Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation
Abstract
1. Introduction
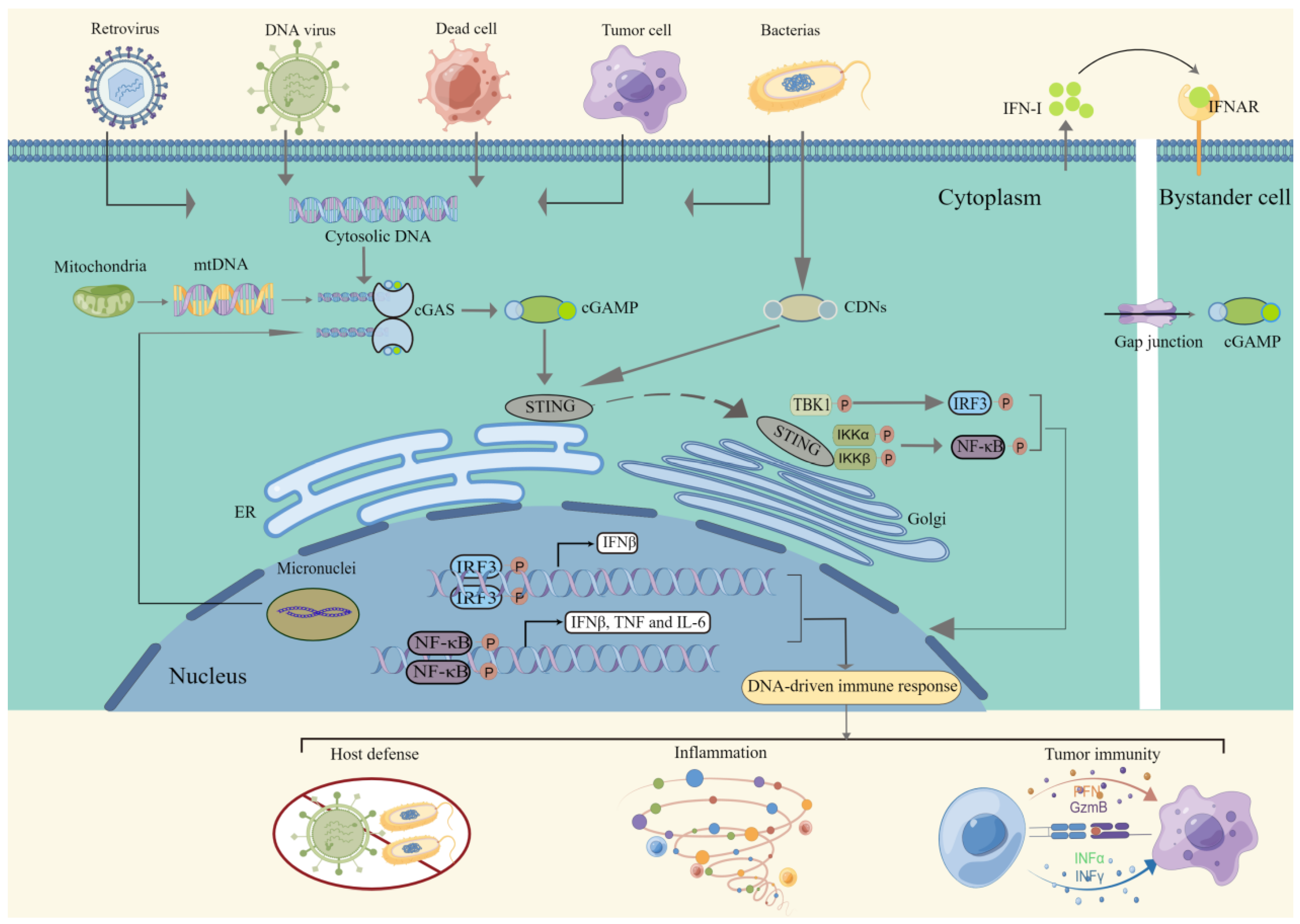
2. Signaling of the cGAS-STING Pathway
2.1. Detection of dsDNA
2.2. Intracellular Signal Transduction
2.3. Immune Response Activation
3. Regulatory Mechanisms of cGAS-STING in the Context of Disease
3.1. Viral and Bacterial Infections
3.2. Cancer
3.3. Autoimmune Diseases
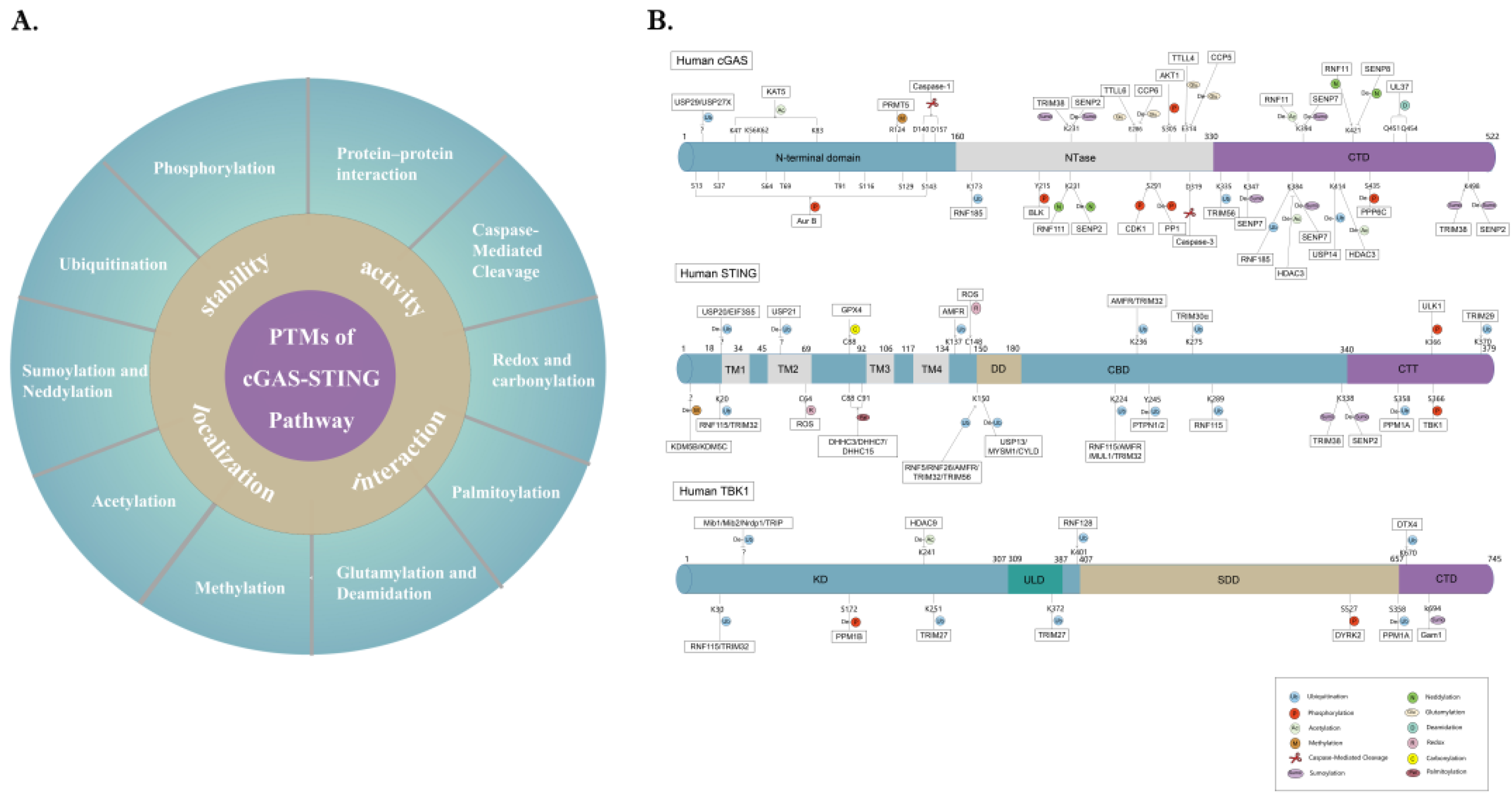
4. PTM Networks Regulating the cGAS-STING Pathway
4.1. Phosphorylation
4.2. Ubiquitination
4.3. SUMOylation and Neddylation
4.4. Acetylation
4.5. Methylation
4.6. Glutamylation and Deamidation
4.7. Palmitoylation
4.8. Redox and Carbonylation
4.9. Caspase-Mediated Cleavage
4.10. Protein–Protein Interaction
5. Treatment with PTMs
6. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef] [PubMed]
- González-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Rotem, Z.; Cox, R.A.; Isaacs, A. Inhibition of Virus Multiplication by Foreign Nucleic Acid. Nature 1963, 197, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Stetson, D.B.; Medzhitov, R. Recognition of Cytosolic DNA Activates an IRF3-Dependent Innate Immune Response. Immunity 2006, 24, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Barrat, F.J.; Elkon, K.B.; Fitzgerald, K.A. Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu. Rev. Med. 2016, 67, 323–336. [Google Scholar] [CrossRef]
- Motwani, M.; Pesiridis, S.; Fitzgerald, K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019, 20, 657–674. [Google Scholar] [CrossRef]
- Deribe, Y.L.; Pawson, T.; Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010, 17, 666–672. [Google Scholar] [CrossRef]
- Li, X.D.; Wu, J.; Gao, D.; Wang, H.; Sun, L.; Chen, Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune ad-juvant effects. Science 2013, 341, 1390–1394. [Google Scholar] [CrossRef]
- Zhang, X.; Bai, X.C.; Chen, Z.J. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 2020, 53, 43–53. [Google Scholar] [CrossRef]
- Civril, F.; Deimling, T.; Mann, C.C.D.O.; Ablasser, A.; Moldt, M.; Witte, G.; Hornung, V.; Hopfner, K.-P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013, 498, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science 2012, 339, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Whiteley, A.T.; de Oliveira Mann, C.C.; Morehouse, B.R.; Nowak, R.P.; Fischer, E.S.; Fischer, E.S.; Gray, N.S.; Mekalanos, J.J.; Kranzusch, P.J. Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 2018, 174, 300–311.e11. [Google Scholar] [CrossRef] [PubMed]
- Luecke, S.; Holleufer, A.; Christensen, M.H.; Jønsson, K.L.; Boni, A.G.; Sørensen, L.K.; Johannsen, M.; Jakobsen, M.R.; Hartmann, R.; Paludan, S.R. cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 2017, 18, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Erttmann, S.F.; Swacha, P.; Aung, K.M.; Brindefalk, B.; Jiang, H.; Hartlova, A.; Uhlin, B.E.; Wai, S.N.; Gekara, N.O. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity 2022, 55, 847–861 e10. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Li, X.; Shu, C.; Yi, G.; Chaton, C.T.; Shelton, C.L.; Diao, J.; Zuo, X.; Kao, C.C.; Herr, A.B.; Li, P. Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization. Immunity 2013, 39, 1019–1031. [Google Scholar] [CrossRef]
- Kato, K.; Omura, H.; Ishitani, R.; Nureki, O. Cyclic GMP–AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Annu. Rev. Biochem. 2017, 86, 541–566. [Google Scholar] [CrossRef]
- Hooy, R.; Massaccesi, G.; Rousseau, K.; Chattergoon, A.M.; Sohn, J. Allosteric coupling between Mn2+ and dsDNA controls the catalytic efficiency and fidelity of cGAS. Nucleic Acids Res. 2020, 48, 4435–4447. [Google Scholar] [CrossRef]
- Wang, C.; Guan, Y.; Lv, M.; Zhang, R.; Guo, Z.; Wei, X.; Du, X.; Yang, J.; Li, T.; Wan, Y.; et al. Manganese Increases the Sensitivity of the cGAS-STING Pathway for Double-Stranded DNA and Is Required for the Host Defense against DNA Viruses. Immunity 2018, 48, 675–687.e7. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, Z.; Wang, B.; Guan, Y.; Su, X.-D.; Jiang, Z. Mn2+ Directly Activates cGAS and Structural Analysis Suggests Mn2+ Induces a Noncanonical Catalytic Synthesis of 2′3′-cGAMP. Cell Rep. 2020, 32, 108053. [Google Scholar] [CrossRef] [PubMed]
- Lahey, L.J.; Mardjuki, R.E.; Wen, X.; Hess, G.T.; Ritchie, C.; Carozza, J.A.; Böhnert, V.; Maduke, M.; Bassik, M.C.; Li, L. LRRC8A:C/E Heteromeric Channels Are Ubiquitous Transporters of cGAMP. Mol. Cell 2020, 80, 578–591.e5. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, C.; Cordova, A.F.; Hess, G.T.; Bassik, M.C.; Li, L. SLC19A1 Is an Importer of the Immunotransmitter cGAMP. Mol. Cell 2019, 75, 372–381.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chen, X.; Planells-Cases, R.; Chu, J.; Wang, L.; Cao, L.; Li, Z.; López-Cayuqueo, K.I.; Xie, Y.; Ye, S.; et al. Transfer of cGAMP into Bystander Cells via LRRC8 Vol-ume-Regulated Anion Channels Augments STING-Mediated Interferon Responses and Anti-viral Immunity. Immunity 2020, 52, 767–781.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, H.; Wu, J.; Zhang, X.; Sun, L.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP containing mixed phosphodiester linkages is an en-dogenous high-affinity ligand for STING. Mol. Cell 2013, 51, 226–235. [Google Scholar] [CrossRef]
- Gao, P.; Ascano, M.; Zillinger, T.; Wang, W.; Dai, P.; Serganov, A.A.; Gaffney, B.L.; Shuman, S.; Jones, R.A.; Deng, L.; et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 2013, 154, 748–762. [Google Scholar] [CrossRef]
- Gui, X.; Yang, H.; Li, T.; Tan, X.; Shi, P.; Li, M.; Du, F.; Chen, Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019, 567, 262–266. [Google Scholar] [CrossRef]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef]
- Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.-C.; Chen, Z.J. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef]
- Tanaka, Y.; Chen, Z.J. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci. Signal. 2012, 5, ra20. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.-T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef] [PubMed]
- Agalioti, T.; Lomvardas, S.; Parekh, B.; Yie, J.; Maniatis, T.; Thanos, D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 2000, 103, 667–678. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Hao, M.; Chang, W.; Imamichi, T. The Role of Ku70 as a Cytosolic DNA Sensor in Innate Immunity and Beyond. Front. Cell Infect. Microbiol. 2021, 11, 761983. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Zhou, M.; Imamichi, H.; Jiao, X.; Sherman, B.T.; Lane, H.C.; Imamichi, T. STING is an essential mediator of the Ku70-mediated pro-duction of IFN-lambda1 in response to exogenous DNA. Sci. Signal. 2017, 10, eaah5054. [Google Scholar] [CrossRef]
