Distinct Mechanisms of Target Search by Endonuclease VIII-like DNA Glycosylases
Abstract
1. Introduction
2. Materials and Methods
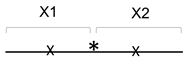
2.1. Oligonucleotides and Substrate Preparation
2.2. Enzymes
2.3. Steady-State Kinetics
2.4. Correlated Cleavage Assay
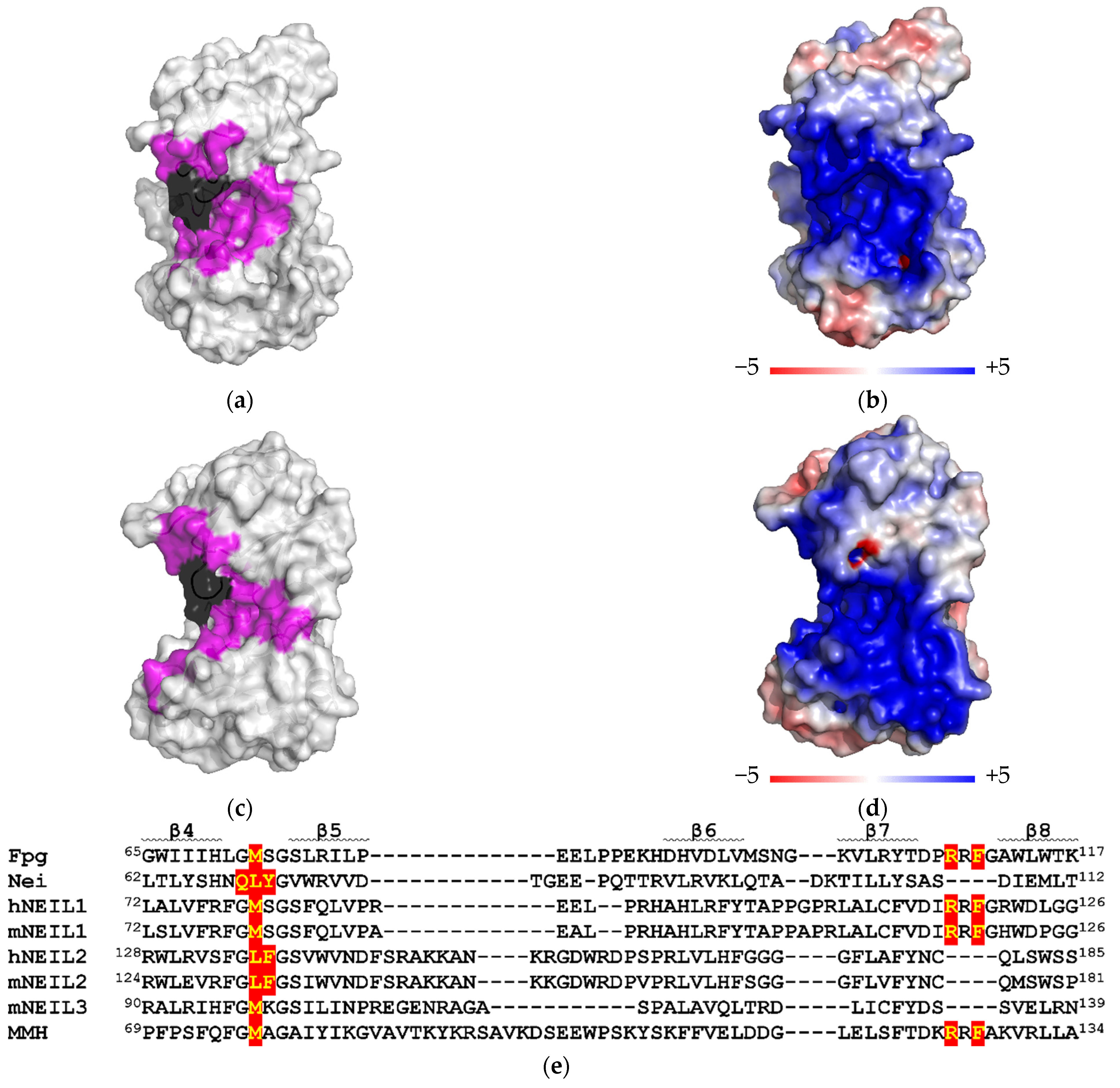
2.5. Computational Analysis
3. Results
3.1. Steady-State Kinetics of Nei, NEIL1 and NEIL2 on Individual Target Sites
3.2. Correlated Cleavage of Substrates
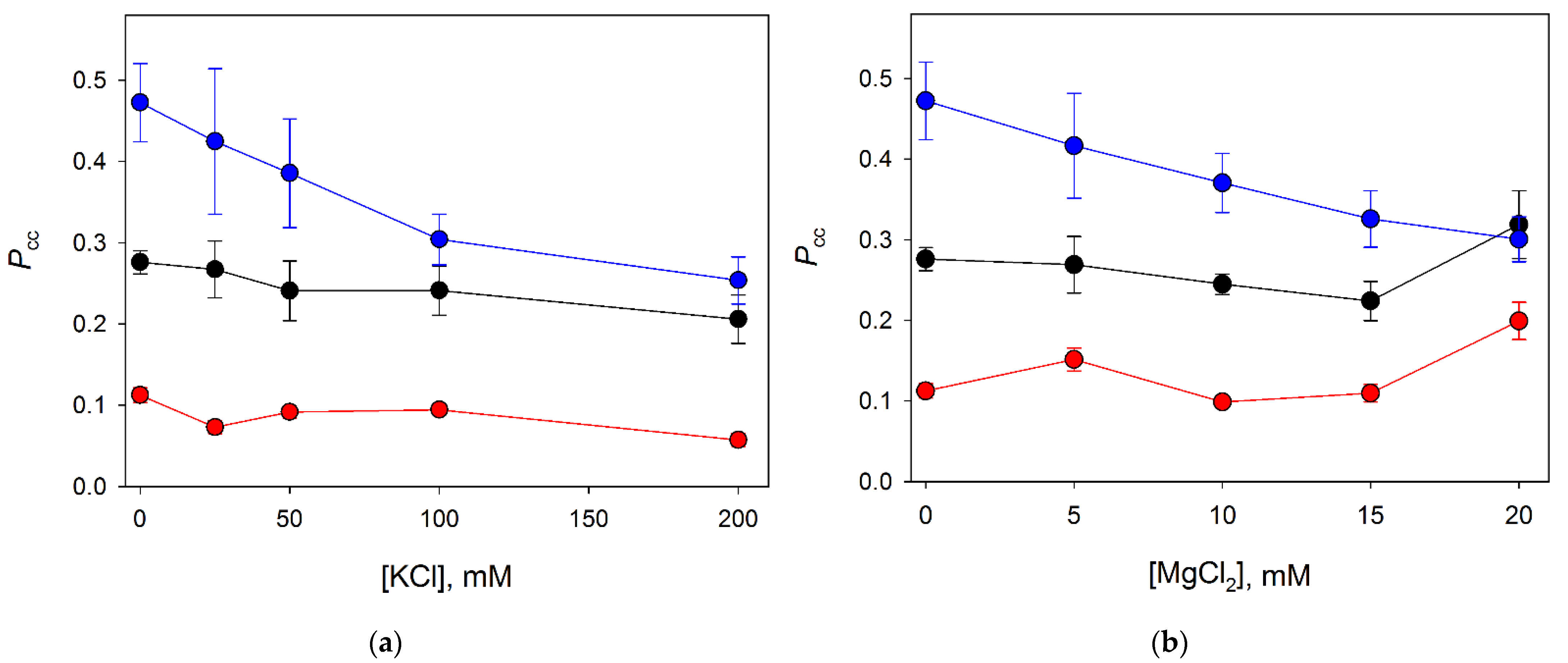
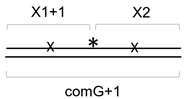
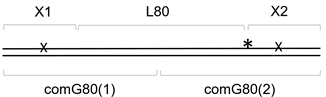
3.3. Distance Dependence of the Correlated Cleavage
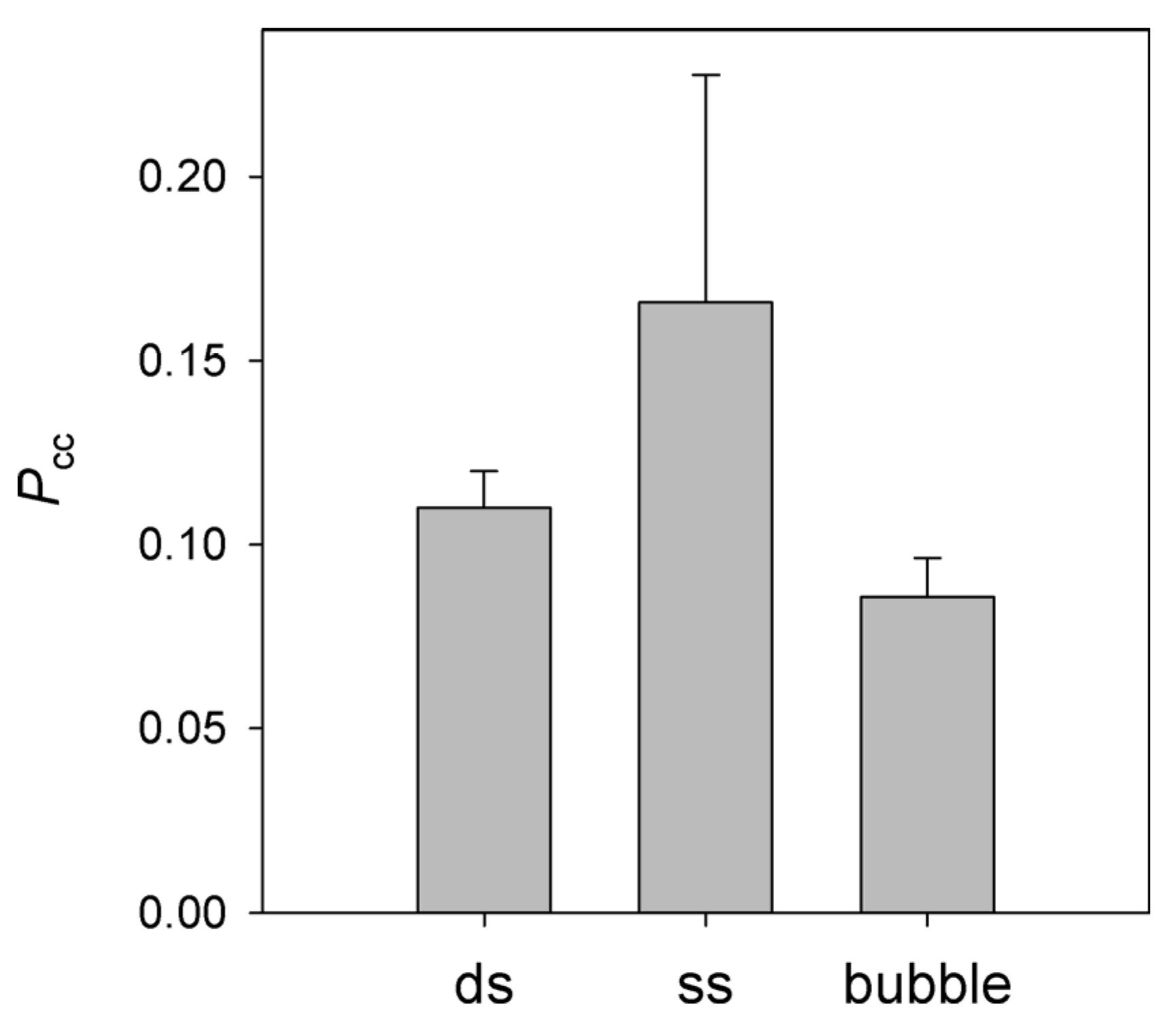
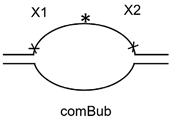
3.4. Processivity of NEIL2 on Substrates of Different Structure
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zharkov, D.O.; Grollman, A.P. The DNA trackwalkers: Principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005, 577, 24–54. [Google Scholar] [CrossRef] [PubMed]
- Berg, O.G.; Winter, R.B.; von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 1981, 20, 6929–6948. [Google Scholar] [CrossRef] [PubMed]
- Winter, R.B.; von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor–operator interaction: Equilibrium measurements. Biochemistry 1981, 20, 6948–6960. [Google Scholar] [CrossRef] [PubMed]
- Winter, R.B.; Berg, O.G.; von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: Kinetic measurements and conclusions. Biochemistry 1981, 20, 6961–6977. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D.; Schultz, R.A.; Ellenberger, T. DNA Repair and Mutagenesis; ASM Press: Washington, DC, USA, 2006; 1118p. [Google Scholar]
- Porecha, R.H.; Stivers, J.T. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl. Acad. Sci. USA 2008, 105, 10791–10796. [Google Scholar] [CrossRef] [PubMed]
- Sidorenko, V.S.; Mechetin, G.V.; Nevinsky, G.A.; Zharkov, D.O. Correlated cleavage of single- and double-stranded substrates by uracil-DNA glycosylase. FEBS Lett. 2008, 582, 410–414. [Google Scholar] [CrossRef]
- Schonhoft, J.D.; Stivers, J.T. Timing facilitated site transfer of an enzyme on DNA. Nat. Chem. Biol. 2012, 8, 205–210. [Google Scholar] [CrossRef]
- Schonhoft, J.D.; Kosowicz, J.G.; Stivers, J.T. DNA translocation by human uracil DNA glycosylase: Role of DNA phosphate charge. Biochemistry 2013, 52, 2526–2535. [Google Scholar] [CrossRef]
