Captopril Alleviates Chondrocyte Senescence in DOCA-Salt Hypertensive Rats Associated with Gut Microbiome Alteration
Abstract
1. Introduction
2. Materials and Methods
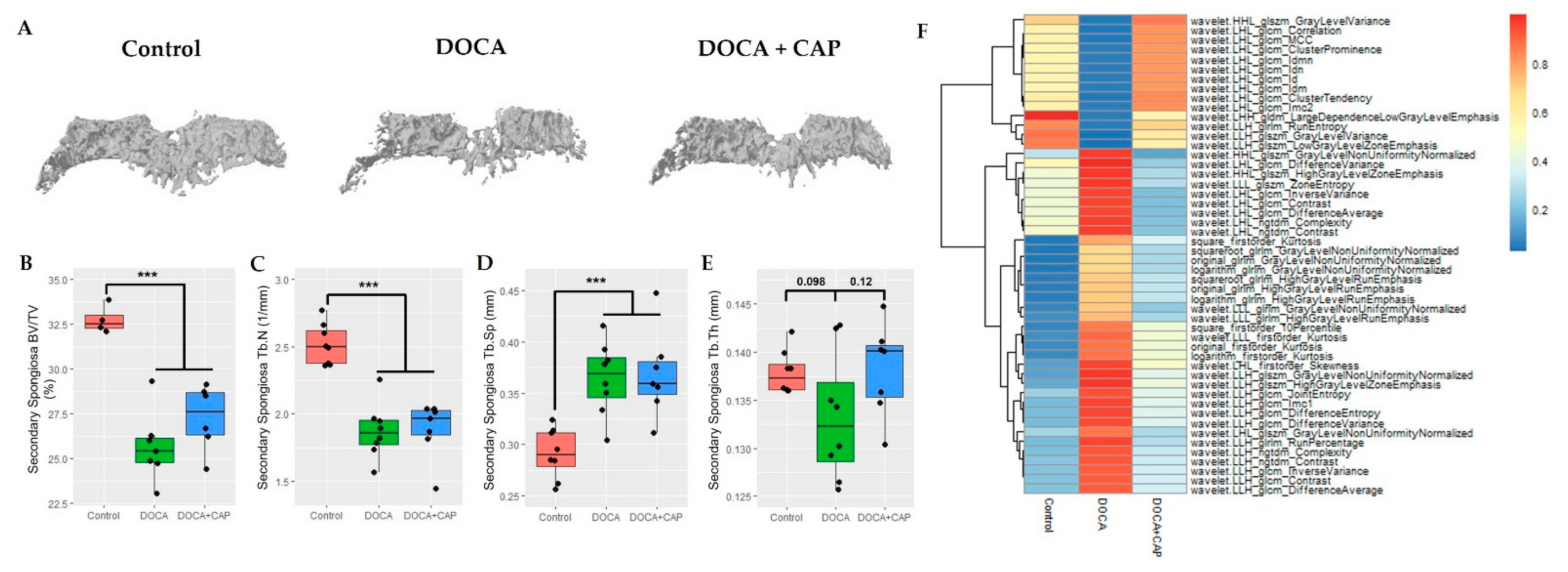
2.1. Micro-CT Analysis
2.2. Histology