- McKnight, K.L.; Swanson, K.V.; Austgen, K.; Richards, C.; Mitchell, J.K.; McGivern, D.R.; Fritch, E.; Johnson, J.; Remlinger, K.; Magid-Slav, M.; et al. Stimulator of interferon genes (STING) is an essential proviral host factor for human rhinovirus species A and C. Proc. Natl. Acad. Sci. USA 2020, 117, 27598–27607. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.M.; Maniatis, T. IKKepsilon and TBK1 are essential compo-nents of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef]
- Gonugunta, V.K.; Sakai, T.; Pokatayev, V.; Yang, K.; Wu, J.; Dobbs, N.; Yan, N. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Rep. 2017, 21, 3234–3242. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Hu, J.; Zhang, H.; Xu, F.; He, W.; Wang, X.; Li, M.; Lu, W.; Zeng, G.; et al. cGAS/STING axis mediates a topoisomerase II inhibitor–induced tumor immunogenicity. J. Clin. Investig. 2019, 129, 4850–4862. [Google Scholar] [CrossRef]
- Burdette, D.L.; Vance, E.R. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2012, 14, 19–26. [Google Scholar] [CrossRef]
- Li, A.; Yi, M.; Qin, S.; Song, Y.; Chu, Q.; Wu, K. Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Majdoul, S.; Compton, A.A. Lessons in self-defence: Inhibition of virus entry by intrinsic immunity. Nat. Rev. Immunol. 2021, 22, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Damania, B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef]
- Ma, Z.; Ni, G.; Damania, B. Innate Sensing of DNA Virus Genomes. Annu. Rev. Virol. 2018, 5, 341–362. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Wu, J.; Wu, Y.-T.; Du, F.; Aroh, C.; Yan, N.; Sun, L.; Chen, Z.J. Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science 2013, 341, 903–906. [Google Scholar] [CrossRef]
- Webb, L.G.; Fernandez-Sesma, A. RNA viruses and the cGAS-STING pathway: Reframing our understanding of innate immune sensing. Curr. Opin. Virol. 2022, 53, 101206. [Google Scholar] [CrossRef]
- Fan, Y.M.; Zhang, Y.L.; Luo, H.; Mohamud, Y. Crosstalk between RNA viruses and DNA sensors: Role of the cGAS-STING signal-ling pathway. Rev. Med. Virol. 2022, 32, e2343. [Google Scholar] [CrossRef]
- Deschamps, T.; Kalamvoki, M. Extracellular Vesicles Released by Herpes Simplex Virus 1-Infected Cells Block Virus Replica-tion in Recipient Cells in a STING-Dependent Manner. J. Virol. 2018, 92, e01102-18. [Google Scholar] [CrossRef]
- Pan, S.; Liu, X.; Ma, Y.; Cao, Y.; He, B. Herpes Simplex Virus 1 γ 1 34.5 Protein Inhibits STING Activation That Restricts Viral Replication. J. Virol. 2018, 92, e01015-18. [Google Scholar] [CrossRef] [PubMed]
- Kalamvoki, M.; Roizman, B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc. Natl. Acad. Sci. USA 2014, 111, E611–E617. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Zheng, S.; Huang, Z.; Lin, Y.; Shen, Q.; Zheng, C. Herpes Simplex Virus 1 Tegument Protein UL46 Inhibits TANK-Binding Kinase 1-Mediated Signaling. mBio 2019, 10, e00919-19. [Google Scholar] [CrossRef]
- Ye, R.; Su, C.; Xu, H.; Zheng, C. Herpes Simplex Virus 1 Ubiquitin-Specific Protease UL36 Abrogates NF-kappaB Activation in DNA Sensing Signal Pathway. J. Virol. 2017, 91, 11851–11860. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Zheng, C. Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J. Virol. 2017, 91, e02414-16. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, T.; Kalamvoki, M. Evasion of the STING DNA-Sensing Pathway by VP11/12 of Herpes Simplex Virus 1. J. Virol. 2017, 91, e00535-17. [Google Scholar] [CrossRef]
- Huang, J.; You, H.; Su, C.; Li, Y.; Chen, S.; Zheng, C. Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity. J. Virol. 2018, 92, e00841-18. [Google Scholar] [CrossRef]
- Bodda, C.; Reinert, L.S.; Fruhwürth, S.; Richardo, T.; Sun, C.; Zhang, B.-C.; Kalamvoki, M.; Pohlmann, A.; Mogensen, T.H.; Bergström, P.; et al. HSV1 VP1-2 deubiquitinates STING to block type I interferon expression and promote brain infection. J. Exp. Med. 2020, 217, e20191422. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Xu, S.; Li, J.; He, S.; Zeng, Y.; Xie, L.; Xie, N.; Liu, T.; Lee, K.; et al. Species-Specific Deamidation of cGAS by Herpes Simplex Virus UL37 Protein Facilitates Viral Replication. Cell Host Microbe 2018, 24, 234–248.e5. [Google Scholar] [CrossRef]
- Xu, G.; Liu, C.; Zhou, S.; Li, Q.; Feng, Y.; Sun, P.; Feng, H.; Gao, Y.; Zhu, J.; Luo, X.; et al. Viral tegument proteins restrict cGAS-DNA phase separation to mediate immune evasion. Mol. Cell 2021, 81, 2823–2837.e9. [Google Scholar] [CrossRef]
- Liu, X.; Main, D.; Ma, Y.; He, B. Herpes Simplex Virus 1 Inhibits TANK-Binding Kinase 1 through Formation of the Us11-Hsp90 Complex. J. Virol. 2018, 92, e00402-18. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Sumner, R.P.; Rasaiyaah, J.; Tan, C.P.; Rodriguez-Plata, M.T.; Van Tulleken, C.; Fink, D.; Zuliani-Alvarez, L.; Thorne, L.; Stirling, D.; et al. HIV-1 Vpr antagonizes innate im-mune activation by targeting karyopherin-mediated NF-kappaB/IRF3 nuclear transport. eLife 2020, 9, e60821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chan, B.; Samarina, N.; Abere, B.; Weidner-Glunde, M.; Buch, A.; Pich, A.; Brinkmann, M.M.; Schulz, T.F. Cytoplasmic isoforms of Kaposi sarcoma her-pesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. USA 2016, 113, E1034–E1043. [Google Scholar] [PubMed]
- Wu, J.-J.; Li, W.; Shao, Y.; Avey, D.; Fu, B.; Gillen, J.; Hand, T.; Ma, S.; Liu, X.; Miley, W.; et al. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 2015, 18, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Jacobs, S.R.; West, J.A.; Stopford, C.; Zhang, Z.; Davis, Z.; Barber, G.N.; Glaunsinger, B.A.; Dittmer, D.P.; Damania, B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. USA 2015, 112, E4306–E4315. [Google Scholar] [CrossRef]
- Lin, R.; Genin, P.; Mamane, Y.; Sgarbanti, M.; Battistini, A.; Harrington, W.J., Jr.; Barber, G.N.; Hiscott, J. HHV-8 encoded vIRF-1 represses the inter-feron antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 2001, 20, 800–811. [Google Scholar] [CrossRef]
- Lefort, S.; Soucy-Faulkner, A.; Grandvaux, N.; Flamand, L. Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to inter-feron-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 2007, 81, 10950–10960. [Google Scholar] [CrossRef]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef]
- Sun, C.; Schattgen, S.; Pisitkun, P.; Jorgensen, J.P.; Hilterbrand, A.T.; Wang, L.J.; West, J.A.; Hansen, K.; Horan, K.A.; Jakobsen, M.R.; et al. Evasion of Innate Cytosolic DNA Sensing by a Gammaherpesvirus Facilitates Establishment of Latent Infection. J. Immunol. 2015, 194, 1819–1831. [Google Scholar] [CrossRef]