- Schonhoft, J.D.; Stivers, J.T. DNA translocation by human uracil DNA glycosylase: The case of single-stranded DNA and clustered uracils. Biochemistry 2013, 52, 2536–2544. [Google Scholar] [CrossRef]
- Cravens, S.L.; Schonhoft, J.D.; Rowland, M.M.; Rodriguez, A.A.; Anderson, B.G.; Stivers, J.T. Molecular crowding enhances facilitated diffusion of two human DNA glycosylases. Nucleic Acids Res. 2015, 43, 4087–4097. [Google Scholar] [CrossRef]
- Cravens, S.L.; Stivers, J.T. Comparative effects of ions, molecular crowding, and bulk DNA on the damage search mechanisms of hOGG1 and hUNG. Biochemistry 2016, 55, 5230–5242. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.W.; David, S.S. Escherichia coli MutY and Fpg utilize a processive mechanism for target location. Biochemistry 2003, 42, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C.; Luo, G.; Kou, S.C.; Mangel, W.F.; Verdine, G.L.; Bagchi, B.; Xie, X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009, 16, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.R.; Kathe, S.D.; Hilzinger, T.S.; Averill, A.M.; Warshaw, D.M.; Wallace, S.S.; Lee, A.J. Single molecule glycosylase studies with engineered 8-oxoguanine DNA damage sites show functional defects of a MUTYH polyposis variant. Nucleic Acids Res. 2019, 47, 3058–3071. [Google Scholar] [CrossRef]
- Buechner, C.N.; Maiti, A.; Drohat, A.C.; Tessmer, I. Lesion search and recognition by thymine DNA glycosylase revealed by single molecule imaging. Nucleic Acids Res. 2015, 43, 2716–2729. [Google Scholar] [CrossRef]
- Hedglin, M.; O’Brien, P.J. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry 2008, 47, 11434–11445. [Google Scholar] [CrossRef]
- Zhang, Y.; O’Brien, P.J. Repair of alkylation damage in eukaryotic chromatin depends on searching ability of alkyladenine DNA glycosylase. ACS Chem. Biol. 2015, 10, 2606–2615. [Google Scholar] [CrossRef]
- Hendershot, J.M.; O’Brien, P.J. Search for DNA damage by human alkyladenine DNA glycosylase involves early intercalation by an aromatic residue. J. Biol. Chem. 2017, 292, 16070–16080. [Google Scholar] [CrossRef]
- Blainey, P.C.; van Oijen, A.M.; Banerjee, A.; Verdine, G.L.; Xie, X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 5752–5757. [Google Scholar] [CrossRef]