2.3. Gut Microbiota Analysis
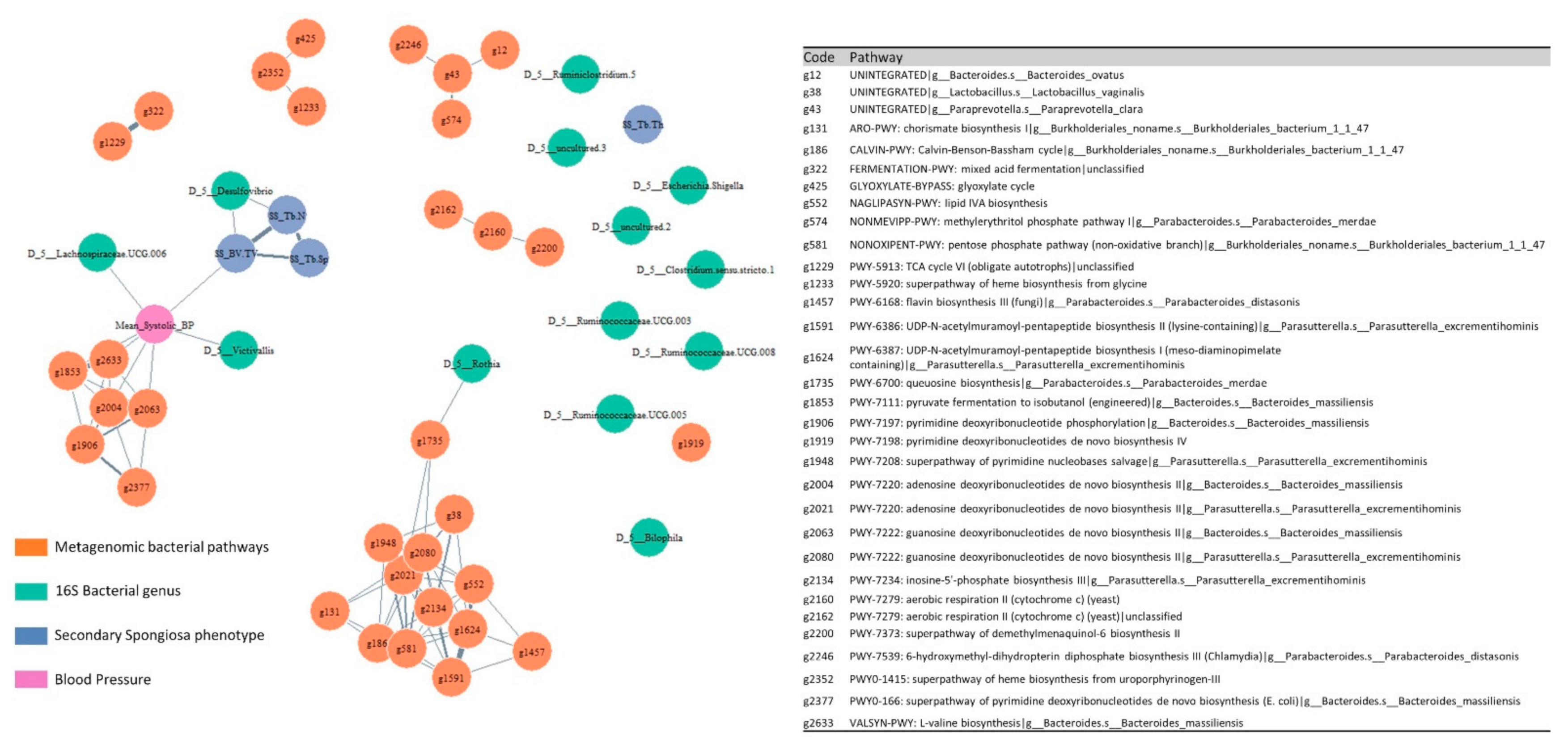
2.4. Multi-Omic Correlation Network
2.5. Statistical Analysis
3. Results
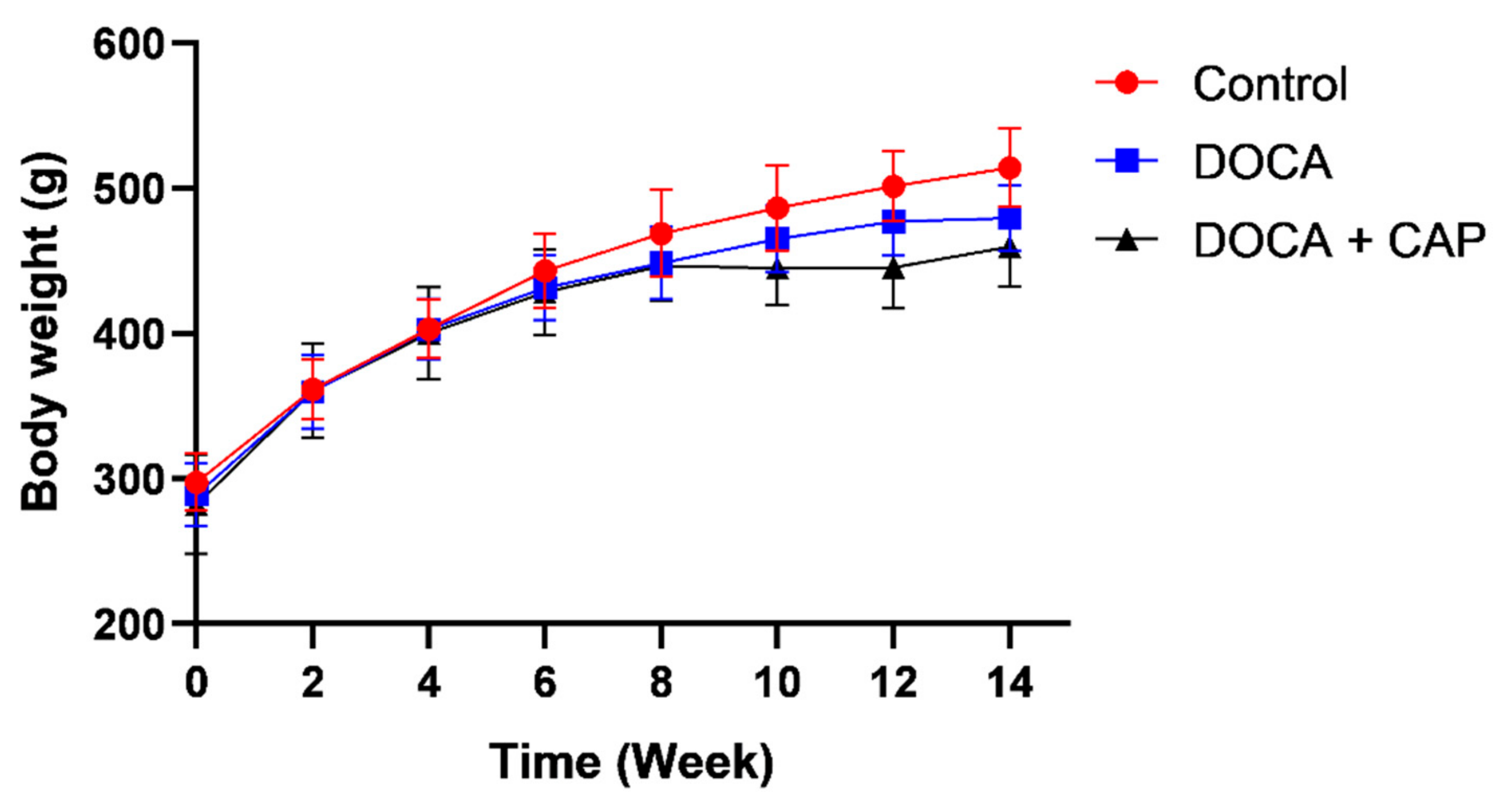
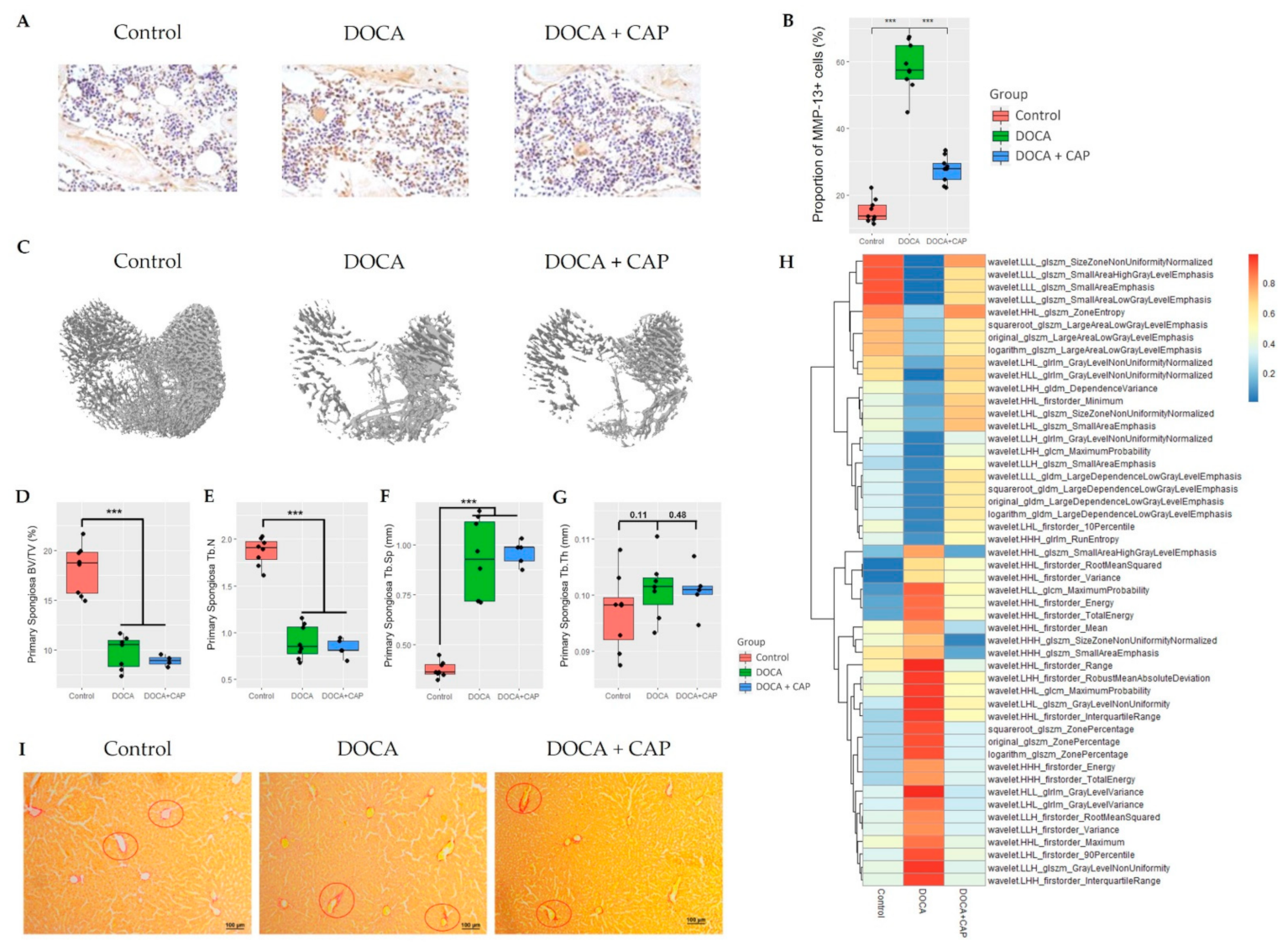
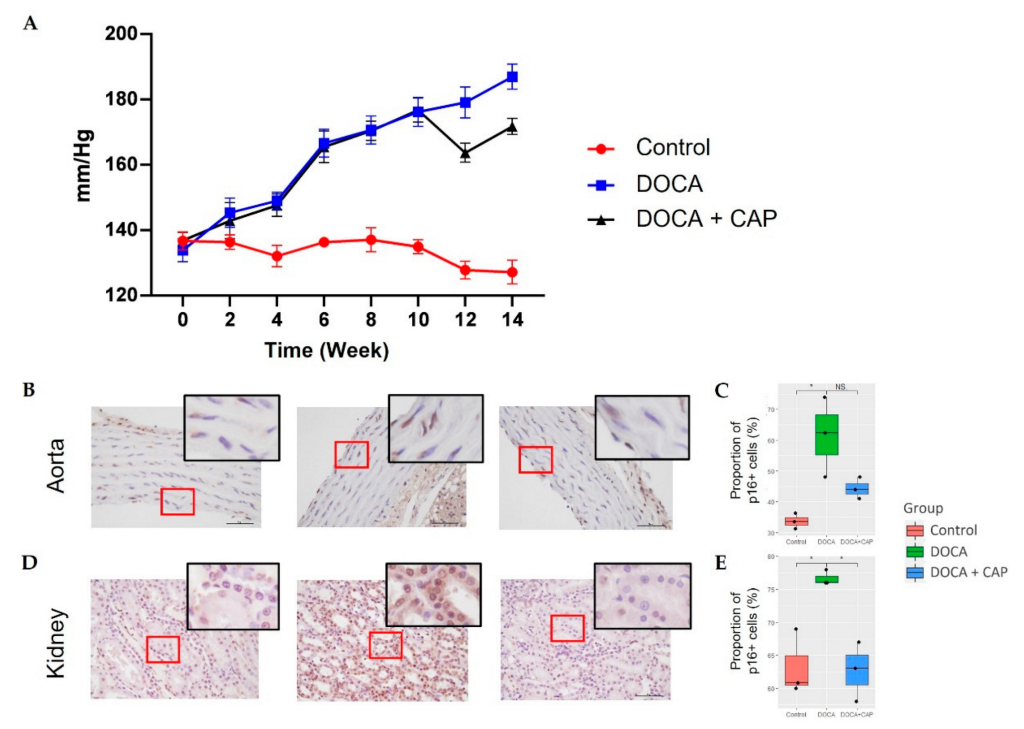