- Kang, H.-R.; Cheong, W.-C.; Park, J.-E.; Ryu, S.; Cho, H.-J.; Youn, H.; Ahn, J.-H.; Song, M.J. Murine Gammaherpesvirus 68 Encoding Open Reading Frame 11 Targets TANK Binding Kinase 1 To Negatively Regulate the Host Type I Interferon Response. J. Virol. 2014, 88, 6832–6846. [Google Scholar] [CrossRef][Green Version]
- Hwang, S.; Kim, K.S.; Flano, E.; Wu, T.-T.; Tong, L.M.; Park, A.N.; Song, M.J.; Sanchez, D.J.; O’Connell, R.M.; Cheng, G.; et al. Conserved Herpesviral Kinase Promotes Viral Persistence by Inhibiting the IRF-3-Mediated Type I Interferon Response. Cell Host Microbe 2009, 5, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.; Sommer, M.; Che, X.; White, K.; Ruyechan, W.T.; Arvin, A.M. Varicella-zoster virus immediate-early protein 62 blocks in-terferon regulatory factor 3 (IRF3) phosphorylation at key serine residues: A novel mechanism of IRF3 inhibition among herpesviruses. J. Virol. 2010, 84, 9240–9253. [Google Scholar] [CrossRef] [PubMed]
- Vandevenne, P.; Lebrun, M.; El Mjiyad, N.; Ote, I.; Di Valentin, E.; Habraken, Y.; Dortu, E.; Piette, J.; Sadzot-Delvaux, C. The varicella-zoster virus ORF47 kinase in-terferes with host innate immune response by inhibiting the activation of IRF3. PLoS ONE 2011, 6, e16870. [Google Scholar] [CrossRef] [PubMed]
- Whitmer, T.; Malouli, D.; Uebelhoer, L.S.; DeFilippis, V.R.; Fruh, K.; Verweij, M.C. The ORF61 Protein Encoded by Simian Varicella Virus and Varicella-Zoster Virus Inhibits NF-kappaB Signaling by Interfering with IkappaBalpha Degradation. J. Virol. 2015, 89, 8687–8700. [Google Scholar] [CrossRef]
- Li, K.; Liu, Y.; Xu, Z.; Zhang, Y.; Luo, D.; Gao, Y.; Qian, Y.; Bao, C.; Liu, C.; Zhang, Y.; et al. Avian oncogenic herpesvirus antagonizes the cGAS-STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog. 2019, 15, e1007999. [Google Scholar] [CrossRef]
- Gao, L.; Li, K.; Zhang, Y.; Liu, Y.; Liu, C.; Zhang, Y.; Gao, Y.; Qi, X.; Cui, H.; Wang, Y.; et al. Inhibition of DNA-Sensing Pathway by Marek’s Disease Virus VP23 Pro-tein through Suppression of Interferon Regulatory Factor 7 Activation. J. Virol. 2019, 93, e01934-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ning, S. New Look of EBV LMP1 Signaling Landscape. Cancers 2021, 13, 5451. [Google Scholar] [CrossRef]
- Phelan, T.; Little, M.A.; Brady, G. Targeting of the cGAS-STING system by DNA viruses. Biochem. Pharmacol. 2020, 174, 113831. [Google Scholar]
- Uhlorn, B.L.; Jackson, R.; Li, S.; Bratton, S.M.; Van Doorslaer, K.; Campos, S.K. Vesicular trafficking permits evasion of cGAS/STING surveillance during initial human papillomavirus infection. PLoS Pathog. 2020, 16, e1009028. [Google Scholar] [CrossRef]
- Luo, X.; Donnelly, C.; Gong, W.; Heath, B.R.; Hao, Y.; Donnelly, L.A.; Moghbeli, T.; Tan, Y.S.; Lin, X.; Bellile, E.; et al. HPV16 drives cancer immune escape via NLRX1-mediated degradation of STING. J. Clin. Investig. 2020, 130, 1635–1652. [Google Scholar] [CrossRef]
- Wu, L.; Cao, J.; Cai, W.L.; Lang, S.M.; Horton, J.R.; Jansen, D.J.; Liu, Z.Z.; Chen, J.F.; Zhang, M.; Mott, B.T.; et al. KDM5 histone demethylases repress immune response via sup-pression of STING. PLoS Biol. 2018, 16, e2006134. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.; Gray, E.E.; Brunette, R.L.; Stetson, D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Stein, S.; Falck-Pedersen, E. Adenovirus Detection by the cGAS/STING/TBK1 DNA Sensing Cascade. J. Virol. 2014, 88, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach-Rivière, L.; Bergez, M.; Mönch, S.; Qu, B.; Riess, M.; Vondran, F.W.R.; Liese, J.; Hornung, V.; Urban, S.; König, R. Hepatitis B Virus DNA is a Substrate for the cGAS/STING Pathway but is not Sensed in Infected Hepatocytes. Viruses 2020, 12, 592. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Chen, J.; Li, Y.; Wang, W.; Du, X.; Song, W.; Zhang, W.; Lin, L.; Yuan, Z. Hepatitis B Virus Polymerase Disrupts K63-Linked Ubiquitination of STING To Block Innate Cytosolic DNA-Sensing Pathways. J. Virol. 2015, 89, 2287–2300. [Google Scholar] [CrossRef]
- Paijo, J.; Doring, M.; Spanier, J.; Grabski, E.; Nooruzzaman, M.; Schmidt, T.; Witte, G.; Messerle, M.; Hornung, V.; Kaever, V.; et al. cGAS Senses Human Cytomegalovirus and In-duces Type I Interferon Responses in Human Monocyte-Derived Cells. PLoS Pathog. 2016, 12, e1005546. [Google Scholar] [CrossRef]
- Dell’Oste, V.; Gatti, D.; Gugliesi, F.; De Andrea, M.; Bawadekar, M.; Lo Cigno, I.; Biolatti, M.; Vallino, M.; Marschall, M.; Gariglio, M.; et al. Innate Nuclear Sensor IFI16 Translocates into the Cytoplasm during the Early Stage of In Vitro Human Cytomegalovirus Infection and Is Entrapped in the Egressing Virions during the Late Stage. J. Virol. 2014, 88, 6970–6982. [Google Scholar] [CrossRef]
- Kumari, P.; Saha, I.; Narayanan, A.; Narayanan, S.; Takaoka, A.; Kumar, N.S.; Tailor, P.; Kumar, H. Essential role of HCMV deubiquitinase in pro-moting oncogenesis by targeting anti-viral innate immune signaling pathways. Cell Death Dis. 2017, 8, e3078. [Google Scholar] [CrossRef]
- Fu, Y.-Z.; Su, S.; Gao, Y.-Q.; Wang, P.-P.; Huang, Z.-F.; Hu, M.-M.; Luo, W.-W.; Li, S.; Luo, M.-H.; Wang, Y.-Y.; et al. Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe 2017, 21, 231–243. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, Y.E.; Stinski, M.F.; Ahn, J.H.; Song, Y.J. Human Cytomegalovirus IE2 86 kDa Protein Induces STING Degradation and Inhibits cGAMP-Mediated IFN-beta Induction. Front. Microbiol. 2017, 8, 1854. [Google Scholar]
- Biolatti, M.; Dell’Oste, V.; Pautasso, S.; Gugliesi, F.; von Einem, J.; Krapp, C.; Jakobsen, M.R.; Borgogna, C.; Gariglio, M.; De Andrea, M.; et al. Human Cytomegalovirus Tegument Protein pp65 (pUL83) Dampens Type I Interferon Production by Inactivating the DNA Sensor cGAS without Affecting STING. J. Virol. 2018, 92, e01774-17. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Park, A.; Kang, S.; Lee, E.; Lee, T.A.; Ra, E.A.; Lee, J.; Lee, S.; Park, B. Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat. Commun. 2018, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Nukui, M.; Roche, K.L.; Jia, J.; Fox, P.L.; Murphy, E.A. Protein S-Nitrosylation of Human Cytomegalovirus pp71 Inhibits Its Ability To Limit STING Antiviral Responses. J. Virol. 2020, 94, e00033-20. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, W.; Wu, X.; Li, X.; Yang, X.; Ran, Y.; Wu, J.; Li, H. Human Cytomegalovirus UL23 Attenuates Signal Transducer and Activator of Transcription 1 Phosphorylation and Type I Interferon Response. Front. Microbiol. 2021, 12, 692515. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.Y.; He, X.B.; Jia, H.J.; Chen, G.H.; Jin, Q.W.; Long, Z.L.; Jing, Z.Z. The cGas-Sting Signaling Pathway Is Required for the Innate Immune Response Against Ectromelia Virus. Front. Immunol. 2018, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Hopfner, K.P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [PubMed]