- Sidorenko, V.S.; Zharkov, D.O. Correlated cleavage of damaged DNA by bacterial and human 8-oxoguanine-DNA glycosylases. Biochemistry 2008, 47, 8970–8976. [Google Scholar] [CrossRef]
- Rowland, M.M.; Schonhoft, J.D.; McKibbin, P.L.; David, S.S.; Stivers, J.T. Microscopic mechanism of DNA damage searching by hOGG1. Nucleic Acids Res. 2014, 42, 9295–9303. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.R.; Kad, N.M.; Nelson, S.R.; Warshaw, D.M.; Wallace, S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011, 39, 7487–7498. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.R.; Dunn, A.R.; Kathe, S.D.; Warshaw, D.M.; Wallace, S.S. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. USA 2014, 111, E2091–E2099. [Google Scholar] [CrossRef] [PubMed]
- Zharkov, D.O.; Shoham, G.; Grollman, A.P. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair 2003, 2, 839–862. [Google Scholar] [CrossRef]
- Boiteux, S.; Coste, F.; Castaing, B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 2017, 107, 179–201. [Google Scholar] [CrossRef]
- Hazra, T.K.; Izumi, T.; Boldogh, I.; Imhoff, B.; Kow, Y.W.; Jaruga, P.; Dizdaroglu, M.; Mitra, S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. USA 2002, 99, 3523–3528. [Google Scholar] [CrossRef]
- Bandaru, V.; Sunkara, S.; Wallace, S.S.; Bond, J.P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair 2002, 1, 517–529. [Google Scholar] [CrossRef]
- Hazra, T.K.; Kow, Y.W.; Hatahet, Z.; Imhoff, B.; Boldogh, I.; Mokkapati, S.K.; Mitra, S.; Izumi, T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002, 277, 30417–30420. [Google Scholar] [CrossRef]
- Morland, I.; Rolseth, V.; Luna, L.; Rognes, T.; Bjørås, M.; Seeberg, E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002, 30, 4926–4936. [Google Scholar] [CrossRef]
- Takao, M.; Kanno, S.-i.; Kobayashi, K.; Zhang, Q.-M.; Yonei, S.; van der Horst, G.T.J.; Yasui, A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 2002, 277, 42205–42213. [Google Scholar] [CrossRef]
- Rosenquist, T.A.; Zaika, E.; Fernandes, A.S.; Zharkov, D.O.; Miller, H.; Grollman, A.P. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair 2003, 2, 581–591. [Google Scholar] [CrossRef]
- Guan, X.; Bai, H.; Shi, G.; Theriot, C.A.; Hazra, T.K.; Mitra, S.; Lu, A.-L. The human checkpoint sensor Rad9–Rad1–Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007, 35, 2463–2472. [Google Scholar] [CrossRef] [PubMed]