3.1. Senescent Cells Accumulation with the Onset of Metabolic OA after DOCA Induction
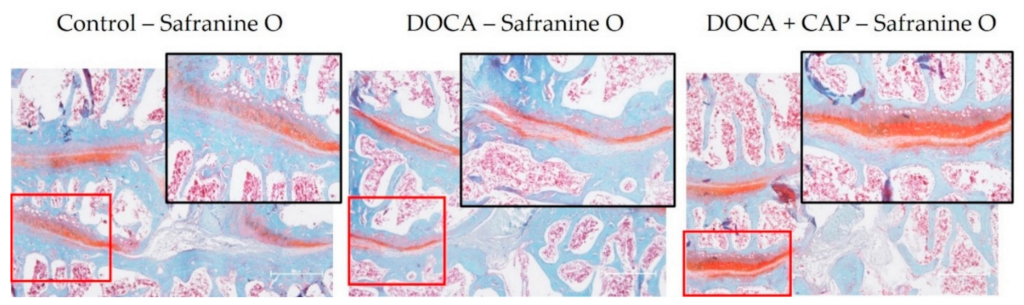
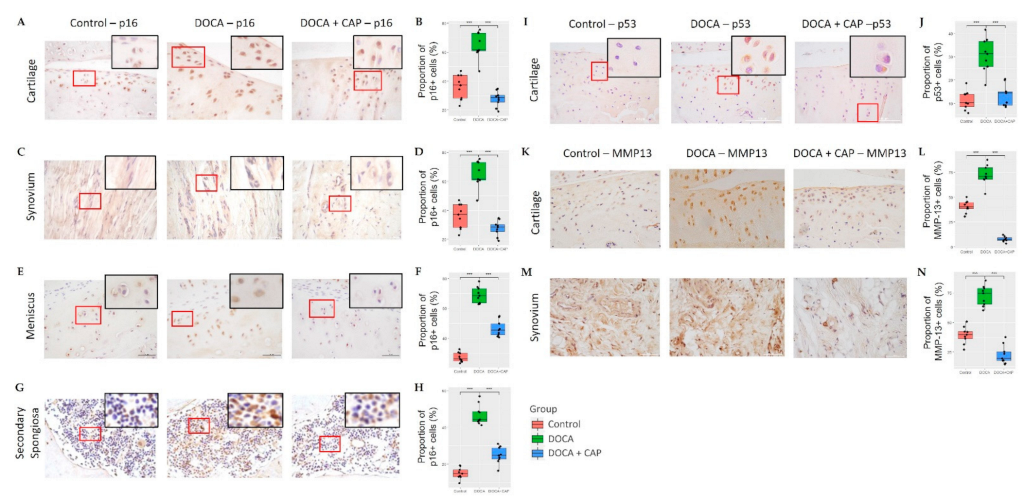
3.2. Captopril Reduced Senescent Cells and Mitigated Joint Deterioration