- Eaglesham, J.B.; Pan, Y.; Kupper, T.S.; Kranzusch, P.J. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 2019, 566, 259–263. [Google Scholar]
- Fang, R.; Jiang, Q.; Guan, Y.; Gao, P.; Zhang, R.; Zhao, Z.; Jiang, Z. Golgi apparatus-synthesized sulfated glycosaminoglycans mediate polymerization and activation of the cGAMP sensor STING. Immunity 2021, 54, 962–975 e8. [Google Scholar] [CrossRef]
- Georgana, I.; Sumner, R.P.; Towers, G.J.; de Motes, C.M. Virulent Poxviruses Inhibit DNA Sensing by Preventing STING Activation. J. Virol. 2018, 92, e02145-17. [Google Scholar] [CrossRef]
- Meade, N.; Furey, C.; Li, H.; Verma, R.; Chai, Q.; Rollins, M.; DiGiuseppe, S.; Naghavi, M.H.; Walsh, D. Poxviruses Evade Cytosolic Sensing through Disruption of an mTORC1-mTORC2 Regulatory Circuit. Cell 2018, 174, 1143–1157.e17. [Google Scholar] [CrossRef]
- Unterholzner, L.; Sumner, R.P.; Baran, M.; Ren, H.; Mansur, D.; Bourke, N.; Randow, F.; Smith, G.L.; Bowie, A.G. Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7. PLoS Pathog. 2011, 7, e1002247. [Google Scholar] [CrossRef] [PubMed]
- Peters, N.; Ferguson, B.; Mazzon, M.; Fahy, A.S.; Krysztofinska, E.; Arribas-Bosacoma, R.; Pearl, L.; Ren, H.; Smith, G.L. A Mechanism for the Inhibition of DNA-PK-Mediated DNA Sensing by a Virus. PLoS Pathog. 2013, 9, e1003649. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.H.; Wong, E.B.; Rubio, D.; Roscoe, F.; Ma, X.; Nair, S.; Remakus, S.; Schwendener, R.; John, S.; Shlomchik, M.; et al. Sequential Activation of Two Pathogen-Sensing Pathways Re-quired for Type I Interferon Expression and Resistance to an Acute DNA Virus Infection. Immunity 2015, 43, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Belmonte, R.; Perez-Nunez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, J.; Wu, Y.; Chen, H.; Zhang, S.; Li, J.; Xin, T.; Jia, H.; Hou, S.; Jiang, Y.; et al. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 2018, 506, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, J.; Ni, J.; Jiang, S.; Xia, N.; Guo, Y.; Shao, Q.; Cao, Q.; Zheng, W.; Chen, N.; et al. The African swine fever virus protease pS273R inhibits DNA sensing cGAS-STING pathway by targeting IKKepsilon. Virulence 2022, 13, 740–756. [Google Scholar] [CrossRef]
- Sun, M.; Yu, S.; Ge, H.; Wang, T.; Li, Y.; Zhou, P.; Pan, L.; Han, Y.; Yang, Y.; Sun, Y.; et al. The A137R Protein of African Swine Fever Virus Inhibits Type I Interferon Production via the Autophagy-Mediated Lysosomal Degradation of TBK1. J. Virol. 2022, 96, e01957-21. [Google Scholar] [CrossRef]
- Hong, J.; Chi, X.; Yuan, X.; Wen, F.; Rai, K.R.; Wu, L.; Song, Z.; Wang, S.; Guo, G.; Chen, J.-L. I226R Protein of African Swine Fever Virus Is a Suppressor of Innate An-tiviral Responses. Viruses 2022, 14, 575. [Google Scholar] [CrossRef]
- Herzner, A.-M.; Hagmann, C.A.; Goldeck, M.; Wolter, S.; Kübler, K.; Wittmann, S.; Gramberg, T.; Andreeva, L.; Hopfner, K.-P.; Mertens, C.; et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 2015, 16, 1025–1033. [Google Scholar] [CrossRef]
- Xu, S.; Ducroux, A.; Ponnurangam, A.; Vieyres, G.; Franz, S.; Musken, M.; Zillinger, T.; Malassa, A.; Ewald, E.; Hornung, V.; et al. cGAS-Mediated Innate Immunity Spreads Intercel-lularly through HIV-1 Env-Induced Membrane Fusion Sites. Cell Host Microbe 2016, 20, 443–457. [Google Scholar] [CrossRef]
- Xu, S.; Ducroux, A.; Ponnurangam, A.; Vieyres, G.; Franz, S.; Musken, M.; Zillinger, T.; Malassa, A.; Ewald, E.; Hornung, V.; et al. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell 2018, 175, 488–501.e22. [Google Scholar]
- Sumner, R.P.; Harrison, L.; Touizer, E.; Peacock, T.P.; Spencer, M.; Zuliani-Alvarez, L.; Towers, G.J. Disrupting HIV-1 capsid formation causes cGAS sensing of viral DNA. EMBO J. 2020, 39, e103958. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Rui, Y.; Lou, M.; Yin, L.; Xiong, H.; Zhou, Z.; Shen, S.; Chen, T.; Zhang, Z.; Zhao, N.; et al. HIV-2/SIV Vpx targets a novel functional domain of STING to selectively inhibit cGAS-STING-mediated NF-kappaB signalling. Nat. Microbiol. 2019, 4, 2552–2564. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef]
- Aguirre, S.; Luthra, P.; Sanchez-Aparicio, M.T.; Maestre, A.M.; Patel, J.; Lamothe, F.; Fredericks, A.C.; Tripathi, S.; Zhu, T.; Pintado-Silva, J.; et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2017, 2, 17037. [Google Scholar] [CrossRef]
- Zevini, A.; Olagnier, D.; Hiscott, J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017, 38, 194–205. [Google Scholar] [CrossRef]
- Yi, G.; Wen, Y.; Shu, C.; Han, Q.; Konan, K.V.; Li, P.; Kao, C.C. Hepatitis C Virus NS4B Can Suppress STING Accumulation To Evade Innate Immune Responses. J. Virol. 2016, 90, 254–265. [Google Scholar] [CrossRef]
- Ding, Q.; Cao, X.; Lu, J.; Huang, B.; Liu, Y.-J.; Kato, N.; Shu, H.-B.; Zhong, J. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 2013, 59, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Sakamoto, N.; Nakagawa, M.; Kakinuma, S.; Mishima, K.; Kusano-Kitazume, A.; Kiyohashi, K.; Murakawa, M.; Nishimura-Sakurai, Y.; Azuma, S.; et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 2012, 57, 46–58. [Google Scholar] [CrossRef]
- Li, M.; Ferretti, M.; Ying, B.; Descamps, H.; Lee, E.; Dittmar, M.; Lee, J.S.; Whig, K.; Kamalia, B.; Dohnalova, L.; et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 2021, 6, eabi9007. [Google Scholar] [CrossRef]
- Liu, X.; Wei, L.; Xu, F.; Zhao, F.; Huang, Y.; Fan, Z.; Mei, S.; Hu, Y.; Zhai, L.; Guo, J.; et al. SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response. Sci. Signal. 2022, 15, eabg8744. [Google Scholar] [CrossRef] [PubMed]
- Neufeldt, C.J.; Cerikan, B.; Cortese, M.; Frankish, J.; Lee, J.Y.; Plociennikowska, A.; Heigwer, F.; Prasad, V.; Joecks, S.; Burkart, S.S.; et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-kappaB. Commun. Biol. 2022, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Su, J.; Shen, S.; Hu, Y.; Huang, D.; Zheng, W.; Lou, M.; Shi, Y.; Wang, M.; Chen, S.; et al. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal. Transduct. Target Ther. 2021, 6, 123. [Google Scholar] [CrossRef]