- Dou, H.; Theriot, C.A.; Das, A.; Hegde, M.L.; Matsumoto, Y.; Boldogh, I.; Hazra, T.K.; Bhakat, K.K.; Mitra, S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen: The potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 2008, 283, 3130–3140. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.A.; Hegde, M.L.; Hazra, T.K.; Mitra, S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair 2010, 9, 643–652. [Google Scholar] [CrossRef]
- Yamamoto, R.; Ohshiro, Y.; Shimotani, T.; Yamamoto, M.; Matsuyama, S.; Ide, H.; Kubo, K. Hypersensitivity of mouse NEIL1-knockdown cells to hydrogen peroxide during S phase. J. Radiat. Res. 2014, 55, 707–712. [Google Scholar] [CrossRef]
- Albelazi, M.S.; Martin, P.R.; Mohammed, S.; Mutti, L.; Parsons, J.L.; Elder, R.H. The biochemical role of the human NEIL1 and NEIL3 DNA glycosylases on model DNA replication forks. Genes 2019, 10, 315. [Google Scholar] [CrossRef]
- Dou, H.; Mitra, S.; Hazra, T.K. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003, 278, 49679–49684. [Google Scholar] [CrossRef]
- Banerjee, D.; Mandal, S.M.; Das, A.; Hegde, M.L.; Das, S.; Bhakat, K.K.; Boldogh, I.; Sarkar, P.S.; Mitra, S.; Hazra, T.K. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011, 286, 6006–6016. [Google Scholar] [CrossRef]
- Chakraborty, A.; Wakamiya, M.; Venkova-Canova, T.; Pandita, R.K.; Aguilera-Aguirre, L.; Sarker, A.H.; Singh, D.K.; Hosoki, K.; Wood, T.G.; Sharma, G.; et al. Neil2-null mice accumulate oxidized DNA bases in the transcriptionally active sequences of the genome and are susceptible to innate inflammation. J. Biol. Chem. 2015, 290, 24636–24648. [Google Scholar] [CrossRef]
- Makasheva, K.A.; Endutkin, A.V.; Zharkov, D.O. Requirements for DNA bubble structure for efficient cleavage by helix–two-turn–helix DNA glycosylases. Mutagenesis 2020, 35, 119–128. [Google Scholar] [CrossRef]
- Liu, M.; Bandaru, V.; Bond, J.P.; Jaruga, P.; Zhao, X.; Christov, P.P.; Burrows, C.J.; Rizzo, C.J.; Dizdaroglu, M.; Wallace, S.S. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 4925–4930. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Imamura, K.; Averill, A.M.; Wallace, S.S.; Doublié, S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure 2013, 21, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Semlow, D.R.; Zhang, J.; Budzowska, M.; Drohat, A.C.; Walter, J.C. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 2016, 167, 498–511.e414. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.R.; Couvé, S.; Zutterling, C.; Albelazi, M.S.; Groisman, R.; Matkarimov, B.T.; Parsons, J.L.; Elder, R.H.; Saparbaev, M.K. The human DNA glycosylases NEIL1 and NEIL3 excise psoralen-induced DNA-DNA cross-links in a four-stranded DNA structure. Sci. Rep. 2017, 7, 17438. [Google Scholar] [CrossRef]
- Kolbanovskiy, M.; Aharonoff, A.; Sales, A.H.; Geacintov, N.E.; Shafirovich, V. Recognition and repair of oxidatively generated DNA lesions in plasmid DNA by a facilitated diffusion mechanism. Biochem. J. 2021, 478, 2359–2370. [Google Scholar] [CrossRef] [PubMed]
- Mechetin, G.V.; Zharkov, D.O. Mechanism of translocation of uracil-DNA glycosylase from Escherichia coli between distributed lesions. Biochem. Biophys. Res. Commun. 2011, 414, 425–430. [Google Scholar] [CrossRef]
- Conlon, K.A.; Miller, H.; Rosenquist, T.A.; Zharkov, D.O.; Berrios, M. The murine DNA glycosylase NEIL2 (mNEIL2) and human DNA polymerase β bind microtubules in situ and in vitro. DNA Repair 2005, 4, 419–431. [Google Scholar] [CrossRef]
- Rieger, R.A.; McTigue, M.M.; Kycia, J.H.; Gerchman, S.E.; Grollman, A.P.; Iden, C.R. Characterization of a cross-linked DNA-endonuclease VIII repair complex by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2000, 11, 505–515. [Google Scholar] [CrossRef]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef]
- Zhu, C.; Lu, L.; Zhang, J.; Yue, Z.; Song, J.; Zong, S.; Liu, M.; Stovicek, O.; Gao, Y.Q.; Yi, C. Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair. Proc. Natl. Acad. Sci. USA 2016, 113, 7792–7797. [Google Scholar] [CrossRef]
- Zharkov, D.O.; Golan, G.; Gilboa, R.; Fernandes, A.S.; Gerchman, S.E.; Kycia, J.H.; Rieger, R.A.; Grollman, A.P.; Shoham, G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002, 21, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Hatahet, Z.; Melamede, R.J.; Kow, Y.W.; Wallace, S.S. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997, 272, 32230–32239. [Google Scholar] [CrossRef] [PubMed]