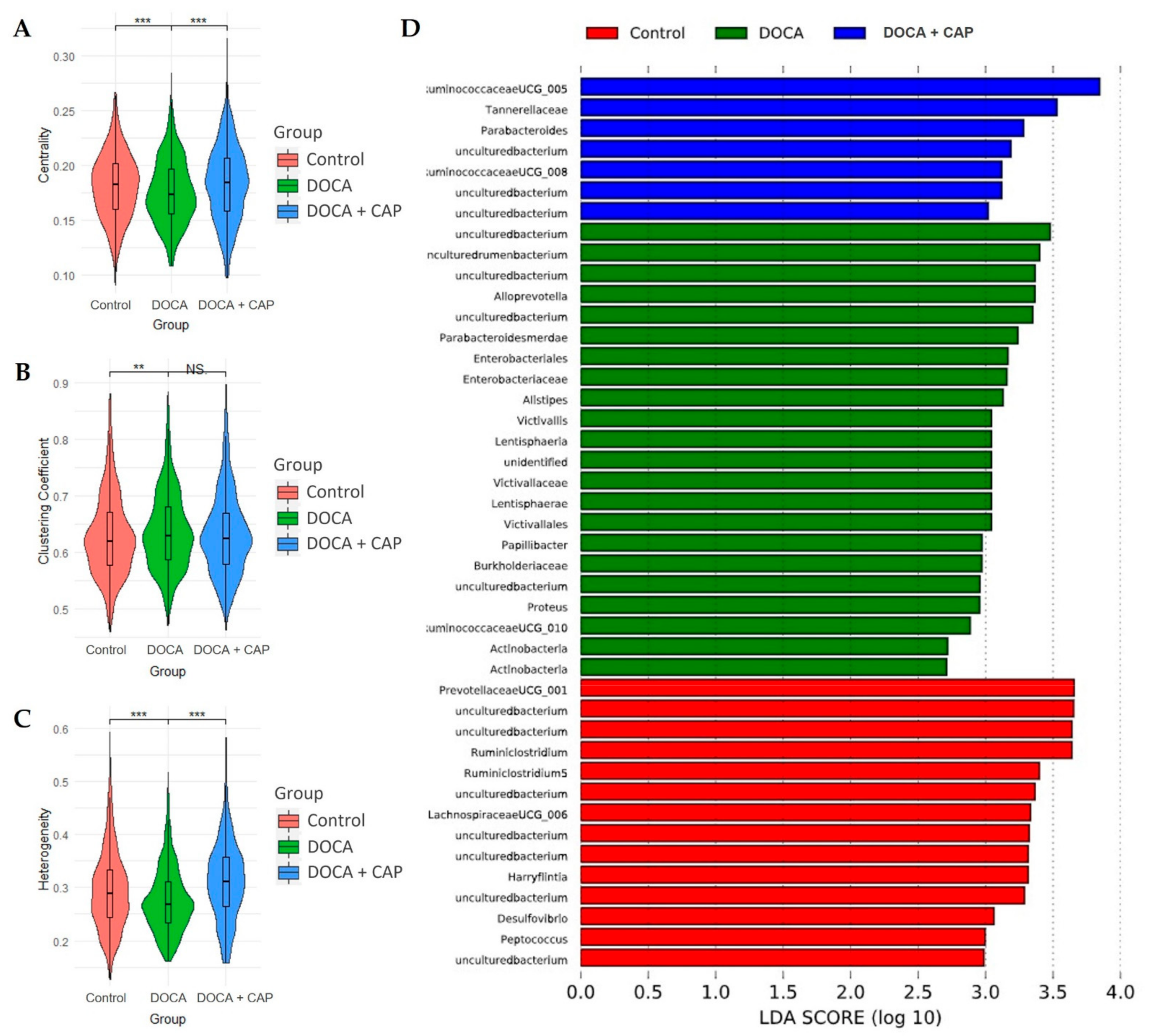
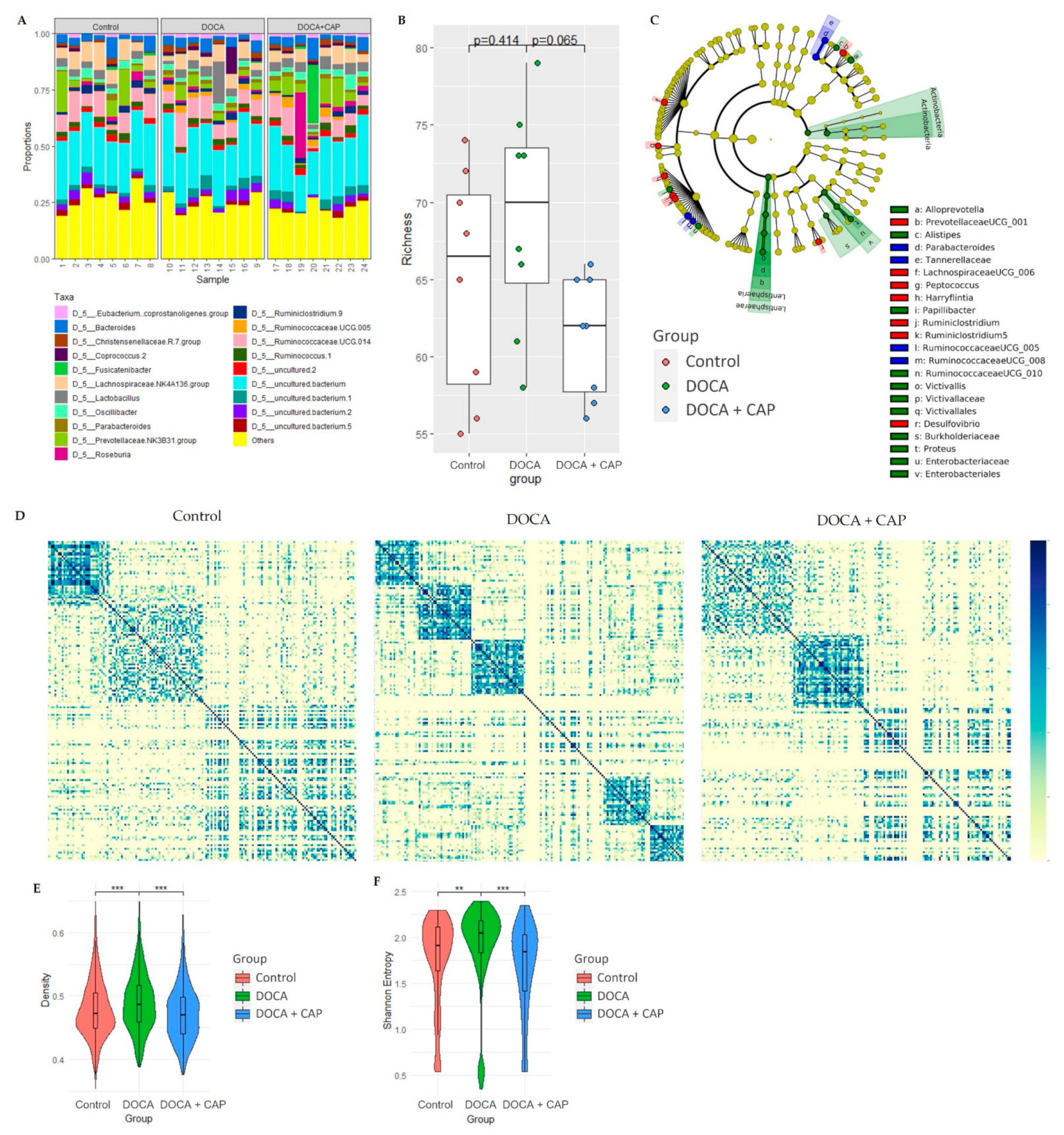
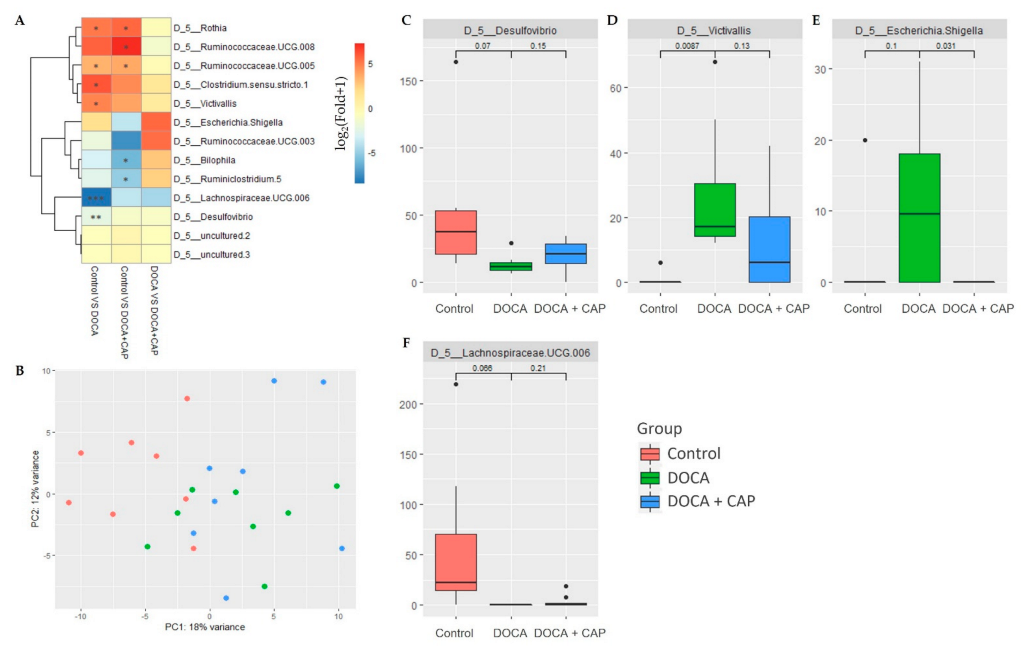
3.3. Rebalancing Effect of Captopril on Gut Microbiota in DOCA-Induced Hypertensive Rats
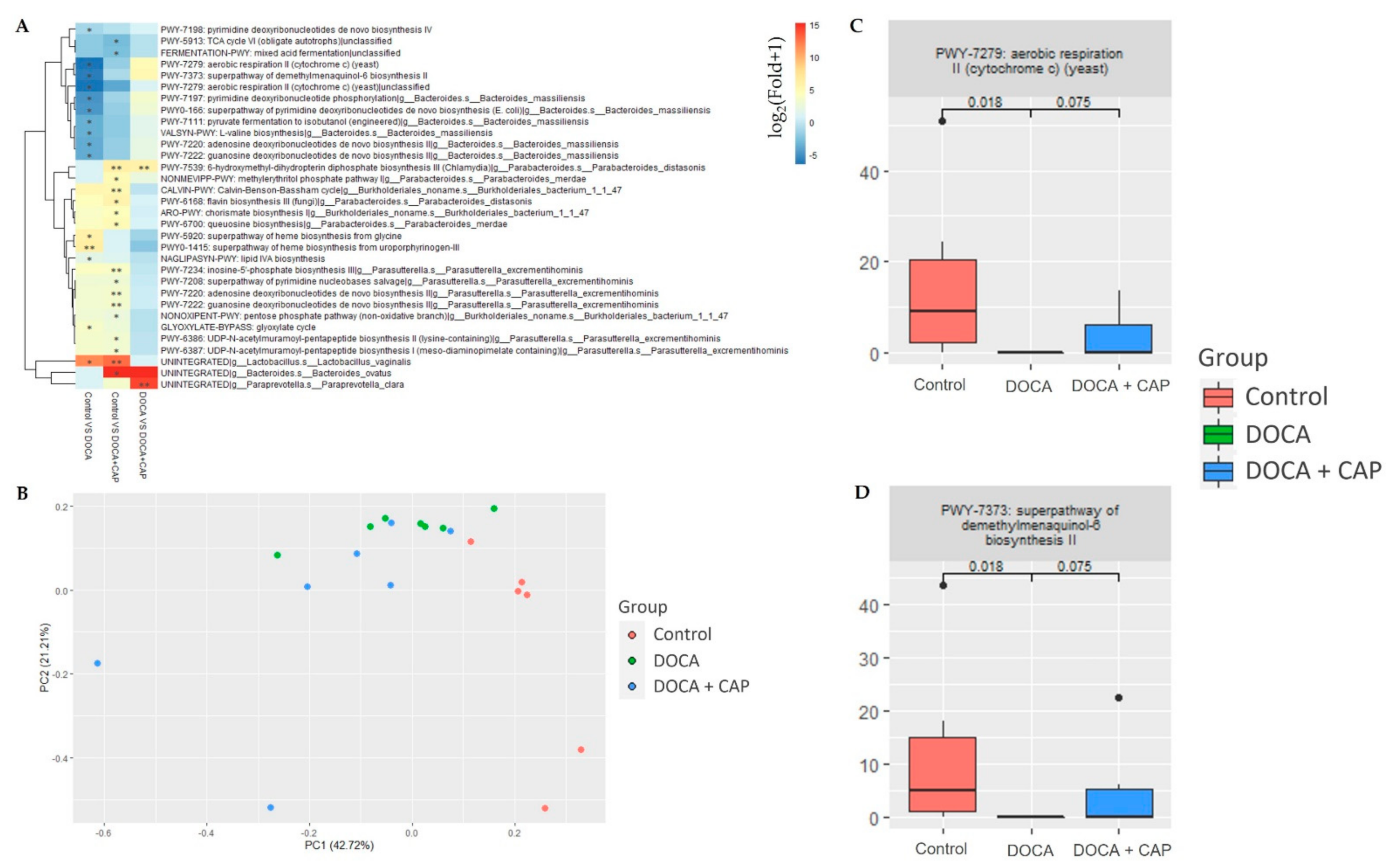
3.4. Identification of Differentially Activated Pathways of Gut Microbiota Associated with Senescent Cells Removal Using Captopril
3.5. Identification of Differentially Activated Pathways of Gut Microbiota Associated with Senescent Cells Removal Using Captopril
3.6. Multi-Omic Correlation Analysis Unraveled Associations between Microbiota and Bone under the Hypertensive and Normotensive Conditions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Experimental Details
- Animals
- 2.
