Abstract
Chimeric RNAs (chiRNAs) play many previously unrecognized roles in different diseases including cancer. They can not only be used as biomarkers for diagnosis and prognosis of various diseases but also serve as potential therapeutic targets. In order to better understand the roles of chiRNAs in pathogenesis, we inserted human sequences into mouse genome and established a knockin mouse model of the tamoxifen-inducible expression of ASTN2-PAPPA antisense chimeric RNA (A-PaschiRNA). Mice carrying the A-PaschiRNA knockin gene do not display any apparent abnormalities in growth, fertility, histological, hematopoietic, and biochemical indices. Using this model, we dissected the role of A-PaschiRNA in chemical carcinogen 4-nitroquinoline 1-oxide (4NQO)-induced carcinogenesis of esophageal squamous cell carcinoma (ESCC). To our knowledge, we are the first to generate a chiRNA knockin mouse model using the Cre-loxP system. The model could be used to explore the roles of chiRNA in pathogenesis and potential targeted therapies.
1. Introduction
Chimeric RNAs (chiRNAs) are known as fusion transcripts because they comprise RNA fragments derived from different genes [1,2]. Canonical chiRNAs are transcripts of chromosomally rearranged DNA, while non-canonical chiRNAs are formed via DNA-independent mechanism such as trans-splicing or transcription readthrough. With the development of advanced deep sequencing technologies, more non-canonical chiRNAs have been discovered [1,3,4]. Although the precise mechanisms underlying the formation of non-canonical chiRNA have not been completely elucidated [5,6], accumulating evidence indicates that chiRNAs play important roles in a broad spectrum of diseases, including cancer [2,7]. Given their unique expression in the tissues/cells, a subgroup of chiRNAs can serve not only biomarkers for both diagnosis and prognosis [5,8] but also potential therapeutic targets [9].
We have previously investigated the potentials of chiRNAs as biomarkers and molecular targets in cancer [10,11,12]. We examined salivary exosomal GOLM1-NAA35 chiRNA (seG-NchiRNA) in patients with esophageal squamous cell carcinoma (ESCC) and found that seG-NchiRNA can serve as a convenient, reliable, and noninvasive biomarker for the assessment of therapeutic response and recurrence [12]. In addition, ASTN2-PAPPAantisense chimeric RNA (A-PaschiRNA), which is generated by transcription read-through/splicing or trans-splicing, is also highly expressed in ESCC but not the surrounding normal esophagus. Results derived from xenograft models indicate that A-PaschiRNA can promote ESCC cell spread to lymph nodes and enhance stemness through modulating OCT4. Mechanistically, A-PaschiRNA induces cancer stemness by activating extracellular-signal-regulated kinase 5 (ERK5)-mediated non-canonical RNA Polymerase II-Associated Factor 1 Homolog (PAF1) [11]. These findings suggest that chiRNAs could play critical roles in cancer pathogenesis. In order to better understand the role of A-PaschiRNA in pathogenesis as well as the underlying molecular mechanisms, we decided to establish a mouse model conditionally expressing A-PaschiRNA.
The Cre-loxP-mediated recombination system has been widely used to create genetically engineered mouse models [13]. In addition, tamoxifen-induced gene expression has been proven to be helpful for the control of conditional expressions [14,15,16,17]. In this study, we first generated and characterized a tamoxifen-inducible A-PaschiRNA knockin (KI) mouse model and then induced ESCC by 4-nitroquinoline 1-oxide (4NQO). By controlling its expression, we demonstrated the impact of A-PaschiRNA on ESCC development.
