Intrachromosomal Looping and Histone K27 Methylation Coordinately Regulates the lncRNA H19-Fetal Mitogen IGF2 Imprinting Cluster in the Decidual Microenvironment of Early Pregnancy
Abstract
1. Introduction
2. Results
2.1. Identification of H19 as a Recurrent Spontaneous Abortion-Associated lncRNA
2.2. Loss of Genomic Imprinting in Decidual Tissues
2.3. The Role of Altered Epigenotypes in the In Vitro Induced Decidualization Model
2.4. Histone Deacetylase Inhibitor Alters Imprinting in Decidualized Cells
2.5. Loss of Imprinting Is Associated with Aberrant Histone H3 Lysine 27 Methylation
2.6. Aberrant Imprinting Is Accompanied by the Loss of Intrachromosomal Looping
2.7. Loss of Imprinting Is Associated with De Novo DNA Methylation in the Imprinting Control Region
3. Discussion
4. Materials and Methods
4.1. Identification of RSA-Associated lncRNAs Using RNA-Seq Data
4.2. Human Decidual Samples
4.3. Culture of Human Primary Endometrial Stromal Cells
4.4. In Vitro Decidualization
4.5. The Role of Histone Deacetylase Inhibitor VPA in Decidualization
4.6. RT-PCR Quantitation
4.7. Allelic Expression of IGF2 and H19
4.8. DNA Methylation Analysis
4.9. Chromosome Conformation Capture (3C)
4.10. Histone Methylation by Chromatin Immunoprecipitation (ChIP) Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| RSA | Recurrent spontaneous abortion; |
| lncRNAs | longnoncoding RNAs; |
| ICR | imprinting control center; |
| H3K27m3 | trimethylation of lysine 27 on histone H3; |
| IGFBP1 | insulin-like growth factor binding protein1; |
| PRL | prolactin; |
| RSA | recurrent spontaneous abortion; |
| SNPs | single nucleotide polymorphisms; |
| LOI | loss of imprinting; |
| MOI | maintenance of imprinting; |
| KEGG | The Kyoto Encyclopedia of Genes and Genomes; |
| E2 | β-Estradiol; |
| P4 | Progesterone; |
| 8-Br-cAMP | 8-Bromoadenosine 3′:5′-Cyclic Monophosphate; |
| PCR2 | polycomb repressive complex 2; |
| 3C | chromosome conformation capture; |
| ChIP | chromatin immunoprecipitation |
References