- Han, L.; Zhuang, M.W.; Deng, J.; Zheng, Y.; Zhang, J.; Nan, M.L.; Zhang, X.J.; Gao, C.; Wang, P.H. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J. Med. Virol. 2021, 93, 5376–5389. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef]
- Clementz, M.A.; Chen, Z.; Banach, B.S.; Wang, Y.; Sun, L.; Ratia, K.; Baez-Santos, Y.M.; Wang, J.; Takayama, J.; Ghosh, A.K.; et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010, 84, 4619–4629. [Google Scholar] [CrossRef]
- Xing, Y.; Chen, J.; Tu, J.; Zhang, B.; Chen, X.; Shi, H.; Baker, S.C.; Feng, L.; Chen, Z. The papain-like protease of porcine epidemic diarrhea virus nega-tively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013, 94 Pt 7, 1554–1567. [Google Scholar] [CrossRef]
- Hu, X.; Tian, J.; Kang, H.; Guo, D.; Liu, J.; Liu, D.; Jiang, Q.; Li, Z.; Qu, J.; Qu, L. Transmissible Gastroenteritis Virus Papain-Like Protease 1 Antagonizes Production of Interferon-beta through Its Deubiquitinase Activity. Biomed. Res. Int. 2017, 2017, 7089091. [Google Scholar] [CrossRef]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus papain-like proteases negatively regulate anti-viral innate immune response through disruption of STING-mediated signaling. PLoS ONE 2012, 7, e30802. [Google Scholar]
- Webb, L.G.; Veloz, J.; Pintado-Silva, J.; Zhu, T.; Rangel, M.V.; Mutetwa, T.; Zhang, L.; Bernal-Rubio, D.; Figueroa, D.; Carrau, L.; et al. Chikungunya virus antagonizes cGAS-STING mediated type-I interferon responses by degrading cGAS. PLoS Pathog. 2020, 16, e1008999. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Lin, T.; Yang, D.; Harrison, A.G.; Vella, A.T.; Fikrig, E.; Wang, P. A Critical Role for STING Signaling in Limiting Patho-genesis of Chikungunya Virus. J. Infect. Dis. 2021, 223, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- McGuckin Wuertz, K.; Treuting, P.M.; Hemann, E.A.; Esser-Nobis, K.; Snyder, A.G.; Graham, J.B.; Daniels, B.P.; Wilkins, C.; Snyder, J.M.; Voss, K.M.; et al. STING is required for host defense against neuropathological West Nile virus infection. PLoS Pathog. 2019, 15, e1007899. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Gaska, J.M.; Douam, F.; Wei, L.; Kim, D.; Balev, M.; Heller, B.; Ploss, A. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci USA 2018, 115, E6310–E6318. [Google Scholar] [CrossRef]
- Dalrymple, N.A.; Cimica, V.; Mackow, E.R. Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant. mBio 2015, 6, e00553-15. [Google Scholar] [CrossRef]
- Yu, P.; Miao, Z.; Li, Y.; Bansal, R.; Peppelenbosch, M.P.; Pan, Q. cGAS-STING effectively restricts murine norovirus infection but antagonizes the antiviral action of N-terminus of RIG-I in mouse macrophages. Gut Microbes 2021, 13, 1959839. [Google Scholar] [CrossRef]
- Li, S.; Qian, N.; Jiang, C.; Zu, W.; Liang, A.; Li, M.; Elledge, S.J.; Tan, X. Gain-of-function genetic screening identifies the antiviral function of TMEM120A via STING activation. Nat. Commun. 2022, 13, 105. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Klionsky, D.J. Inflammatory-dependent Sting activation induces antiviral autophagy to limit zika virus in the Drosophila brain. Autophagy 2018, 15, 1–3. [Google Scholar] [CrossRef]
- Liu, Y.; Cherry, S. Zika virus infection activates sting-dependent antiviral autophagy in the Drosophila brain. Autophagy 2018, 15, 174–175. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, Q.; Wu, Y.; Ma, L.; Zhang, Z.; Liu, T.; Jin, S.; She, Y.; Li, Y.P.; Cui, J. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J. 2018, 37, e99347. [Google Scholar] [CrossRef]
- Iampietro, M.; Dumont, C.; Mathieu, C.; Spanier, J.; Robert, J.; Charpenay, A.; Dupichaud, S.; Dhondt, K.P.; Aurine, N.; Pelissier, R.; et al. Activation of cGAS/STING pathway upon paramyxovirus infection. iScience 2021, 24, 102519. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Hoshi, M.; Ikeda, F.; Fujiyuki, T.; Yoneda, M.; Kai, C. Downregulation of mitochondrial biogenesis by virus infection triggers antiviral responses by cyclic GMP-AMP synthase. PLoS Pathog. 2021, 17, e1009841. [Google Scholar] [CrossRef] [PubMed]
- Franz, K.M.; Neidermyer, W.J.; Tan, Y.-J.; Whelan, S.P.J.; Kagan, J.C. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl. Acad. Sci. USA 2018, 115, E2058–E2067. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, J.; Zhu, Y.; Ji, X.; Wang, K.; Jiang, S.; Luo, J.; Wang, H.; Zheng, W.; Chen, N.; et al. Chicken DNA Sensing cGAS-STING Signal Pathway Mediates Broad Spectrum Antiviral Functions. Vaccines 2020, 8, 369. [Google Scholar] [CrossRef]
- Cui, S.; Yu, Q.; Chu, L.; Cui, Y.; Ding, M.; Wang, Q.; Wang, H.; Chen, Y.; Liu, X.; Wang, C. Nuclear cGAS Functions Non-canonically to Enhance Antiviral Im-munity via Recruiting Methyltransferase Prmt5. Cell Rep. 2020, 33, 108490. [Google Scholar] [CrossRef]
- Holm, C.K.; Rahbek, S.H.; Gad, H.H.; Bak, R.O.; Jakobsen, M.R.; Jiang, Z.; Hansen, A.L.; Jensen, S.K.; Sun, C.; Thomsen, M.K.; et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 2016, 7, 10680. [Google Scholar] [CrossRef]
- Rodríguez-García, E.; Olagüe, C.; Ríus-Rocabert, S.; Ferrero, R.; Llorens, C.; Larrea, E.; Fortes, P.; Prieto, J.; González-Aseguinolaza, G.; Nistal-Villan, E. TMEM173 Alternative Spliced Isoforms Modulate Viral Replication through the STING Pathway. Immunohorizons 2018, 2, 363–376. [Google Scholar] [CrossRef]
- Moriyama, M.; Koshiba, T.; Ichinohe, T. Influenza A virus M2 protein triggers mitochondrial DNA-mediated antiviral immune responses. Nat. Commun. 2019, 10, 4624. [Google Scholar] [CrossRef]
- Seo, G.J.; Kim, C.; Shin, W.-J.; Sklan, E.H.; Eoh, H.; Jung, J.U. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 2018, 9, 613. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Y.; Sun, S.; Luo, J.; Jiang, S.; Zhang, J.; Liu, X.; Shao, Q.; Cao, Q.; Zheng, W.; et al. The Innate Immune DNA Sensing cGAS-STING Signaling Pathway Mediates Anti-PRRSV Function. Viruses 2021, 13, 1829. [Google Scholar] [CrossRef]
- Liu, B.-Y.; Yu, X.-J.; Zhou, C.-M. SAFA initiates innate immunity against cytoplasmic RNA virus SFTSV infection. PLoS Pathog. 2021, 17, e1010070. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, X.; Luo, W.; Xiao, J.; Song, X.; Wang, Y.; Shuai, H.; Ren, Z.; Wang, Y. Roles of Emerging RNA-Binding Activity of cGAS in Innate Anti-viral Response. Front. Immunol. 2021, 12, 741599. [Google Scholar] [CrossRef] [PubMed]
- Holm, C.; Jensen, S.B.; Jakobsen, M.R.; Cheshenko, N.; Horan, A.K.; Moeller, H.B.; Gonzalez-Dosal, R.; Rasmussen, S.B.; Christensen, M.H.; Yarovinsky, O.T.; et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 2012, 13, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Triantafilou, M.; Ramanjulu, J.; Booty, L.M.; Jimenez-Duran, G.; Keles, H.; Saunders, K.; Nevins, N.; Koppe, E.; Modis, L.K.; Pesiridis, G.S.; et al. Human rhinovirus promotes STING trafficking to replication organelles to promote viral replication. Nat. Commun. 2022, 13, 1406. [Google Scholar] [CrossRef]
