2020, 9369268. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A.; Joshi, P.; Contreras, E.; Gama, V. Remodeling of mitochondrial morphology and function: An emerging hallmark of cellular reprogramming. Cell Stress 2019, 3, 181–194. [Google Scholar] [CrossRef]
- Choi, H.W.; Kim, J.H.; Chung, M.K.; Hong, Y.J.; Jang, H.S.; Seo, B.J.; Jung, T.H.; Kim, J.S.; Chung, H.M.; Byun, S.J.; et al. Mitochondrial and metabolic remodeling during reprogramming and differentiation of the reprogrammed cells. Stem Cells Dev. 2015, 24, 1366–1373. [Google Scholar] [CrossRef]
- Choi, J.; Seo, B.J.; La, H.; Yoon, S.H.; Hong, Y.J.; Lee, J.H.; Chung, H.M.; Hong, K.; Do, J.T. Comparative analysis of the mitochondrial morphology, energy metabolism, and gene expression signatures in three types of blastocyst-derived stem cells. Redox Biol. 2020, 30, 101437. [Google Scholar] [CrossRef] [PubMed]
- Tsogtbaatar, E.; Landin, C.; Minter-Dykhouse, K.; Folmes, C.D.L. Energy Metabolism Regulates Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 87. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, S.A.; Prasain, N.; Bae, D.; Kang, H.; Ha, T.; Kim, J.S.; Hong, K.S.; Mantel, C.; Moon, S.H.; et al. Metabolome Profiling of Partial and Fully Reprogrammed Induced Pluripotent Stem Cells. Stem Cells Dev. 2017, 26, 734–742. [Google Scholar] [CrossRef]
- Liu, B.; Chen, S.; Xu, Y.; Lyu, Y.; Wang, J.; Du, Y.; Sun, Y.; Liu, H.; Zhou, H.; Lai, W.; et al. Chemically defined and xeno-free culture condition for human extended pluripotent stem cells. Nat. Commun. 2021, 12, 3017. [Google Scholar] [CrossRef]
- Han, D.W.; Do, J.T.; Gentile, L.; Stehling, M.; Lee, H.T.; Scholer, H.R. Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells 2008, 26, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Do, J.T.; Han, D.W.; Gentile, L.; Sobek-Klocke, I.; Stehling, M.; Scholer, H.R. Enhanced reprogramming of Xist by induced upregulation of Tsix and Dnmt3a. Stem Cells 2008, 26, 2821–2831. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Guo, R.; Ji, T.; Zhang, J.; Xu, J.; Li, Y.; Sheng, Y.; Wang, Y.; Fang, K.; Wen, Y.; et al. YY1 safeguard multidimensional epigenetic landscape associated with extended pluripotency. Nucleic Acids Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.; Markov, G.J.; Brady, J.J.; Palla, A.; Zeng, H.; Sebastiano, V.; Blau, H.M. NKX3-1 is required for induced pluripotent stem cell reprogramming and can replace OCT4 in mouse and human iPSC induction. Nat. Cell Biol. 2018, 20, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.E.; Fisher, A.G. Reprogramming lineage identity through cell-cell fusion. Curr. Opin. Genet. Dev. 2021, 70, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Do, J.T.; Choi, H.W.; Choi, Y.; Scholer, H.R. Pluripotent hybrid cells contribute to extraembryonic as well as embryonic tissues. Stem Cells Dev. 2011, 20, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Posfai, E.; Schell, J.P.; Janiszewski, A.; Rovic, I.; Murray, A.; Bradshaw, B.; Yamakawa, T.; Pardon, T.; El Bakkali, M.; Talon, I.; et al. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 2021, 23, 49–60. [Google Scholar] [CrossRef]
- Seo, B.J.; Yoon, S.H.; Do, J.T. Mitochondrial Dynamics in Stem Cells and Differentiation. Int. J. Mol. Sci. 2018, 19, 3893. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Choi, M.; Margineantu, D.; Margaretha, L.; Hesson, J.; Cavanaugh, C.; Blau, C.A.; Horwitz, M.S.; Hockenbery, D.; Ware, C.; et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012, 31, 2103–2116. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef]
- Jiang, B.H.; Tseng, W.L.; Li, H.Y.; Wang, M.L.; Chang, Y.L.; Sung, Y.J.; Chiou, S.H. Poly(ADP-Ribose) Polymerase 1: Cellular Pluripotency, Reprogramming, and Tumorogenesis. Int. J. Mol. Sci. 2015, 16, 15531–15545. [Google Scholar] [CrossRef]
- Ogino, H.; Nozaki, T.; Gunji, A.; Maeda, M.; Suzuki, H.; Ohta, T.; Murakami, Y.; Nakagama, H.; Sugimura, T.; Masutani, M. Loss of Parp-1 affects gene expression profile in a genome-wide manner in ES cells and liver cells. BMC Genom. 2007, 8, 41. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.-H.; Choi, J.; Hong, Y.-J.; La, H.; Hong, T.-K.; Hong, K.; Do, J.-T. Developmental Potency and Metabolic Traits of Extended Pluripotency Are Faithfully Transferred to Somatic Cells via Cell Fusion-Induced Reprogramming. Cells 2022, 11, 3266. https://doi.org/10.3390/cells11203266
Song J-H, Choi J, Hong Y-J, La H, Hong T-K, Hong K, Do J-T. Developmental Potency and Metabolic Traits of Extended Pluripotency Are Faithfully Transferred to Somatic Cells via Cell Fusion-Induced Reprogramming. Cells. 2022; 11(20):3266. https://doi.org/10.3390/cells11203266
Chicago/Turabian StyleSong, Jae-Hoon, Joonhyuk Choi, Yean-Ju Hong, Hyeonwoo La, Tae-Kyung Hong, Kwonho Hong, and Jeong-Tae Do. 2022. "Developmental Potency and Metabolic Traits of Extended Pluripotency Are Faithfully Transferred to Somatic Cells via Cell Fusion-Induced Reprogramming" Cells 11, no. 20: 3266. https://doi.org/10.3390/cells11203266
APA StyleSong, J.-H., Choi, J., Hong, Y.-J., La, H., Hong, T.-K., Hong, K., & Do, J.-T. (2022). Developmental Potency and Metabolic Traits of Extended Pluripotency Are Faithfully Transferred to Somatic Cells via Cell Fusion-Induced Reprogramming. Cells, 11(20), 3266. https://doi.org/10.3390/cells11203266