- How, J.; Leiva, O.; Bogue, T.; Fell, G.G.; Bustoros, M.W.; Connell, N.T. Pregnancy outcomes, risk factors, and cell count trends in pregnant women with essential thrombocythemia. Leuk. Res. 2020, 98, 106459. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Gimenez, C.; Alijotas-Reig, J. Recurrent miscarriage: Causes, evaluation and management. Postgrad. Med. J. 2015, 91, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Pinar, M.H.; Gibbins, K.; He, M.; Kostadinov, S.; Silver, R. Early Pregnancy Losses: Review of Nomenclature, Histopathology, and Possible Etiologies. Fetal. Pediatr. Pathol. 2018, 37, 191–209. [Google Scholar] [CrossRef]
- McPherson, E. Recurrence of stillbirth and second trimester pregnancy loss. Am. J. Med. Genet. A 2016, 170A, 1174–1180. [Google Scholar] [CrossRef]
- Ticconi, C.; Pietropolli, A.; Di Simone, N.; Piccione, E.; Fazleabas, A. Endometrial Immune Dysfunction in Recurrent Pregnancy Loss. Int. J. Mol. Sci. 2019, 20, 5332. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; Tsuzuki, T.; Murata, H. Decidualization of the human endometrium. Reprod. Med. Biol. 2018, 17, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, R.R.P.; Aberkane, A.; Polanski, L.; Maruyama, Y.; Baumgarten, M.; Lucas, E.S.; Quenby, S.; Chan, J.K.Y.; Raine-Fenning, N.; Brosens, J.J.; et al. Deregulation of the endometrial stromal cell secretome precedes embryo implantation failure. Mol. Hum. Reprod. 2017, 23, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.A.; Simitsidellis, I.; Cousins, F.L.; Critchley, H.O.; Saunders, P.T. Intracrine Androgens Enhance Decidualization and Modulate Expression of Human Endometrial Receptivity Genes. Sci. Rep. 2016, 6, 19970. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Wang, C.; Hu, J.F.; Li, W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front. Genet. 2020, 11, 277. [Google Scholar] [CrossRef]
- Patty, B.J.; Hainer, S.J. Non-Coding RNAs and Nucleosome Remodeling Complexes: An Intricate Regulatory Relationship. Biology 2020, 9, 213. [Google Scholar] [CrossRef]
- Huang, H.; Sun, J.; Sun, Y.; Wang, C.; Gao, S.; Li, W.; Hu, J.F. Long noncoding RNAs and their epigenetic function in hematological diseases. Hematol. Oncol. 2019, 37, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S. The possible role of long non-coding RNAs in recurrent miscarriage. Mol. Biol. Rep. 2022, 49, 9687–9697. [Google Scholar] [CrossRef] [PubMed]
- Pope, C.; Mishra, S.; Russell, J.; Zhou, Q.; Zhong, X.B. Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases 2017, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, W.A.; Mann, M.R.W. Long noncoding RNA functionality in imprinted domain regulation. PLoS Genet. 2020, 16, e1008930. [Google Scholar] [CrossRef]