- Bonvin, A.M.J.J. Localisation and dynamics of sodium counterions around DNA in solution from molecular dynamics simulation. Eur. Biophys. J. 2000, 29, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Subirana, J.A.; Soler-López, M. Cations as hydrogen bond donors: A view of electrostatic interactions in DNA. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Stanford, N.P.; Szczelkun, M.D.; Marko, J.F.; Halford, S.E. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000, 19, 6546–6557. [Google Scholar] [CrossRef]
- Belotserkovskii, B.P.; Zarling, D.A. Analysis of a one-dimensional random walk with irreversible losses at each step: Applications for protein movement on DNA. J. Theor. Biol. 2004, 226, 195–203. [Google Scholar] [CrossRef]
- Aamann, M.D.; Hvitby, C.; Popuri, V.; Muftuoglu, M.; Lemminger, L.; Skeby, C.K.; Keijzers, G.; Ahn, B.; Bjørås, M.; Bohr, V.A.; et al. Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity. Mech. Ageing Dev. 2014, 135, 1–14. [Google Scholar] [CrossRef]
- Cao, C.; Jiang, Y.L.; Stivers, J.T.; Song, F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat. Struct. Mol. Biol. 2004, 11, 1230–1236. [Google Scholar] [CrossRef]
- Parker, J.B.; Bianchet, M.A.; Krosky, D.J.; Friedman, J.I.; Amzel, L.M.; Stivers, J.T. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature 2007, 449, 433–437. [Google Scholar] [CrossRef]
- Banerjee, A.; Santos, W.L.; Verdine, G.L. Structure of a DNA glycosylase searching for lesions. Science 2006, 311, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Spong, M.C.; Nam, K.; Banerjee, A.; Jiralerspong, S.; Karplus, M.; Verdine, G.L. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 2009, 462, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Bergonzo, C.; Campbell, A.J.; Li, H.; Mechetin, G.V.; de los Santos, C.; Grollman, A.P.; Fedorova, O.S.; Zharkov, D.O.; Simmerling, C. Active destabilization of base pairs by a DNA glycosylase wedge initiates damage recognition. Nucleic Acids Res. 2015, 43, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Endutkin, A.V.; Bergonzo, C.; Campbell, A.J.; de los Santos, C.; Grollman, A.; Zharkov, D.O.; Simmerling, C. A dynamic checkpoint in oxidative lesion discrimination by formamidopyrimidine–DNA glycosylase. Nucleic Acids Res. 2016, 44, 683–694. [Google Scholar] [CrossRef]
- Fromme, J.C.; Verdine, G.L. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 2002, 9, 544–552. [Google Scholar] [CrossRef]