2. Materials and Methods
2.1. Establishment and Characterization of A-PaschiRNA Conditional Knockin Model
Mice used in this study were generated from crossing A-PaschiRNAflox/flox mice with CAG-CreER mice on C57BL/6 background. CAG-CreER mice were originally generated by Jackson Model Animal Laboratory and provided by Cyagen Biosciences (Guangzhou, China). The “CAG promoter-loxP-3*polyA-loxP-A-Pas sequence-polyA” cassette was inserted into intron 1 of ROSA26. The targeting vector was obtained from Cyagen Biosciences (Guangzhou, China). In the targeting vector, the Neo (neomycin resistance gene) cassette was flanked by SDA (self-deletion anchor) sites, and DTA (diphtheria toxin A) was used for negative selection. Correctly inserted constructs were confirmed by digestions with different restriction enzymes and DNA sequencing (Supplementary Table S1). 3-month-old male and female mice were administered tamoxifen (Cat. T-5648, Sigma, St. Louis, MO, USA) for 5 consecutive days (50 mg/kg body weight; intraperitoneal injection). Mice were genotyped for floxed and Cre alleles as well as the excised allele after tamoxifen induction using the following primers: F4 (5′-GGAAAGAACCAGCTGGGGGATATC-3′), R4 (5′-GCCATTTAAGCCATGGGAAGTTAG-3′), and F5 (5′-TGGACAGAGGAGCCATAACTGCAG-3′) for targeted allele; Primers F1 (5′-CATATTGGCAGAACGAAAACGC-3′) and R1 (5′-CCTGTTTCACTATCCAGGTTACGG-3′) for Cre transgene. PCR products were separated on a 2% agarose gel to verify the DNA size [18]. All mice were housed under strictly controlled daily lighting (12 h light/dark) at 20–22 °C and 50–55% humidity and were provided ultra-filtered water and food ad libitum. All animal procedures were approved by the Animal Care and Use Committee of Jinan University (IACUC-20191210-03).
2.2. Targeting A-PaschiRNA to Embryonic Stem Cells (ES Cells)
The ROSA26 targeting construct was linearized by restriction enzyme NotI (R3189L, New England Biolabs, Ipswich, MA, USA), followed by phenol/chloroform extraction and ethanol precipitation. The linearized vector was transfected into C57BL/6 ES cells by electroporation, and the transfected ES cells were subject to G418 selection (200 μg/mL) for 24 h. The following primers were used for screening potentially targeted clones: primers P3 (5′-CAAAGCTGAAAGCTAAGTCTGCAG-3′) and P4 (5′-GGGCCATTTACCGTAAGTTATGTAACG-3′) for KI1 PCR; primers P5 (5′-CAACGTGCTGGTTATTGTGCTGTCT-3′) and P6 (5′-TGGTGGCCACGTGTAGTGGATCC-3′) for KI2 PCR. The positive clones (1A1, 1A8, 1A10, 1E3, 1G3, and 1H2) were expanded and further characterized by Southern blot analysis (Supplementary Figure S2F). DNAs were digested by endonucleases, separated by electrophoresis, transferred to a nitrocellulose membrane, and hybridized with labeled DNA probes. All six expanded clones were confirmed to be correctly targeted.
2.3. Generation of Mice Expressing chiRNA and Breeding Scheme
Targeted ES cell clone 1E3 was injected into C57BL/6 albino embryos, which were then re-implanted into CD-1 pseudo-pregnant females. Founder animals were identified by their coat color, and their germline transmission was confirmed by breeding with C57BL/6 females and subsequent genotyping of the offspring. The Neo cassette is self-deleted in germ cells so the offsprings were Neo cassette-free. F1 offspring were identified by PCR for the presence of the A-Pasflox/+ allele using genomic DNA isolated from tail tissue. The following primers were used for screening for KI1, KI2, KI3, and Neo deletion: F1 (5′-CATATTGGCAGAACGAAAACGC-3′) and R1 (5′-CCTGTTTCACTATCCAGGTTACGG-3′) for KI1; F2 (5′-GCATCCTCAAGGACACCAAAATCAC-3′) and R2 (5′-GATATCCCCCAGCTGGTTCTTTCC-3′) for KI2; F3 (5′-CAACGTGCTGGTTATTGTGCTGTCT-3′) and R3 (5′-TGGTGGCCACGTGTAGTGGATCC-3′) for KI3; and F4 (5′-GGAAAGAACCAGCTGGGGGATATC-3′), R4 (5′-GCCATTTAAGCCATGGGAAGTTAG-3′), and F5 (5′-TGGACAGAGGAGCCATAACTGCAG-3′) for neo deletion (Supplementary Figure S3A). A-Pasflox/flox, CAG-Cre mice were generated via crossing A-Pasflox/+ mice with CAG-CreER mice. After tamoxifen induction, A-PaschiRNA expressions in vital organs (liver, intestines, ovary, heart, brain, muscle, kidney, skin, tongue, stomach, lung, spleen, and esophagus) were detected by real-time quantitative PCR (RT-qPCR) [18]. The experiment process of RT-qPCR is described as follows. Total RNA was isolated from mice tissues by the TRIzol reagent (Cat. 15596-018, ThermoFisher, Waltham, MA, USA) and reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Cat. 4368813, Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s instructions. cDNA was amplified and quantified in the CFX Connect system (Cat. 