- Jenson, J.; Chen, Z.J. Bacteria sting viral invaders. Nature 2020, 586, 363–364. [Google Scholar] [CrossRef]
- Guimarães, E.S.; Marinho, F.V.; de Queiroz, N.M.G.P.; Antunes, M.M.; Oliveira, S.C. Impact of STING Inflammatory Signaling during Intracellular Bacterial Infections. Cells 2021, 11, 74. [Google Scholar] [CrossRef]
- Liu, N.; Pang, X.; Zhang, H.; Ji, P. The cGAS-STING Pathway in Bacterial Infection and Bacterial Immunity. Front. Immunol. 2021, 12, 814709. [Google Scholar] [CrossRef]
- Witte, C.E.; Archer, K.A.; Rae, C.S.; Sauer, J.-D.; Woodward, J.J.; Portnoy, D.A. Innate Immune Pathways Triggered by Listeria monocytogenes and Their Role in the Induction of Cell-Mediated Immunity. Adv. Immunol. 2012, 113, 135–156. [Google Scholar] [CrossRef]
- Andrade, W.A.; Agarwal, S.; Mo, S.; Shaffer, S.A.; Dillard, J.P.; Schmidt, T.; Hornung, V.; Fitzgerald, K.A.; Kurt-Jones, E.A.; Golenbock, D.T. Type I Interferon Induction by Neisseria gonor-rhoeae: Dual Requirement of Cyclic GMP-AMP Synthase and Toll-like Receptor 4. Cell Rep. 2016, 15, 2438–2448. [Google Scholar] [CrossRef]
- Marinho, F.V.; Benmerzoug, S.; Oliveira, S.C.; Ryffel, B.; Quesniaux, V. The Emerging Roles of STING in Bacterial Infections. Trends Microbiol. 2017, 25, 906–918. [Google Scholar] [CrossRef]
- Wein, T.; Sorek, R. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat. Rev. Immunol. 2022, 22, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Solimini, N.L.; Elledge, S.J. Principles of Cancer Therapy: Oncogene and Non-oncogene Addiction. Cell 2009, 136, 823–837. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Khoury-Hanold, W.; Staron, M.; Tal, M.C.; Pineda, C.M.; Lang, S.M.; Bestwick, M.; Duguay, B.A.; Raimundo, N.; MacDuff, D.A.; et al. Mitochondrial DNA stress primes the anti-viral innate immune response. Nature 2015, 520, 553–557. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage ena-bles pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, K.J.; Carroll, P.; Martin, C.-A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef]
- Gentili, M.; Kowal, J.; Tkach, M.; Satoh, T.; Lahaye, X.; Conrad, C.; Boyron, M.; Lombard, B.; Durand, S.; Kroemer, G.; et al. Transmission of innate immune signaling by packag-ing of cGAMP in viral particles. Science 2015, 349, 1232–1236. [Google Scholar] [CrossRef]
- Demaria, O.; De Gassart, A.; Coso, S.; Gestermann, N.; Di Domizio, J.; Flatz, L.; Gaide, O.; Michielin, O.; Hwu, P.; Petrova, T.V.; et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 15408–15413. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Wu, X.; Ma, D.; Wu, J.; Wang, L.; Jiang, Y.; Fei, Y.; Zhu, C.; Tan, R.; et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018, 563, 131–136. [Google Scholar] [CrossRef]
- Woo, S.R.; Fuertes, M.B.; Corrales, L.; Spranger, S.; Furdyna, M.J.; Leung, M.Y.; Duggan, R.; Wang, Y.; Barber, G.N.; Fitzgerald, K.A.; et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014, 41, 830–842. [Google Scholar] [CrossRef]
- Deng, L.; Liang, H.; Xu, M.; Yang, X.; Burnette, B.; Arina, A.; Li, X.D.; Mauceri, H.; Beckett, M.; Darga, T.; et al. STING-Dependent Cytosolic DNA Sensing Promotes Radia-tion-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 2014, 41, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; McAllister, D.; Vonderhaar, E.P.; Palen, K.; Riese, M.J.; Gershan, J.; Johnson, B.D.; Dwinell, M.B. STING agonist inflames the pancreatic cancer immune microenvironment and reduces tumor burden in mouse models. J. Immunother. Cancer 2019, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Bacher, N.; Raker, V.; Hofmann, C.; Graulich, E.; Schwenk, M.; Baumgrass, R.; Bopp, T.; Zechner, U.; Merten, L.; Becker, C.; et al. Interferon-alpha suppresses cAMP to dis-arm human regulatory T cells. Cancer Res. 2013, 73, 5647–5656. [Google Scholar] [CrossRef]
- Cunha, L.D.; Yang, M.; Carter, R.; Guy, C.; Harris, L.; Crawford, J.C.; Quarato, G.; Boada-Romero, E.; Kalkavan, H.; Johnson, M.D.L.; et al. LC3-Associated Phagocytosis in Myeloid Cells Pro-motes Tumor Immune Tolerance. Cell 2018, 175, 429–441.e16. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Qi, J.; Zhao, Q.; Wu, Q.-N.; Wei, D.-L.; Wei, X.-L.; Liu, J.; Chen, J.; Zeng, Z.-L.; Ju, H.-Q.; et al. Targeting the STING pathway in tumor-associated macrophages regulates innate immune sensing of gastric cancer cells. Theranostics 2020, 10, 498–515. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.; Mao, A.J.; Lensink-Vasan, M.; Wang, L.; Vance, R.E.; Raulet, D.H. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 2018, 49, 754–763.e4. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.; Ghosh, K.; Vizioli, M.G.; Zhu, J.; Sen, P.; Wangensteen, K.J.; Simithy, J.; Lan, Y.; Lin, Y.; Zhou, Z.; et al. Cytoplasmic chromatin triggers inflammation in se-nescence and cancer. Nature 2017, 550, 402–406. [Google Scholar] [CrossRef]
- Glück, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.-W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017, 19, 1061–1070. [Google Scholar] [CrossRef]
- Leon, K.E.; Buj, R.; Lesko, E.; Dahl, E.S.; Chen, C.-W.; Tangudu, N.K.; Imamura-Kawasawa, Y.; Kossenkov, A.V.; Hobbs, R.P.; Aird, K.M. DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. J. Cell Biol. 2021, 220, e202008101. [Google Scholar] [CrossRef] [PubMed]
- Gulen, M.F.; Koch, U.; Haag, S.M.; Schuler, F.; Apetoh, L.; Villunger, A.; Radtke, F.; Ablasser, A. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 2017, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Tong, J.; Yang, L.; Wei, L.; Stolz, D.B.; Yu, J.; Zhang, J.; Zhang, L. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc. Natl. Acad. Sci. USA 2018, 115, 3930–3935. [Google Scholar] [CrossRef] [PubMed]
- Petrasek, J.; Iracheta-Vellve, A.; Csak, T.; Satishchandran, A.; Kodys, K.; Kurt-Jones, E.A.; Fitzgerald, K.A.; Szabo, G. STING-IRF3 pathway links en-doplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl. Acad. Sci. USA 2013, 110, 16544–16549. [Google Scholar] [CrossRef] [PubMed]