- Gilboa, R.; Zharkov, D.O.; Golan, G.; Fernandes, A.S.; Gerchman, S.E.; Matz, E.; Kycia, J.H.; Grollman, A.P.; Shoham, G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002, 277, 19811–19816. [Google Scholar] [CrossRef]
- Duclos, S.; Aller, P.; Jaruga, P.; Dizdaroglu, M.; Wallace, S.S.; Doublié, S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair 2012, 11, 714–725. [Google Scholar] [CrossRef]
- Eckenroth, B.E.; Cao, V.B.; Averill, A.M.; Dragon, J.A.; Doublié, S. Unique structural features of mammalian NEIL2 DNA glycosylase prime its activity for diverse DNA substrates and environments. Structure 2021, 29, 29–42.e24. [Google Scholar] [CrossRef]
- Prakash, A.; Eckenroth, B.E.; Averill, A.M.; Imamura, K.; Wallace, S.S.; Doublié, S. Structural investigation of a viral ortholog of human NEIL2/3 DNA glycosylases. DNA Repair 2013, 12, 1062–1071. [Google Scholar] [CrossRef]
- Golan, G.; Zharkov, D.O.; Feinberg, H.; Fernandes, A.S.; Zaika, E.I.; Kycia, J.H.; Grollman, A.P.; Shoham, G. Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res. 2005, 33, 5006–5016. [Google Scholar] [CrossRef]
- Zhdanova, P.V.; Ishchenko, A.A.; Chernonosov, A.A.; Zharkov, D.O.; Koval, V.V. Dynamics and conformational changes in human NEIL2 DNA glycosylase analyzed by hydrogen/deuterium exchange mass spectrometry. J. Mol. Biol. 2022, 434, 167334. [Google Scholar] [CrossRef]
- Doublié, S.; Bandaru, V.; Bond, J.P.; Wallace, S.S. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 10284–10289. [Google Scholar] [CrossRef] [PubMed]
- Landová, B.; Šilhán, J. Conformational changes of DNA repair glycosylase MutM triggered by DNA binding. FEBS Lett. 2020, 594, 3032–3044. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, M.; Mikawa, T.; Kumasaka, T.; Yamamoto, M.; Kato, R.; Fukuyama, K.; Inoue, Y.; Kuramitsu, S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000, 19, 3857–3869. [Google Scholar] [CrossRef] [PubMed]
- Gruskin, E.A.; Lloyd, R.S. Molecular analysis of plasmid DNA repair within ultraviolet-irradiated Escherichia coli. I. T4 endonuclease V-initiated excision repair. J. Biol. Chem. 1988, 263, 12728–12737. [Google Scholar] [CrossRef]
- Esadze, A.; Rodriguez, G.; Weiser, B.P.; Cole, P.A.; Stivers, J.T. Measurement of nanoscale DNA translocation by uracil DNA glycosylase in human cells. Nucleic Acids Res. 2017, 45, 12413–12424. [Google Scholar] [CrossRef] [PubMed]