1855200, BIO-RAD, Hercules, CA, USA) by using SYBR Green Master (Cat. 08733/09121, CWBIO, China). Reactions were performed using a total volume of 20 μL, which contained 1 μL of cDNA template (corresponding to 5 ng of the starting amount of RNA), 0.2 mM each primer, and 10 μL 2× SYBR Premix Ex Taq II. PCR cycling conditions were as follows: 94 °C for 30 s, followed by 40 cycles of 94 °C for 10 s, 55–62 °C for 10 s, and 72 °C for 10 s in a 96-well reaction plate, and the annealing temperature was based on the Tm value of primers. The melting curve was recorded after 40 cycles to verify primer specificity by heating from 65 °C to 95 °C. Each RT-qPCR reaction was performed in triplicate (technical replicates) on samples from three individual plants (biological replicates). cDNA was subjected to RT-qPCR with the following primers: A-PaschiRNA forward: 5′-CAACGTGCTGGTTATTGTGCTGTCT-3′ and reverse: 5′-TGGTGGCCACGTGTAGTGGATCC-3′; β-actin forward: 5′-GAACCCCAAGGCCAACCGCGAGA-3′ and reverse: 5′-TGACCCCGTCACCGGAGTCCATC-3′. β-actin served as a reference gene for normalization.
2.4. Induction the Expression of ESCC Model by 4NQO with or without Expression of chiRNA
Wild type (WT) and A-PaschiRNA KI mice (3 males and 3 females per group) weighing 20–22 g were randomly selected 4 weeks after the tamoxifen injection and anesthetized with 50 mg/kg body weight of avertin (2, 2, 2, -Tribromoethanol, 20 mg/mL, Sigma, St. Louis, MO, USA). Blood was collected from the canthal vein and subjected to cytological and biochemical tests. Small pieces of liver, kidney, spleen, lung, heart, and brain were fixed in formalin for 24 h and embedded in paraffin. After deparaffinization in xylene, the tissue sections (5 μm) were stained with Hematoxylin and Eosin (HE) and examined by an experienced veterinary pathologist. Six-week-old WT and A-PaschiRNA KI mice treated with tamoxifen were given 100 μg/mL carcinogen 4NQO (Cat. N8141, Sigma, St. Louis, MO, USA) in drinking water for 16 weeks [19]. Whole esophagi were longitudinally dissected and pictured. The gross lesions (>1 mm diameter) of esophagi were counted and fixed in formalin. The bodyweight of the mice was measured weekly.
2.5. Statistical Analysis
Student’s t-test or analysis of variance (ANOVA) followed by appropriate multiple comparison tests using SPSS statistics 20.0 software. When p < 0.05, it is considered statistically significant.
3. Results
3.1. Construction of the Targeting Vector
Rosa26 (NCBI Reference Sequence: NR_027008.1) is the most commonly used “safe harbor” locus because Rosa26 encodes a nonessential nuclear RNA expressed in almost all tissues. The conditional expression of an exogenous gene will result when a loxP-3*stop-loxP sequence is inserted upstream of the exogenous sequence at the Rosa26 locus, and this model was crossed with a Cre deleter. As illustrated in Supplementary Figure S1A, we inserted the “CAG promoter-loxP-3*polyA-loxP-A-Pas sequence-polyA” cassette into intron 1 of ROSA26 to build gene-edited mouse models. We used high-fidelity Taq DNA polymerase to amplify mouse genomic fragments containing homology arms (Has) from BAC (bacterial artificial chromosome) clone and assembled into a targeting vector together with recombination sites and selection markers (Supplementary Figure S1B). Next, the targeting vector was linearized with restriction enzymes and used as a PCR template. Supplementary Figure S1C showed expected PCR bands indicating that the targeting clone had been successfully constructed.
3.2. Generation and Screening of ES Cells
The ROSA26 targeting construct was linearized by restriction enzyme NotI, followed by phenol/chloroform extraction and ethanol precipitation. Linearized DNA was transfected into C57BL/6 ES cells by electroporation, and transfected ES cells were subject to G418 selection (200 μg/mL) 24 h post electroporation. One-hundred and eighty-six G418-resistant clones were amplified in 96-well plates. Two 96-well plates were made, with one of them frozen at −80 °C and the other was used for DNA isolation and subsequence PCR screening for homologous recombination. The PCR screening identified 37 potential clones (Supplementary Figure S2A–D). The positive clones (1A1, 1A8, 1A10, 1E3, 1G3, and 1H2) were expanded and further characterized by Southern blot analyses. DNA was digested with either EcoNI or EcoRV and hybridized using a Neo probe, which was expected to detect the following DNA fragment from the targeted allele: ~15.13 kb (with EcoNI digestion) and ~11.01 kb (with EcoRV digestion) (Supplementary Figure S2E,F). These results suggest all clones are correctly targeted.