- Zierhut, C.; Yamaguchi, N.; Paredes, M.; Luo, J.D.; Carroll, T.; Funabiki, H. The Cytoplasmic DNA Sensor cGAS Promotes Mi-totic Cell Death. Cell 2019, 178, 302–315.e23. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Liu, P.; Zhao, F.; Liu, J.; Hong, Z.; Ren, H.; Gu, G.; Wang, G.; Wu, X.; Zheng, T.; et al. STING-mediated Syk Signaling Attenuates Tumorigenesis of Coli-tisassociated Colorectal Cancer Through Enhancing Intestinal Epithelium Pyroptosis. Inflamm. Bowel Dis. 2022, 28, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Nassour, J.; Radford, R.; Correia, A.; Fuste, J.M.; Schoell, B.; Jauch, A.; Shaw, R.J.; Karlseder, J. Autophagic cell death restricts chromosomal insta-bility during replicative crisis. Nature 2019, 565, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Sandoval, T.A.; Chae, C.S.; Chopra, S.; Tan, C.; Rutkowski, M.R.; Raundhal, M.; Chaurio, R.A.; Payne, K.K.; Konrad, C.; et al. IRE1alpha-XBP1 controls T cell function in ovar-ian cancer by regulating mitochondrial activity. Nature 2018, 562, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Bhatelia, K.; Singh, K.; Prajapati, P.; Sripada, L.; Roy, M.; Singh, R. MITA modulated autophagy flux promotes cell death in breast cancer cells. Cell. Signal. 2017, 35, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, Y.-J.; Dobbs, N.; Sakai, T.; Liou, J.; Miner, J.J.; Yan, N. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 2019, 216, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, N.; Neven, B.; Gentili, M.; Callebaut, I.; Maschalidi, S.; Stolzenberg, M.-C.; Goudin, N.; Frémond, M.-L.; Nitschke, P.; Molina, T.; et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Investig. 2014, 124, 5516–5520. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Barber, G.N. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr. Opin. Immunol. 2014, 31, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Sliter, D.A.; Martinez, J.; Hao, L.; Chen, X.; Sun, N.; Fischer, T.D.; Burman, J.L.; Li, Y.; Zhang, Z.; Narendra, D.P.; et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 2018, 561, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Lee-Kirsch, M.A.; Gong, M.; Chowdhury, D.; Senenko, L.; Engel, K.; Lee, Y.-A.; De Silva, U.; Bailey, S.L.; Witte, T.; Vyse, T.J.; et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007, 39, 1065–1067. [Google Scholar] [CrossRef] [PubMed]
- Crow, Y.J.; Hayward, B.E.; Parmar, R.; Robins, P.; Leitch, A.; Ali, M.; Black, D.N.; van Bokhoven, H.; Brunner, H.G.; Hamel, B.C.; et al. Mutations in the gene encoding the 3′-5′ DNA exonu-clease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006, 38, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Stamp, G.; Robins, P.; Dulic, A.; Rosewell, I.; Hrivnak, G.; Daly, G.; Lindahl, T.; Barnes, D.E. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell Biol. 2004, 24, 6719–6727. [Google Scholar] [CrossRef]
- Gao, D.; Li, T.; Li, X.-D.; Chen, X.; Li, Q.-Z.; Wight-Carter, M.; Chen, Z.J. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 2015, 112, E5699–E5705. [Google Scholar] [CrossRef]
- Yoshida, H.; Okabe, Y.; Kawane, K.; Fukuyama, H.; Nagata, S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 2005, 6, 49–56. [Google Scholar] [CrossRef]
- Saldanha, R.G.; Balka, K.R.; Davidson, S.; Wainstein, B.K.; Wong, M.; MacIntosh, R.; Loo, C.K.C.; Weber, M.A.; Kamath, V.; Moghaddas, F.; et al. A Mutation Outside the Dimerization Domain Causing Atypical STING-Associated Vasculopathy With Onset in Infancy. Front. Immunol. 2018, 9, 1535. [Google Scholar] [CrossRef]
- Liu, Y.; Jesus, A.A.; Marrero, B.; Yang, D.; Ramsey, S.E.; Montealegre Sanchez, G.A.; Tenbrock, K.; Wittkowski, H.; Jones, O.Y.; Kuehn, H.S.; et al. Activated STING in a Vascular and Pulmonary Syndrome. N. Engl. J. Med. 2014, 371, 507–518. [Google Scholar] [CrossRef]
- King, K.R.; Aguirre, A.D.; Ye, Y.X.; Sun, Y.; Roh, J.D.; Ng, R.P., Jr.; Kohler, R.H.; Arlauckas, S.P.; Iwamoto, Y.; Savol, A.; et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 2017, 23, 1481–1487. [Google Scholar] [CrossRef] [PubMed]
- Luther, J.; Khan, S.; Gala, M.K.; Kedrin, D.; Sridharan, G.; Goodman, R.P.; Garber, J.J.; Masia, R.; Diagacomo, E.; Adams, D.; et al. Hepatic gap junctions amplify alcohol liver inju-ry by propagating cGAS-mediated IRF3 activation. Proc. Natl. Acad. Sci. USA 2020, 117, 11667–11673. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.J.; De Nardo, D.; Moghaddas, F.; Tran, L.S.; Bachem, A.; Nguyen, T.; Hayman, T.; Tye, H.; Vince, J.E.; Bedoui, S.; et al. Posttranslational Modification as a Critical Determinant of Cytoplasmic Innate Immune Recognition. Physiol. Rev. 2017, 97, 1165–1209. [Google Scholar] [CrossRef] [PubMed]
- Proud, C.G. Phosphorylation and Signal Transduction Pathways in Translational Control. Cold Spring Harb. Perspect. Biol. 2018, 11, a033050. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.J.; Yang, A.; Tan, B.; Kim, S.; Liang, Q.; Choi, Y.; Yuan, W.; Feng, P.; Park, H.-S.; Jung, J.U. Akt Kinase-Mediated Checkpoint of cGAS DNA Sensing Pathway. Cell Rep. 2015, 13, 440–449. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, M.M.; Bian, L.J.; Liu, Y.; Chen, Q.; Shu, H.B. Phosphorylation of cGAS by CDK1 impairs self-DNA sensing in mitosis. Cell Discov. 2020, 6, 26. [Google Scholar] [CrossRef]
- Li, T.; Huang, T.; Du, M.; Chen, X.; Du, F.; Ren, J.; Chen, Z.J. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science 2021, 371, eabc5386. [Google Scholar] [CrossRef]
- Li, M.; Shu, H.B. Dephosphorylation of cGAS by PPP6C impairs its substrate binding activity and innate antiviral response. Protein Cell 2020, 11, 584–599. [Google Scholar] [CrossRef]
- Konno, H.; Konno, K.; Barber, G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sus-tained innate immune signaling. Cell 2013, 155, 688–698. [Google Scholar] [CrossRef]
- Li, Z.; Liu, G.; Sun, L.; Teng, Y.; Guo, X.; Jia, J.; Sha, J.; Yang, X.; Chen, D.; Sun, Q. PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation. PLoS Pathog. 2015, 11, e1004783. [Google Scholar] [CrossRef]
- Wang, F.; Alain, T.; Szretter, K.; Stephenson, K.; Pol, J.G.; Atherton, M.J.; Hoang, H.-D.; Fonseca, B.D.; Zakaria, C.; Chen, L.; et al. S6K-STING interaction regulates cytosolic DNA–mediated activation of the transcription factor IRF3. Nat. Immunol. 2016, 17, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Yi, X.M.; Wu, X.; Shang, J.; Shu, H.B. PTPN1/2-mediated dephosphorylation of MITA/STING promotes its 20S proteasomal degradation and attenuates innate antiviral response. Proc. Natl. Acad. Sci. USA 2019, 116, 20063–20069. [Google Scholar] [CrossRef] [PubMed]
- An, T.; Li, S.; Pan, W.; Tien, P.; Zhong, B.; Shu, H.B.; Wu, S. DYRK2 Negatively Regulates Type I Interferon Induction by Promoting TBK1 Degradation via Ser527 Phosphorylation. PLoS Pathog. 2015, 11, e1005179. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liang, L.; Fan, Y.; Sun, S.; An, L.; Shi, Z.; Cheng, J.; Jia, W.; Sun, W.; Mori-Akiyama, Y.; et al. PPM1B negatively regulates antiviral response via dephosphorylating TBK1. Cell Signal. 2012, 24, 2197–2204. [Google Scholar] [CrossRef]