- Marasek, P.; Dzijak, R.; Studenyak, I.; Fiserova, J.; Ulicna, L.; Novak, P.; Hozak, P. Paxillin-dependent regulation of IGF2 and H19 gene cluster expression. J. Cell Sci. 2015, 128, 3106–3116. [Google Scholar] [CrossRef]
- Kasprzak, A.; Adamek, A. Insulin-Like Growth Factor 2 (IGF2) Signaling in Colorectal Cancer-From Basic Research to Potential Clinical Applications. Int. J. Mol. Sci. 2019, 20, 4915. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Webber, A.L.; Brunkow, M.E.; Tilghman, S.M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993, 7, 1663–1673. [Google Scholar] [CrossRef]
- Lewis, A.; Murrell, A. Genomic imprinting: CTCF protects the boundaries. Curr. Biol. 2004, 14, R28426–R28428. [Google Scholar] [CrossRef]
- Hu, J.F.; Hoffman, A.R. Targeting aberrant IGF2 epigenetics as a novel anti-tumor approach. In DNA Methylation: Patterns, Functions and Roles in Disease; Holland, K., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2016; pp. 91–110. [Google Scholar]
- MacDonald, W.A.; Mann, M.R. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol. Reprod. Dev. 2014, 81, 126–140. [Google Scholar] [CrossRef]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002, 3, 662–673. [Google Scholar] [CrossRef]
- Cannarella, R.; Crafa, A.; Condorelli, R.A.; Mongioi, L.M.; La Vignera, S.; Calogero, A.E. Relevance of sperm imprinted gene methylation on assisted reproductive technique outcomes and pregnancy loss: A systematic review. Syst. Biol. Reprod. Med. 2021, 67, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Lanzillotti, C.; Mazziotta, C.; Tognon, M.; Martini, F. Epigenetics of Male Infertility: The Role of DNA Methylation. Front. Cell Dev. Biol. 2021, 9, 689624. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Vu, T.H.; Ulaner, G.A.; Littman, E.; Ling, J.Q.; Chen, H.L.; Hu, J.F.; Behr, B.; Giudice, L.; Hoffman, A.R. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol. Hum. Reprod. 2005, 11, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Barberet, J.; Ducreux, B.; Guilleman, M.; Simon, E.; Bruno, C.; Fauque, P. DNA methylation profiles after ART during human lifespan: A systematic review and meta-analysis. Hum. Reprod. Update 2022, 144, 393. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Selvatici, R.; Di Domenico, M.; Marci, R.; Vesce, F.; Tognon, M.; Martini, F. Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males. Epigenetics 2013, 8, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo-Sanchez, A.; Sanchez-Delgado, M.; Mora, J.R.H.; Santamaria, N.T.; Gratacos, E.; Esteller, M.; de Heredia, M.L.; Nunes, V.; Choux, C.; Fauque, P.; et al. Differences in expression rather than methylation at placenta-specific imprinted loci is associated with intrauterine growth restriction. Clin. Epigenetics 2019, 11, 35. [Google Scholar] [CrossRef]
- Hanna, C.W.; Penaherrera, M.S.; Saadeh, H.; Andrews, S.; McFadden, D.E.; Kelsey, G.; Robinson, W.P. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016, 26, 756–767. [Google Scholar] [CrossRef]
- Marquardt, R.M.; Lee, K.; Kim, T.H.; Lee, B.; DeMayo, F.J.; Jeong, J.W. Interleukin-13 receptor subunit alpha-2 is a target of progesterone receptor and steroid receptor coactivator-1 in the mouse uterusdagger. Biol. Reprod. 2020, 103, 760–768. [Google Scholar] [CrossRef]
- Rytkonen, K.T.; Erkenbrack, E.M.; Poutanen, M.; Elo, L.L.; Pavlicev, M.; Wagner, G.P. Decidualization of Human Endometrial Stromal Fibroblasts is a Multiphasic Process Involving Distinct Transcriptional Programs. Reprod. Sci. 2019, 26, 323–336. [Google Scholar] [CrossRef]
- Hu, J.F.; Hoffman, A.R. Examining histone acetlylation at specific genomic regions. Methods Mol. Biol. 2001, 181, 285–296. [Google Scholar]
- Li, T.; Hu, J.F.; Qiu, X.; Ling, J.; Chen, H.; Wang, S.; Hou, A.; Vu, T.H.; Hoffman, A.R. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex-2 intrachromosomal loop. Mol. Cell. Biol. 2008, 28, 6473–6482. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, G.; Christian, M.; Steel, J.H.; Henriet, P.; Poutanen, M.; Brosens, J.J. Down-regulation of the histone methyltransferase EZH2 contributes to the epigenetic programming of decidualizing human endometrial stromal cells. Mol. Endocrinol. 2011, 25, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, H.; Li, W.; Cui, J.; Wang, G.; Hu, X.; Hoffman, A.R.; Hu, J. Promoter histone H3K27 methylation in the control of IGF2 imprinting in human tumor cell lines. Hum. Mol. Genet. 