3.3. Genotyping of F1 A-PaschiRNAflox/+ Mice
Targeted ES cell clone 1E3 was injected into C57BL/6 albino embryos and re-implanted into CD-1 pseudo-pregnant females. Founder animals were identified by their coat color. Their germline transmission was confirmed by breeding with C57BL/6 females and subsequent genotyping of the offspring. The Neo cassette is self-deleted in germ cells; thus, the offspring is Neo cassette-free. As shown in Supplementary Figure S3A, KI1 (F1 and R1), KI2 (F2 and R2), KI3 (F3 and R3), and Neo (F4 and R4 and F5 and R4) were confirmed with the insertion of fragment pairs and Neo self-removal. Finally, three male and seven female heterozygous mice (F1 generation: #1, #8, #9, #10, #12, #15, #18, #20, #21, and #22) were identified as positive KI1 and KI2 (Supplementary Figure S3B,C). The A-PaschiRNA positive pups were reconfirmed by PCR screening for KI3 and Neo deletion (Supplementary Figure S3D,E).
3.4. Tamoxifen-Inducible Expression of A-PaschiRNA
The schema of structure of A-PaschiRNA KI mice was shown in Figure 1A. We then crossed CAG-CreER with a responsive reporter line, R26-A-PaschiRNA, to generate the A-PaschiRNAflox/flox, CAG-Cre mouse and genotyped by PCR (Figure 1B). The tamoxifen induction strategy is shown in Figure 1C (left panel). The genotypes of mice induced by tamoxifen or vehicle are shown in Figure 1C (right panel). In order to examine A-PaschiRNA expression in adult tissues, we treated 8-week-old to 10-week-old A-PaschiRNAflox/flox, CAG-Cre mice with tamoxifen for 5 days before tissue samples were collected. A-PaschiRNA expression in the vital organs including heart, brain, lung, liver, intestine, stomach, kidney, spleen, ovary, tongue, esophagus, muscle, and skin were significantly increased when A-PaschiRNAflox/flox, CAG-Cre mice were treated with tamoxifen (Figure 1D). Before tamoxifen induction, the expression of A-PaschiRNA could not be detected by RT-qPCR in multiple organs of mice (Supplementary Figure S4A). These data altogether suggest that we have successfully established a KI mouse model with inducible expression of A-PaschiRNA.
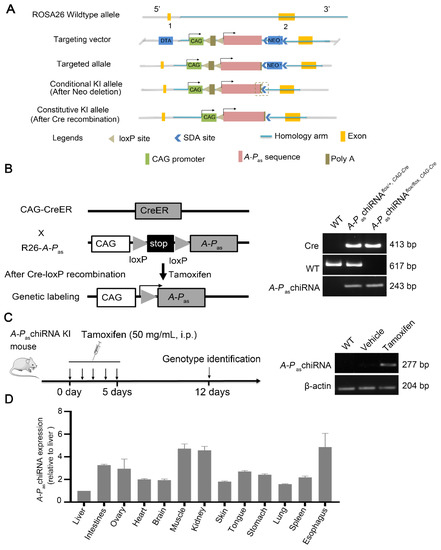
Figure 1.
Genotyping of the A-PaschiRNA flox/flox, CAG-Cre mice. (A) Schema of structure of A-PaschiRNA KI mice. (B) Scheme for genetic lineage tracing strategy (left panel). Wild type (WT), heterozygous (hetero), and homozygous (homo) mice were clearly distinguished according to Supplementary Figure S3A (right panel). Wild type: 617 bp (using primer F5/R4), A-PaschiRNA homozygotes: 243 bp (using primer F4/R4), A-PaschiRNA heterozygotes: 243 bp/617 bp (using primer F4/R4 and F5/R4), and Cre amplicon: 413 bp (using primer F1/R1). (C) Experimental strategy for tamoxifen induction (left panel). A-PaschiRNA expression was detected by PCR after tamoxifen induction. The PCR product of A-PaschiRNA was 277 bp, (using primer F3/R3) (right panel). (D) Quantification of the expression of A-PaschiRNA in different organs. A-PaschiRNA was highly enriched in the adult vital organs. Quantification of the expression of A-PaschiRNA in different organs of the A-PaschiRNAflox/flox, CAG-Cre mice after tamoxifen induction. A-PaschiRNAs were normalized to β-actin (ΔCt = Ct (A-PaschiRNA) − Ct (β-actin)). The results are shown as average fold change relative to liver, the lowest expression of A-PaschiRNA in organ, using 2−ΔΔCt, which ΔΔCt = ΔCt (organ) − ΔCt (liver).