- DOCA-salt hypertensive rat model and captopril treatment
- 3.
- Measurement of body weight and blood pressure
- 4.
- Histology
- 5.
- Micro-CT analysis
- 6.
- Gut microbiota sequencing analysis
- 7.
- Gut microbiota compositional analysis
- 8.
- Gut microbiota weighted correlation network analysis
- 9.
- Differential abundance analysis of microbiota
- 10.
- Differential expression analysis of microbiota metagenomic data analysis
Appendix B
| Pairs | df | Sums of Squares | F Model | R2 | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|---|
| Control vs. DOCA | 1 | 0.115566 | 2.14969 | 0.13311 | 0.006 | 0.018 |
| Control vs. DOCA + CAP | 1 | 0.136479 | 1.943359 | 0.121891 | 0.019 | 0.057 |
| DOCA vs. DOCA + CAP | 1 | 0.080237 | 1.167919 | 0.076999 | 0.227 | 0.681 |
| df | Sums of Squares | Mean Squares | F Model | R2 | p-Value |
|---|---|---|---|---|---|
| 2 | 0.221521 | 0.11076 | 1.724451 | 0.141066 | 0.008 |
| 21 | 1.348818 | 0.064229 | NA | 0.858934 | |
| 23 | 1.570338 | NA | NA | 1 |
References
- Ragonnaud, E.; Biragyn, A. Gut microbiota as the key controllers of “healthy” aging of elderly people. Immun. Ageing 2021, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Wilmanski, T.; Diener, C.; Rappaport, N.; Patwardhan, S.; Wiedrick, J.; Lapidus, J.; Earls, J.C.; Zimmer, A.; Glusman, G.; Robinson, M. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 2021, 3, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef]
- Collins, K.; Paul, H.; Reimer, R.; Seerattan, R.; Hart, D.; Herzog, W. Relationship between inflammation, the gut microbiota, and metabolic osteoarthritis development: Studies in a rat model. Osteoarthr. Cartil. 2015, 23, 1989–1998. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sivan, U.; Tan, S.L.; Lippo, L.; De Angelis, J.; Labella, R.; Singh, A.; Chatzis, A.; Cheuk, S.; Medhghalchi, M. High-resolution 3D imaging uncovers organ-specific vascular control of tissue aging. Sci. Adv. 2021, 7, eabd7819. [Google Scholar] [CrossRef] [PubMed]
- Westhoff, J.H.; Hilgers, K.F.; Steinbach, M.P.; Hartner, A.; Klanke, B.; Amann, K.; Melk, A. Hypertension induces somatic cellular senescence in rats and humans by induction of cell cycle inhibitor p16INK4a. Hypertension 2008, 52, 123–129. [Google Scholar] [CrossRef]
- McCarthy, C.G.; Wenceslau, C.F.; Webb, R.C.; Joe, B. Novel contributors and mechanisms of cellular senescence in hypertension-associated premature vascular aging. Am. J. Hypertens. 2019, 32, 709–719. [Google Scholar] [CrossRef]
- Jeon, O.H.; Kim, C.; Laberge, R.M.; Demaria, M.; Rathod, S.; Vasserot, A.P.; Chung, J.W.; Kim, D.H.; Poon, Y.; David, N.; et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 2017, 23, 775–781. [Google Scholar] [CrossRef]
- Farr, J.N.; Xu, M.; Weivoda, M.M.; Monroe, D.G.; Fraser, D.G.; Onken, J.L.; Negley, B.A.; Sfeir, J.G.; Ogrodnik, M.B.; Hachfeld, C.M. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017, 23, 1072–1079. [Google Scholar] [CrossRef]
- Cao, X.; Luo, P.; Huang, J.; Liang, C.; He, J.; Wang, Z.; Shan, D.; Peng, C.; Wu, S. Intraarticular senescent chondrocytes impair the cartilage regeneration capacity of mesenchymal stem cells. Stem Cell Res. Ther. 2019, 10, 86. [Google Scholar] [CrossRef]
- Kumar, S.; Dietrich, N.; Kornfeld, K. Angiotensin converting enzyme (ACE) inhibitor extends Caenorhabditis elegans life span. PLoS Genet. 2016, 12, e1005866. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, H.; Lam, T.Y.; Shum, T.-F.; Tsai, T.-Y.; Chiou, J. Hypotensive effect of captopril on deoxycorticosterone acetate-salt-induced hypertensive rat is associated with gut microbiota alteration. Hypertens. Res. 2021, 45, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.H.; Kim, I.J.; Ma, S.K.; Kim, S.W. Rosiglitazone prevents the progression of renal injury in DOCA-salt hypertensive rats. Hypertens. Res. 2010, 33, 255–262. [Google Scholar] [CrossRef]