2014, 23, 117–128. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, X.; Liu, X.; Wang, G.; Wen, X.; Zhang, X.; Hoffman, A.R.; Li, W.; Hu, J.F.; Cui, J. Loss of insulin-like growth factor II imprinting is a hallmark associated with enhanced chemo/radiotherapy resistance in cancer stem cells. Oncotarget 2016, 7, 51349–51364. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.F.; Hoffman, A.R. Chromatin looping is needed for iPSC induction. Cell Cycle 2014, 13, 2975–2982. [Google Scholar] [CrossRef]
- Ulaner, G.A.; Yang, Y.; Hu, J.F.; Li, T.; Vu, T.H.; Hoffman, A.R. CTCF binding at the insulin-like growth factor-II (IGF2)/H19 imprinting control region is insufficient to regulate IGF2/H19 expression in human tissues. Endocrinology 2003, 144, 4420–4426. [Google Scholar] [CrossRef][Green Version]
- O’Connor, B.B.; Pope, B.D.; Peters, M.M.; Ris-Stalpers, C.; Parker, K.K. The role of extracellular matrix in normal and pathological pregnancy: Future applications of microphysiological systems in reproductive medicine. Exp. Biol. Med. 2020, 245, 1163–1174. [Google Scholar] [CrossRef]
- Conrad, K.P.; Rabaglino, M.B.; Post Uiterweer, E.D. Emerging role for dysregulated decidualization in the genesis of preeclampsia. Placenta 2017, 60, 119–129. [Google Scholar] [CrossRef]
- Vinketova, K.; Mourdjeva, M.; Oreshkova, T. Human Decidual Stromal Cells as a Component of the Implantation Niche and a Modulator of Maternal Immunity. J. Pregnancy 2016, 2016, 8689436. [Google Scholar] [CrossRef]
- Crespi, B.J. Why and How Imprinted Genes Drive Fetal Programming. Front. Endocrinol. 2019, 10, 940. [Google Scholar] [CrossRef]
- Zhang, K.; Smith, G.W. Maternal control of early embryogenesis in mammals. Reprod. Fertil. Dev. 2015, 27, 880–896. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; Halle, D.; Raveh, E.; Gilon, M.; Sorin, V.; Hochberg, A. The role of the oncofetal H19 lncRNA in tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget 2016, 7, 3748–3765. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W. Placental imprinting: Emerging mechanisms and functions. PLoS Genet. 2020, 16, e1008709. [Google Scholar] [CrossRef]
- Nordin, M.; Bergman, D.; Halje, M.; Engstrom, W.; Ward, A. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014, 47, 189–199. [Google Scholar] [CrossRef]