3.5. Evaluation of the Phenotype Characterization of A-PaschiRNA KI Mice
No significant differences between the WT and A-PaschiRNA KI mice were observed in body weight, fur, and other physiological indices after 8 weeks of tamoxifen treatment (Figure 2A). The fertility of female A-PaschiRNA KI mice was not affected by the expression of tamoxifen-induced A-PaschiRNA since there were no obvious changes in the average number of pregnancies, litter size, and morbidity and mortality in A-PaschiRNA KI mice (Supplementary Table S2). In addition, eight physiological indexes and nine biochemical indexes were also measured. Hematology results indicated that all measured factors were within normal ranges, suggesting that tamoxifen-induced A-PaschiRNA did not induce inflammatory responses (Supplementary Table S3). There was no significant difference in liver and kidney function indexes between the two groups (Supplementary Table S4). In addition, HE staining of the major organs including brain, heart, liver, spleen, lung, and kidney (Figure 2B) showed no significant difference between A-PaschiRNA KI and WT mice. These data altogether indicate that tamoxifen-induced A-PaschiRNA does not affect the general physiology of the mice.
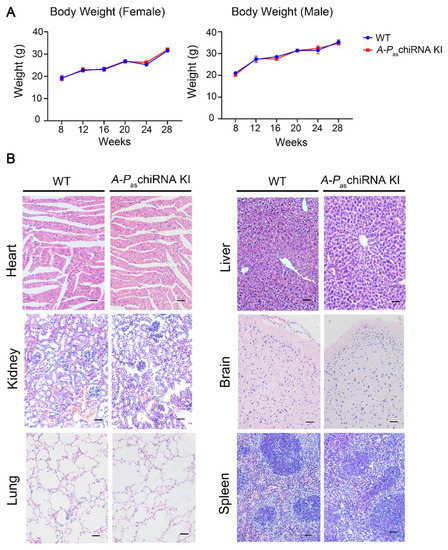
Figure 2.
Body weight and major organs in WT and A-PaschiRNA KI mice. (A) Body weight of WT and A-PaschiRNA KI mice was measured weekly for 20 weeks. Body weight of female (n = 3 per group; left panel) and male mice (n = 3 per group; right panel) did not differ significantly between two groups. (B) HE staining of major organs (i.e., heart, kidney, lung, liver, brain, and spleen). Scale bar: 50 μm.
3.6. Chemical Carcinogen-Induced A-PaschiRNA KI Mice Mimicking Human ESCC
Next, we examined the role of A-PaschiRNA in the carcinogenesis of ESCC when mice were exposed to carcinogen 4NQO (Figure 3A). The mice drank water containing 4NQO (100 μg/mL) for 16 weeks before esophageal tissues were collected. Figure 3B showed that A-PaschiRNA dramatically increased 4NQO-induced ESCC initiation (the number of tumors) compared with WT mice. As previously described, the esophageal wall of WT mice was thick, rough, and lost proper organization at the end of the 16-week 4NQO treatment [20,21]. However, with identical treatment, the esophagi form mice expressing A-PaschiRNA deteriorated rapidly and exhibited multiple visible lesions (>1 mm diameter). A-PaschiRNA KI mice prior to tamoxifen induction were also fed with 4NQO in drinking water to observe tumor formation during this experiment, and it was found that there was no significant difference in tumor formation between WT mice and A-PaschiRNA KI mice untreated with tamoxifen (Supplementary Figure S4B). HE staining of the esophagus of WT and A-PaschiRNA KI mice after 4NQO treatment. The results showed that the esophageal wall of WT mice had a loss of organization of the epithelium (dysplasia), whereas A-PaschiRNA KI mice exhibited the formation of esophageal squamous cell carcinoma and the invasion of neoplastic epithelial cells into subepithelial tissues (invasive carcinoma) (Supplementary Figure S4C). Altogether, these results indicate that A-PaschiRNA is capable of enhancing 4NQO-induced tumorigenesis and driving cancer cells to be more aggressive.