| ID | Sequence (5′→3′) a,b,c |
|---|---|
| X1 | TCCCTTCXCTCCTTTCCTTC |
| X2 | GGACTTCXCTCCTTTCCAGA |
| C1 | GAAGGAAAGGAGCGAAGGGA |
| C2 | TCTGGAAAGGAGCGAAGTCC |
| comG | TCTGGAAAGGAGGGAAGTCCGAAGGAAAGGAGGGAAGGGA |
| X1+1 | TCCCTTCXCTCCTTTCCTTCC |
| comG+1 | TCTGGAAAGGAGGGAAGTCCGGAAGGAAAGGAGGGAAGGGA |
| L40 | pGGACTTTACTTGCGTTAGAGC |
| comG40 | TCTGGAAAGGAGGGAAGTCCGCTCTAACGCAAGTAAAGTCCGAAGGAAAGGAGGGAAGGGA |
| L60 | pGGACCTTTCATTTGTGCGATCTTTCCTCTCGTTCAGACCTC |
| comG60(1) | pGATCGCACAAATGAAAGGTCCGAAGGAAAGGAGGGAAGGGA |
| comG60(2) | TCTGGAAAGGAGGGAAGTCCGAGGTCTGAACGAGAGGAAA |
| L80 | pGGACCTTTCATTTGTGCGATGAGTGAATTTCGGGATTTAGCTTTCCTCTCGTTCAGACCTC |
| comG80(1) | pAAATTCACTCATCGCACAAATGAAAGGTCCGAAGGAAAGGAGGGAAGGGA |
| comG80(2) | TCTGGAAAGGAGGGAAGTCCGAGGTCTGAACGAGAGGAAAGCTAAATCCCG |
| comBub | TCTGGAAAGGAGATGGACTAACGAACCCAAGTAGAAGGGA |
| com30 | AAAGGAGCGAAGTCCGAAGGAAAGGAGCGA |
| Type of the Experiment | |||
|---|---|---|---|
| Steady-state kinetics | Dependence on [K+] and [Mg2+] | Processivity of NEIL2 | Dependence on the distance between the targets |
 |  |  |  |
 |  | ||
 | |||
 | |||
| KM, nM | Vmax, nM × min−1 | Vmax/KM, min−1 | ||
|---|---|---|---|---|
| Nei | X1/C1 | 52 ± 13 | 10 ± 2 | 0.20 ± 0.02 |
| X2/C2 | 56 ± 17 | 11 ± 2 | 0.20 ± 0.02 | |
| NEIL1 | X1/C1 | n/d b | n/d | 0.08 ± 0.01 |
| X2/C2 | n/d | n/d | 0.12 ± 0.01 | |
| NEIL2 | X1/C1 | 101 ± 22 | 17 ± 3 | 0.17 ± 0.01 |
| X2/C2 | 138 ± 37 | 17 ± 3 | 0.15 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diatlova, E.A.; Mechetin, G.V.; Zharkov, D.O. Distinct Mechanisms of Target Search by Endonuclease VIII-like DNA Glycosylases. Cells 2022, 11, 3192. https://doi.org/10.3390/cells11203192
Diatlova EA, Mechetin GV, Zharkov DO. Distinct Mechanisms of Target Search by Endonuclease VIII-like DNA Glycosylases. Cells. 2022; 11(20):3192. https://doi.org/10.3390/cells11203192
Chicago/Turabian StyleDiatlova, Evgeniia A., Grigory V. Mechetin, and Dmitry O. Zharkov. 2022. "Distinct Mechanisms of Target Search by Endonuclease VIII-like DNA Glycosylases" Cells 11, no. 20: 3192. https://doi.org/10.3390/cells11203192
APA StyleDiatlova, E. A., Mechetin, G. V., & Zharkov, D. O. (2022). Distinct Mechanisms of Target Search by Endonuclease VIII-like DNA Glycosylases. Cells, 11(20), 3192. https://doi.org/10.3390/cells11203192