- Blyth, A.J.; Kirk, N.S.; Forbes, B.E. Understanding IGF-II Action through Insights into Receptor Binding and Activation. Cells 2020, 9, 2276. [Google Scholar] [CrossRef]
- Fowden, A.L. The insulin-like growth factors and feto-placental growth. Placenta 2003, 24, 803–812. [Google Scholar] [CrossRef]
- Ratajczak, M.Z. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ’passkey’ to cancerogenesis. Folia Histochem. Cytobiol. 2012, 50, 171–179. [Google Scholar] [CrossRef]
- Argyraki, M.; Damdimopoulou, P.; Chatzimeletiou, K.; Grimbizis, G.F.; Tarlatzis, B.C.; Syrrou, M.; Lambropoulos, A. In-utero stress and mode of conception: Impact on regulation of imprinted genes, fetal development and future health. Hum. Reprod. Update 2019, 25, 777–801. [Google Scholar] [CrossRef]
- Matouk, I.J.; Halle, D.; Gilon, M.; Hochberg, A. The non-coding RNAs of the H19-IGF2 imprinted loci: A focus on biological roles and therapeutic potential in Lung Cancer. J. Transl. Med. 2015, 13, 113. [Google Scholar] [CrossRef]
- Ivanova, E.; Canovas, S.; Garcia-Martinez, S.; Romar, R.; Lopes, J.S.; Rizos, D.; Sanchez-Calabuig, M.J.; Krueger, F.; Andrews, S.; Perez-Sanz, F.; et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenetics 2020, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- Marcho, C.; Cui, W.; Mager, J. Epigenetic dynamics during preimplantation development. Reproduction 2015, 150, R109–R120. [Google Scholar] [CrossRef] [PubMed]
- Ankolkar, M.; Patil, A.; Warke, H.; Salvi, V.; Mokashi, N.K.; Pathak, S.; Balasinor, N.H. Methylation analysis of idiopathic recurrent spontaneous miscarriage cases reveals aberrant imprinting at H19 ICR in normozoospermic individuals. Fertil. Steril. 2012, 98, 1186–1192. [Google Scholar] [CrossRef] [PubMed]
- Aygun, D.; Bjornsson, H.T. Clinical epigenetics: A primer for the practitioner. Dev. Med. Child. Neurol. 2020, 62, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Pianka, M.A.; McIntosh, A.T.; Patel, S.D.; Bakhshi, P.R.; Jung, M. Close yet so far away: A look into the management strategies of genetic imprinting disorders. Am. J. Stem. Cells 2018, 7, 72–81. [Google Scholar] [PubMed]
- Jelinic, P.; Shaw, P. Loss of imprinting and cancer. J. Pathol. 2007, 211, 261–268. [Google Scholar] [CrossRef]