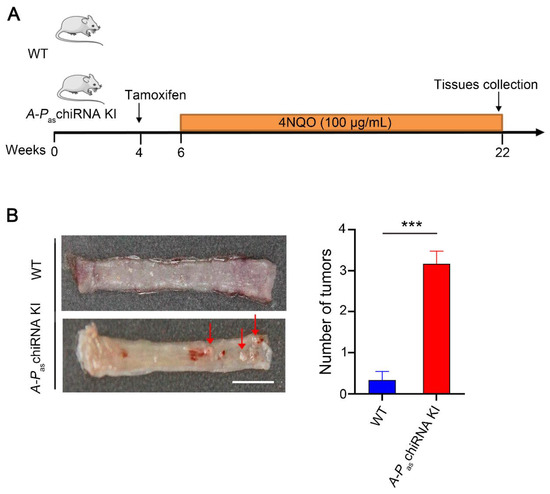
Figure 3.
4NQO-induced ESCC tumors in WT and A-PaschiRNA KI mice. (A) The protocol of chemical-induction of ESCC in WT and A-PaschiRNA KI mice. Mice were given 4NQO (100 μg/mL) in drinking water for 16 weeks. (B) The left panel demonstrates the gross anatomy of the representative esophagus from WT and A-PaschiRNA KI mice. Red arrows indicate esophageal tumors. The number of tumors per mouse in the esophagus from WT and A-PaschiRNA KI mice was shown in the right panel. n = 6 per group; *** p < 0.001; Scale bar: 5 mm.
4. Discussion
Discovery of chiRNAs has added a novel layer of complex RNA universe and provided insights into molecular mechanisms of human diseases. Results from numerous studies suggest that chiRNAs not only play important roles in cancer pathogenesis but also can serve as biomarkers and therapeutic targets [10,11,12,22,23,24,25]. Different tools tailored explicitly to chiRNA research have been developed [7]. However, applicable in vivo mouse models for studying chiRNA in physiological state are lacking. Using the Cre-loxP system, we established an in vivo model to control the expression of A-PaschiRNA by tamoxifen in C57BL/6 mice with a few advantages. In fact, A-PaschiRNA KI mice are born without any embryonic lethal, and there is no obvious pathological change in organs nor abnormal metabolism. Since A-PaschiRNA can be expressed at the desired time point, this model can be used in different experiments to address specific scientific questions that other available models could not answer. As shown in this study, controlling the expression of chiRNA enabled us to dissect the role of A-PaschiRNA in 4NQO-induced ESCC. Additionally, since controlled expression of A-PaschiRNA can occur in many organs other than the esophagus including brain, heart, lungs, liver, stomach, colon, and spleen tissues, this model can broaden our understanding of A-PaschiRNA in physiological and pathological settings.
Without the tamoxifen-inducible A-PaschiRNA KI mouse model, we were able to show that A-PaschiRNA is highly enriched in ESCC [11], while whether the enriched expression of A-PaschiRNA is the causation or resultant of ESCC pathogenesis is still unknown. The inducible model enabled us to show the role of A-PaschiRNA in 4NQO-induced ESCC initiation, evidenced by the increased number of the tumor compared with WT mice. Previously, we demonstrated that A-PaschiRNA can induce cancer stemness by activating ERK5-mediated non-canonical PAF1 [11]. With this inducible model, we further showed the enhancing effect of A-PaschiRNA on tumor progression because cancer cells become more malignant in the presence of A-PaschiRNA. These findings not only support our previous reports but enable us to understand the underlying molecular mechanism at different levels.
One limitation of our study was that the expression of A-PaschiRNA induced by tamoxifen occurs in multiple organs, we cannot exclude the possible effect from overexpressed A-PaschiRNA in the other organs. Nevertheless, studies using this animal model with a combination of different research tools will investigate unique questions that otherwise not be answered. To our knowledge, we are the first to establish a chiRNA KI mouse model. The methodology used in this report will be valuable for establishing knockin animal models of other chiRNAs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11020277/s1, Table S1: Sequence of the final targeting vector, Table S2: The fertility of two groups of female mice, Table S3: Hematological counts of two groups of mice, Table S4: Serum biochemical indices of two groups of mice. Figure S1: Experimental design for generation of targeting vector, Figure S2: Characterization of the ES clones, Figure S3: Genotype identification of F1 A-PaschiRNA flox/+ mice, Figure S4: The characteristic of A-PaschiRNA in mice.