- Dhand, N.K.; Khatkar, M.S. Statulator: An online statistical calculator. Sample Size Calc. Comp. Two Indep. Means 2014. [Google Scholar]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Philipot, D.; Guerit, D.; Platano, D.; Chuchana, P.; Olivotto, E.; Espinoza, F.; Dorandeu, A.; Pers, Y.M.; Piette, J.; Borzi, R.M.; et al. p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res. Ther. 2014, 16, R58. [Google Scholar] [CrossRef]
- Xu, M.; Bradley, E.W.; Weivoda, M.M.; Hwang, S.M.; Pirtskhalava, T.; Decklever, T.; Curran, G.L.; Ogrodnik, M.; Jurk, D.; Johnson, K.O.; et al. Transplanted Senescent Cells Induce an Osteoarthritis-Like Condition in Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Aquino, V.; Lobaton, G.O.; Li, H.; Colon-Perez, L.; Goel, R.; Qi, Y.; Zubcevic, J.; Febo, M.; Richards, E.M. Sustained captopril-induced reduction in blood pressure is associated with alterations in gut-brain axis in the spontaneously hypertensive rat. J. Am. Heart Assoc. 2019, 8, e010721. [Google Scholar] [CrossRef]
- Yan, Q.; Gu, Y.; Li, X.; Yang, W.; Jia, L.; Chen, C.; Han, X.; Huang, Y.; Zhao, L.; Li, P. Alterations of the gut microbiome in hypertension. Front. Cell. Infect. Microbiol. 2017, 7, 381. [Google Scholar] [CrossRef]
- Taylor, W.R.; Takemiya, K. Hypertension opens the flood gates to the gut microbiota. Am. Heart Assoc. 2017, 120, 249–251. [Google Scholar] [CrossRef]
- Rodriguez, J.; Hiel, S.; Neyrinck, A.M.; Le Roy, T.; Pötgens, S.A.; Leyrolle, Q.; Pachikian, B.D.; Gianfrancesco, M.A.; Cani, P.D.; Paquot, N. Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut 2020, 69, 1975–1987. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.H.; Lenz, K.L.; Pollitt, E.N.; Ferguson, D.; Hutson, I.; Springer, L.E.; Oestreich, A.K.; Tang, R.; Choi, Y.-R.; Meyer, G.A. Adipose tissue is a critical regulator of osteoarthritis. Proc. Natl. Acad. Sci. USA 2021, 118, e2021096118. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, C.; Zhang, Y.; Zhang, W.; Doherty, M.; Yang, T.; Zhai, G.; Obotiba, A.D.; Lyu, H.; Zeng, C. Association between gut microbiota and symptomatic hand osteoarthritis: Data from the Xiangya Osteoarthritis Study. Arthritis Rheumatol. 2021, 73, 1656–1662. [Google Scholar] [CrossRef]
- Yu, X.-H.; Yang, Y.-Q.; Cao, R.-R.; Bo, L.; Lei, S.-F. The causal role of gut microbiota in development of osteoarthritis. Osteoarthr. Cartil. 2021, 29, 1741–1750. [Google Scholar] [CrossRef]
- Nunberg, M.; Werbner, N.; Neuman, H.; Bersudsky, M.; Braiman, A.; Ben-Shoshan, M.; Ben Izhak, M.; Louzoun, Y.; Apte, R.N.; Voronov, E. Interleukin 1α-deficient mice have an altered gut microbiota leading to protection from dextran sodium sulfate-induced colitis. MSystems 2018, 3, e00213–e00217. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, L.C.; Zhang, Y.; Kuang, X.; Koohi-Moghadam, M.; Wu, H.; Lam, T.Y.C.; Chiou, J.; Wen, C. Captopril Alleviates Chondrocyte Senescence in DOCA-Salt Hypertensive Rats Associated with Gut Microbiome Alteration. Cells 2022, 11, 3173. https://doi.org/10.3390/cells11193173
Chan LC, Zhang Y, Kuang X, Koohi-Moghadam M, Wu H, Lam TYC, Chiou J, Wen C. Captopril Alleviates Chondrocyte Senescence in DOCA-Salt Hypertensive Rats Associated with Gut Microbiome Alteration. Cells. 2022; 11(19):3173. https://doi.org/10.3390/cells11193173
Chicago/Turabian StyleChan, Lok Chun, Yuqi Zhang, Xiaoqing Kuang, Mohamad Koohi-Moghadam, Haicui Wu, Theo Yu Chung Lam, Jiachi Chiou, and Chunyi Wen. 2022. "Captopril Alleviates Chondrocyte Senescence in DOCA-Salt Hypertensive Rats Associated with Gut Microbiome Alteration" Cells 11, no. 19: 3173. https://doi.org/10.3390/cells11193173
APA StyleChan, L. C., Zhang, Y., Kuang, X., Koohi-Moghadam, M., Wu, H., Lam, T. Y. C., Chiou, J., & Wen, C. (2022). Captopril Alleviates Chondrocyte Senescence in DOCA-Salt Hypertensive Rats Associated with Gut Microbiome Alteration. Cells, 11(19), 3173. https://doi.org/10.3390/cells11193173