- Court, F.; Tayama, C.; Romanelli, V.; Martin-Trujillo, A.; Iglesias-Platas, I.; Okamura, K.; Sugahara, N.; Simon, C.; Moore, H.; Harness, J.V.; et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014, 24, 554–569. [Google Scholar] [CrossRef]
- McMinn, J.; Wei, M.; Schupf, N.; Cusmai, J.; Johnson, E.B.; Smith, A.C.; Weksberg, R.; Thaker, H.M.; Tycko, B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 2006, 27, 540–549. [Google Scholar] [CrossRef]
- Romanelli, V.; Nakabayashi, K.; Vizoso, M.; Moran, S.; Iglesias-Platas, I.; Sugahara, N.; Simon, C.; Hata, K.; Esteller, M.; Court, F.; et al. Variable maternal methylation overlapping the nc886/vtRNA2-1 locus is locked between hypermethylated repeats and is frequently altered in cancer. Epigenetics 2014, 9, 783–790. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, Y.; Xiong, J.; Liu, J.; Zha, Y.; Kang, Q.; Zhi, P.; Wang, Q.; Wang, H.; Zeng, W.; et al. Activated gammadelta T Cells With Higher CD107a Expression and Inflammatory Potential During Early Pregnancy in Patients With Recurrent Spontaneous Abortion. Front. Immunol. 2021, 12, 724662. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, P.; Domnina, A.; Gorelova, I.; Rulev, M.; Petrosyan, M.; Nikolsky, N.; Borodkina, A. "All-In-One" Genetic Tool Assessing Endometrial Receptivity for Personalized Screening of Female Sex Steroid Hormones. Front. Cell Dev. Biol. 2021, 9, 624053. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Li, J.; Miao, B.; Wang, S.; Dong, W.; Xu, H.; Si, C.; Wang, W.; Duan, S.; Lou, J.; Bao, Z.; et al. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief Bioinform. 2022, 23. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, J.F.; Wang, H.; Cui, J.; Gao, S.; Hoffman, A.R.; Li, W. CRISPR Cas9-guided chromatin immunoprecipitation identifies miR483 as an epigenetic modulator of IGF2 imprinting in tumors. Oncotarget 2017, 8, 34177–34190. [Google Scholar] [CrossRef]
- Chen, N.; Yan, X.; Zhao, G.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218. [Google Scholar] [CrossRef]
- Pian, L.; Wen, X.; Kang, L.; Li, Z.; Nie, Y.; Du, Z.; Yu, D.; Zhou, L.; Jia, L.; Chen, N.; et al. Targeting the IGF1R Pathway in Breast Cancer Using Antisense lncRNA-Mediated Promoter cis Competition. Mol. Ther. Nucleic Acids 2018, 12, 105–117. [Google Scholar] [CrossRef]