Author Contributions
H.Z. conceived and supervised the research. Y.L., L.D. and Z.Y. participated in data analysis and figure preparation. Y.L., J.Z. and F.L. (Fan Liu) collected the samples and performed the experiments. K.L., L.D., Z.Y. and F.L. (Feifei Li) participated in the initial phase of the project and some data collection. H.Z. and Z.Y. wrote the manuscript. R.P.C., D.Z., S.G. and Y.P. interpreted data, contributed to discussion, and reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by a grant in part by the National Natural Science Foundation of China (82072683, 81773087, 81071736, 81572876 and 30973508 to H.Z.); the Natural Science Foundation of Guangdong Province of China (2021A1515012522 and 9151018004000000 to H.Z.); the Science and Technology Planning Project of Guangdong Province of China (2019A030317024 to H.Z.); Special Project on the Integration of Industry, Education and Research of Guangdong Province (2011A090100024 to H.Z.); and flagship specialty construction project General surgery (Funding no. 711003).
Institutional Review Board Statement
The study was approved by the Animal Care and Use Committee of Jinan University (approval no. IACUC-20191210-03).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study will be made available upon reasonable request.
Acknowledgments
We acknowledge members of H. Zhang’s laboratory for technical help and discussion. The authors thank Laising Yen for valuable discussions.
Conflicts of Interest
The authors declare no competing interest.
References
- Wang, Y.; Zou, Q.; Li, F.; Zhao, W.; Xu, H.; Zhang, W.; Deng, H.; Yang, X. Identification of the cross-strand chimeric RNAs generated by fusions of bi-directional transcripts. Nat. Commun. 2021, 12, 4645. [Google Scholar] [CrossRef] [PubMed]
- Elfman, J.; Li, H. Chimeric RNA in Cancer and Stem Cell Differentiation. Stem Cells Int. 2018, 2018, 3178789. [Google Scholar] [CrossRef] [Green Version]
- Audano, P.A.; Sulovari, A.; Graves-Lindsay, T.A.; Cantsilieris, S.; Sorensen, M.; Welch, A.E.; Dougherty, M.L.; Nelson, B.J.; Shah, A.; Dutcher, S.K.; et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell 2019, 176, 663–675.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zekavat, S.M.; Ruotsalainen, S.; Handsaker, R.E.; Alver, M.; Bloom, J.; Poterba, T.; Seed, C.; Ernst, J.; Chaffin, M.; Engreitz, J.; et al. Publisher Correction: Deep coverage whole genome sequences and plasma lipoprotein(a) in individuals of European and African ancestries. Nat. Commun. 2020, 11, 1715. [Google Scholar] [CrossRef] [Green Version]
- Ke, X.; Xiong, X.; Lin, Y.; Zhang, H. Chimeric RNA and Exosomes-Based Liquid Biopsy. Methods Mol. Biol. 2020, 2079, 211–218. [Google Scholar] [CrossRef]
- Frenkel-Morgenstern, M.; Gorohovski, A.; Lacroix, V.; Rogers, M.; Ibanez, K.; Boullosa, C.; Andres Leon, E.; Ben-Hur, A.; Valencia, A. ChiTaRS: A database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013, 41, D142–D151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Xie, Z.; Li, H. Intergenically Spliced Chimeric RNAs in Cancer. Trends Cancer 2016, 2, 475–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Qin, F.; Kumar, S.; Elfman, J.; Lin, E.; Pham, L.P.; Yang, A.; Li, H. The landscape of chimeric RNAs in non-diseased tissues and cells. Nucleic Acids Res. 2020, 48, 1764–1778. [Google Scholar] [CrossRef] [Green Version]
- Frenkel-Morgenstern, M.; Lacroix, V.; Ezkurdia, I.; Levin, Y.; Gabashvili, A.; Prilusky, J.; Del Pozo, A.; Tress, M.; Johnson, R.; Guigo, R.; et al. Chimeras taking shape: Potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012, 22, 1231–1242. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Lin, W.; Kannan, K.; Luo, L.; Li, J.; Chao, P.W.; Wang, Y.; Chen, Y.P.; Gu, J.; Yen, L. Aberrant chimeric RNA GOLM1-MAK10 encoding a secreted fusion protein as a molecular signature for human esophageal squamous cell carcinoma. Oncotarget 2013, 4, 2135–2143. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xiong, X.; Yao, Z.; Zhu, J.; Lin, Y.; Lin, W.; Li, K.; Xu, X.; Guo, Y.; Chen, Y.; et al. Chimeric RNA ASTN2-PAPPAas aggravates tumor progression and metastasis in human esophageal cancer. Cancer Lett. 2021, 501, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Dong, H.; Deng, W.; Lin, W.; Li, K.; Xiong, X.; Guo, Y.; Zhou, F.; Ma, C.; Chen, Y.; et al. Evaluation of Salivary Exosomal Chimeric GOLM1-NAA35 RNA as a Potential Biomarker in Esophageal Carcinoma. Clin. Cancer Res. 2019, 25, 3035–3045. [Google Scholar] [CrossRef] [PubMed]
- McLellan, M.A.; Rosenthal, N.A.; Pinto, A.R. Cre-loxP-Mediated Recombination: General Principles and Experimental Considerations. Curr. Protoc. Mouse Biol. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Reinert, R.B.; Kantz, J.; Misfeldt, A.A.; Poffenberger, G.; Gannon, M.; Brissova, M.; Powers, A.C. Tamoxifen-Induced Cre-loxP Recombination Is Prolonged in Pancreatic Islets of Adult Mice. PLoS ONE 2012, 7, e33529. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, M.; Im, S.K.; Fang, S. Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 2018, 34, 147–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, M.E.; Zhou, W.; Thangaraj, J.; Kyriakakis, P.; Wu, Y.; Huang, Z.; Ho, P.; Pan, Y.; Limsakul, P.; Xu, X.; et al. An AND-Gated Drug and Photoactivatable Cre-loxP System for Spatiotemporal Control in Cell-Based Therapeutics. ACS Synth. Biol. 2019, 8, 2359–2371. [Google Scholar] [CrossRef]
- Carstens, K.E.; Gloss, B.R.; Alexander, G.M.; Dudek, S.M. Modified adeno-associated virus targets the bacterial enzyme chondroitinase ABC to select mouse neuronal populations in vivo using the Cre-LoxP system. Eur. J. Neurosci. 2021, 53, 4005–4015. [Google Scholar] [CrossRef]
- Wang, L.; Li, K.; Lin, X.; Yao, Z.; Wang, S.; Xiong, X.; Ning, Z.; Wang, J.; Xu, X.; Jiang, Y.; et al. Metformin induces human esophageal carcinoma cell pyroptosis by targeting the miR-497/PELP1 axis. Cancer Lett. 2019, 450, 22–31. [Google Scholar] [CrossRef]
- Wang, L.; Du, L.; Xiong, X.; Lin, Y.; Zhu, J.; Yao, Z.; Wang, S.; Guo, Y.; Chen, Y.; Geary, K.; et al. Repurposing dextromethorphan and metformin for treating nicotine-induced cancer by directly targeting CHRNA7 to inhibit JAK2/STAT3/SOX2 signaling. Oncogene 2021, 40, 1974–1987. [Google Scholar] [CrossRef]
- Wang, S.; Du, Z.; Luo, J.; Wang, X.; Li, H.; Liu, Y.; Zhang, Y.; Ma, J.; Xiao, W.; Wang, Y.; et al. Inhibition of heat shock protein 90 suppresses squamous carcinogenic progression in a mouse model of esophageal cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1405–1416. [Google Scholar] [CrossRef]
- Tang, X.H.; Knudsen, B.; Bemis, D.; Tickoo, S.; Gudas, L.J. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin. Cancer Res. 2004, 10, 301–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Li, H. Prediction, Characterization, and In Silico Validation of Chimeric RNAs. Methods Mol. Biol. 2020, 2079, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Yang, S.; Singh, S.; Qin, F.; Kumar, S.; Wang, L.; Ma, D.; Li, H. The Landscape and Implications of Chimeric RNAs in Cervical Cancer. EBioMedicine 2018, 37, 158–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Singh, S.; Chen, X.; Zheng, Z.; Huang, J.; Lin, T.; Li, H. The landscape of chimeric RNAs in bladder urothelial carcinoma. Int. J. Biochem. Cell Biol. 2019, 110, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Wang, L.; Wang, J.; Ittmann, M.M.; Li, W.; Yen, L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9172–9177. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).