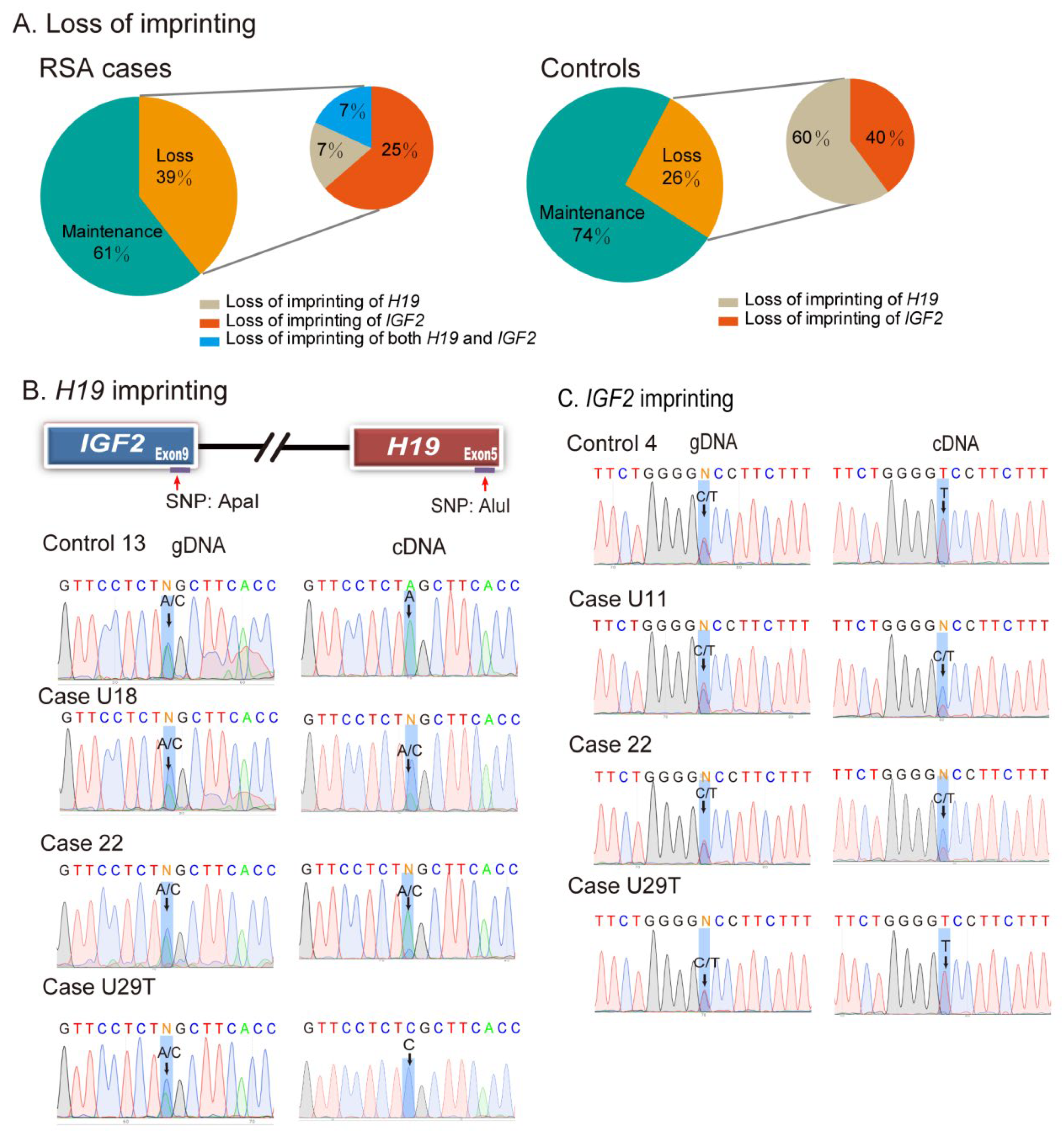

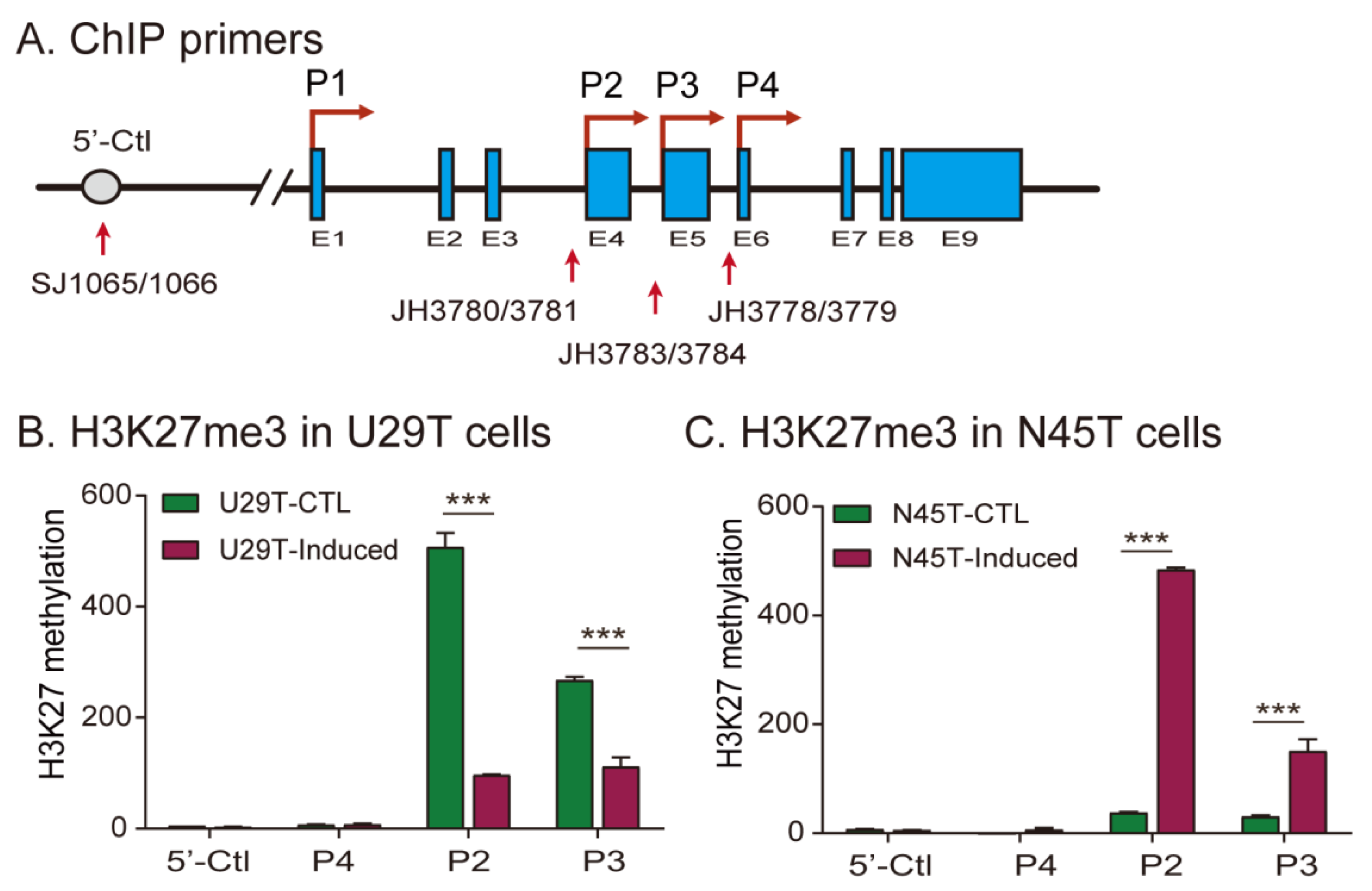
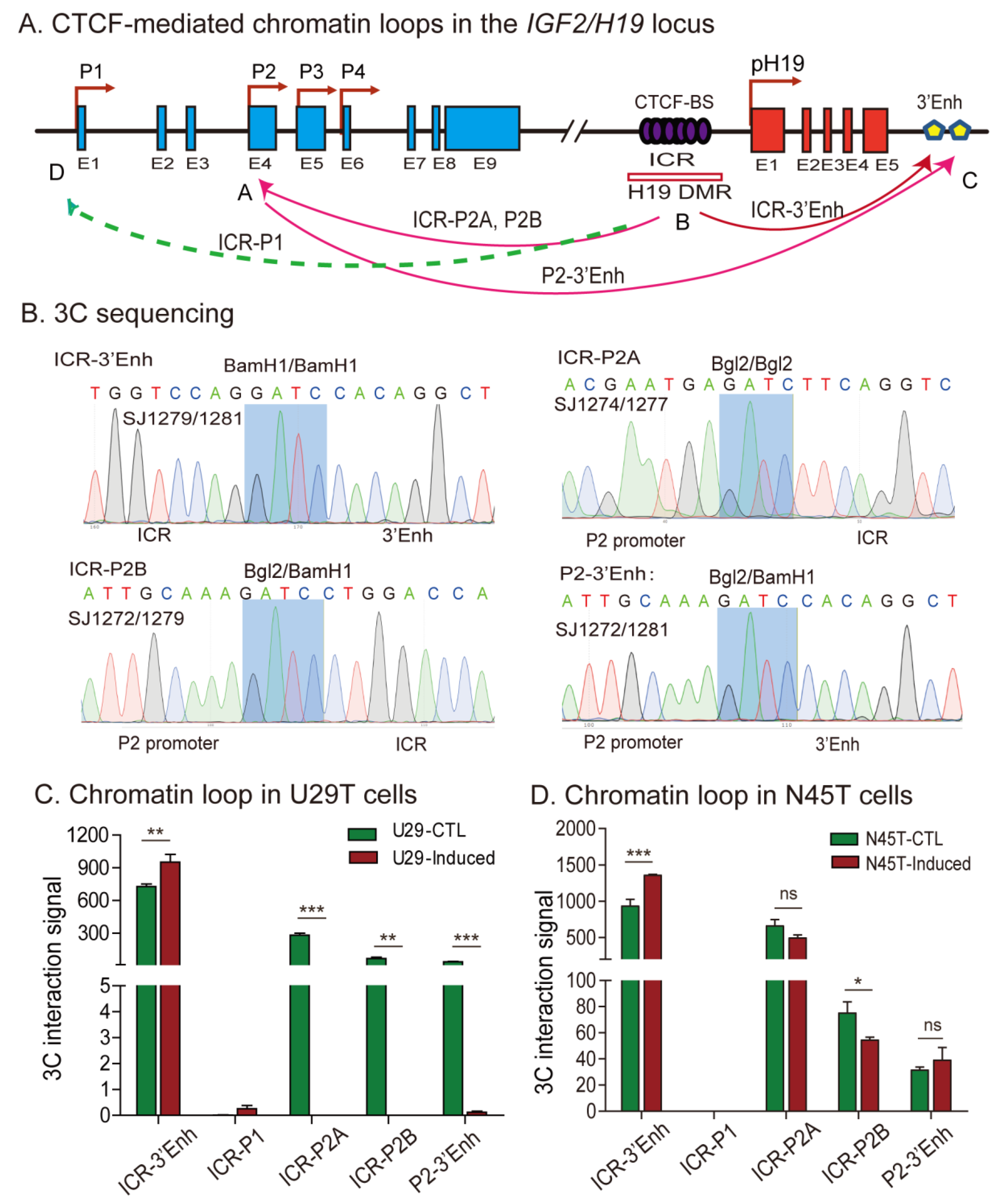
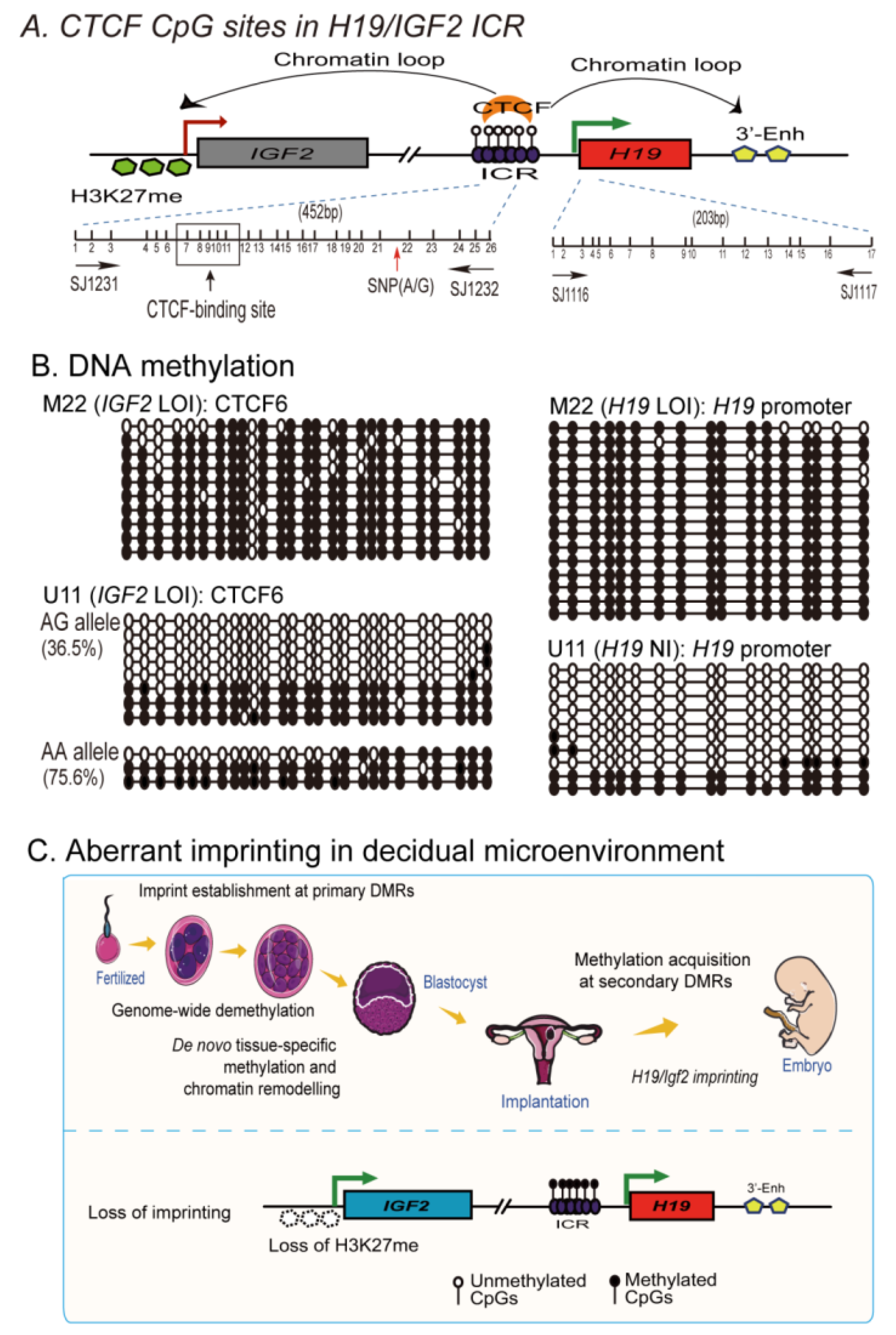
| H19 | IGF2 | ||||
|---|---|---|---|---|---|
| Cases (ID) | Genotype | cDNA | Genotype | cDNA | |
| Loss of imprinting of H19 (9.5%) * | |||||
| 1 | U18 | A/C | a/c | C/T | t |
| 2 | U21 | A/C | a/c | T/T | - |
| Loss of imprinting of IGF2 (35%) ** | |||||
| 1 | 8 | A/C | a | C/T | c/t |
| 2 | E1 | A/C | c | C/T | c/t |
| 3 | E3 | A/A | - | C/T | c/t |
| 4 | E5 | A/C | c | C/T | c/t |
| 5 | U11 | A/A | - | C/T | c/t |
| 6 | U14 | A/A | - | C/T | c/t |
| 7 | U17 | A/A | - | C/T | c/t |
| Loss of imprinting of H19 and IGF2 *** | |||||
| 1 | M22 | A/C | a/c | C/T | c/t |
| 2 | U20 | A/C | a/c | C/T | c/t |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, X.; Zhang, Q.; Zhou, L.; Li, Z.; Wei, X.; Yang, W.; Zhang, J.; Li, H.; Xu, Z.; Cui, X.; et al. Intrachromosomal Looping and Histone K27 Methylation Coordinately Regulates the lncRNA H19-Fetal Mitogen IGF2 Imprinting Cluster in the Decidual Microenvironment of Early Pregnancy. Cells 2022, 11, 3130. https://doi.org/10.3390/cells11193130
Wen X, Zhang Q, Zhou L, Li Z, Wei X, Yang W, Zhang J, Li H, Xu Z, Cui X, et al. Intrachromosomal Looping and Histone K27 Methylation Coordinately Regulates the lncRNA H19-Fetal Mitogen IGF2 Imprinting Cluster in the Decidual Microenvironment of Early Pregnancy. Cells. 2022; 11(19):3130. https://doi.org/10.3390/cells11193130
Chicago/Turabian StyleWen, Xue, Qi Zhang, Lei Zhou, Zhaozhi Li, Xue Wei, Wang Yang, Jiaomei Zhang, Hui Li, Zijun Xu, Xueling Cui, and et al. 2022. "Intrachromosomal Looping and Histone K27 Methylation Coordinately Regulates the lncRNA H19-Fetal Mitogen IGF2 Imprinting Cluster in the Decidual Microenvironment of Early Pregnancy" Cells 11, no. 19: 3130. https://doi.org/10.3390/cells11193130
APA StyleWen, X., Zhang, Q., Zhou, L., Li, Z., Wei, X., Yang, W., Zhang, J., Li, H., Xu, Z., Cui, X., Zhang, S., Wang, Y., Li, W., Hoffman, A. R., Liu, Z., Hu, J.-F., & Cui, J. (2022). Intrachromosomal Looping and Histone K27 Methylation Coordinately Regulates the lncRNA H19-Fetal Mitogen IGF2 Imprinting Cluster in the Decidual Microenvironment of Early Pregnancy. Cells, 11(19), 3130. https://doi.org/10.3390/cells11193130