- Karpova, A.Y.; Trost, M.; Murray, J.M.; Cantley, L.C.; Howley, P.M. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 2002, 99, 2818–2823. [Google Scholar] [CrossRef]
- Li, S.; Zhu, M.; Pan, R.; Fang, T.; Cao, Y.Y.; Chen, S.; Zhao, X.; Lei, C.Q.; Guo, L.; Chen, Y.; et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 2016, 17, 241–249. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, M.; Xue, Y.; Li, Z.; Wen, W.; Liu, X.; Ma, Y.; Zhang, L.; Shen, Z.; Cao, X. Interferon-inducible cytoplasmic lncLrrc55-AS promotes antiviral innate responses by strengthening IRF3 phosphorylation. Cell Res. 2019, 29, 641–654. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, Z.J. The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 2011, 12, 35–48. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, L.; Hong, Z.; Lv, Z.; Mao, Z.; Tang, Y.; Kong, X.; Li, S.; Cui, Y.; Liu, H.; et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017, 13, e1006264. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y. Ubiquitination of cGAS by TRAF6 regulates anti-DNA viral innate immune responses. Biochem. Biophys. Res. Commun. 2019, 514, 659–664. [Google Scholar] [CrossRef]
- Liu, Z.-S.; Zhang, Z.-Y.; Cai, H.; Zhao, M.; Mao, J.; Dai, J.; Xia, T.; Zhang, X.-M.; Li, T. RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci. 2018, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Meng, Q.; Qin, Y.; Liang, P.; Tan, P.; He, L.; Zhou, Y.; Chen, Y.; Huang, J.; Wang, R.F.; et al. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autoph-agy Receptor p62 to Promote Innate Immune Responses. Mol. Cell 2016, 64, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jiang, F.; Kong, L.; Li, B.; Yang, Y.; Zhang, L.; Liu, B.; Zheng, Y.; Gao, C. Cutting Edge: USP27X Deubiquitinates and Stabilizes the DNA Sensor cGAS to Regulate Cytosolic DNA-Mediated Signaling. J. Immunol. 2019, 203, 2049–2054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, Z.; An, R.; Ye, L.; Zhong, B. USP29 maintains the stability of cGAS and promotes cellular antiviral responses and autoimmunity. Cell Res. 2020, 30, 914–927. [Google Scholar] [CrossRef]
- Tsuchida, T.; Zou, J.; Saitoh, T.; Kumar, H.; Abe, T.; Matsuura, Y.; Kawai, T.; Akira, S. The Ubiquitin Ligase TRIM56 Regulates Innate Immune Responses to Intracellular Double-Stranded DNA. Immunity 2010, 33, 765–776. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, M.M.; Wang, Y.Y.; Shu, H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012, 287, 28646–28655. [Google Scholar] [CrossRef]
- Ni, G.; Konno, H.; Barber, G.N. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci. Immunol. 2017, 2, eaah7119. [Google Scholar] [CrossRef]
- Zhang, Z.-D.; Xiong, T.-C.; Yao, S.-Q.; Wei, M.-C.; Chen, M.; Lin, D.; Zhong, B. RNF115 plays dual roles in innate antiviral responses by catalyzing distinct ubiquitination of MAVS and MITA. Nat. Commun. 2020, 11, 5536. [Google Scholar] [CrossRef]
- Tian, M.; Liu, W.; Zhang, Q.; Huang, Y.; Li, W.; Wang, W.; Zhao, P.; Huang, S.; Song, Y.; Shereen, M.A.; et al. MYSM1 Represses Innate Immunity and Autoimmunity through Suppressing the cGAS-STING Pathway. Cell Rep. 2020, 33, 108297. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, X.; Cui, Y.; Tang, Y.; Chen, W.; Li, S.; Yu, H.; Pan, Y.; Wang, C. The E3 Ubiquitin Ligase AMFR and INSIG1 Bridge the Activation of TBK1 Kinase by Modifying the Adaptor STING. Immunity 2014, 41, 919–933. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, Q.; Jing, Y.Y.; Zhang, M.; Wang, H.Y.; Cai, Z.; Liuyu, T.; Zhang, Z.D.; Xiong, T.C.; Wu, Y.; et al. USP13 negatively regulates antiviral responses by deubiqui-tinating STING. Nat. Commun. 2017, 8, 15534. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, L.; Jin, J.; Luan, Y.; Chen, C.; Li, Y.; Chu, H.; Wang, X.; Liao, G.; Yu, Y.; et al. p38 inhibition provides anti–DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J. Exp. Med. 2017, 214, 991–1010. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, A.; Zhang, H.; Wang, J.; Li, X.C.; Zeng, M.-S.; Zhang, Z. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Commun. 2017, 8, 945. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lian, Q.; Yang, B.; Yan, S.; Zhou, H.; He, L.; Lin, G.; Lian, Z.; Jiang, Z.; Sun, B. TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015, 11, e1005012. [Google Scholar] [CrossRef]
- Shi, M.; Deng, W.; Bi, E.; Mao, K.; Ji, Y.; Lin, G.; Wu, X.; Tao, Z.; Li, Z.; Cai, X.; et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 2008, 9, 369–377. [Google Scholar] [CrossRef]
- Zhong, B.; Zhang, L.; Lei, C.; Li, Y.; Mao, A.P.; Yang, Y.; Wang, Y.Y.; Zhang, X.L.; Shu, H.B. The ubiquitin ligase RNF5 regulates antiviral responses by medi-ating degradation of the adaptor protein MITA. Immunity 2009, 30, 397–407. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Y.; Cui, Y.; Song, D.; Zhang, G.; Ma, S.; Liu, Y.; Chen, M.; Chen, F.; Wang, H.; et al. RNF90 negatively regulates cellular antiviral responses by targeting MITA for degradation. PLoS Pathog. 2020, 16, e1008387. [Google Scholar] [CrossRef]
- Qin, Y.; Zhou, M.-T.; Hu, M.-M.; Hu, Y.-H.; Zhang, J.; Guo, L.; Zhong, B.; Shu, H.-B. RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms. PLoS Pathog. 2014, 10, e1004358. [Google Scholar] [CrossRef]
- Luo, W.W.; Li, S.; Li, C.; Lian, H.; Yang, Q.; Zhong, B.; Shu, H.B. iRhom2 is essential for innate immunity to DNA viruses by medi-ating trafficking and stability of the adaptor STING. Nat. Immunol. 2016, 17, 1057–1066. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, M.X.; Zhang, Q.; Zhu, G.F.; Yuan, L.; Zhang, D.E.; Zhu, Q.; Yao, J.; Shu, H.B.; Zhong, B. USP18 recruits USP20 to promote innate antiviral re-sponse through deubiquitinating STING/MITA. Cell Res. 2016, 26, 1302–1319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, N.; Cui, Y.; Hong, Z.; Liu, X.; Wang, Q.; Li, S.; Liu, H.; Yu, H.; Cai, Y.; et al. The deubiquitinase CYLD is a specific checkpoint of the STING antiviral signaling pathway. PLoS Pathog. 2018, 14, e1007435. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, S.; Liu, L.; Wang, H.; Yang, B. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res. 2017, 232, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.K.; Nandakumar, R.; Stadler, D.; Malo, A.; Valls, R.M.; Wang, F.; Reinert, L.S.; Dagnaes-Hansen, F.; Hollensen, A.K.; Mikkelsen, J.G.; et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 2016, 64, 746–759. [Google Scholar] [CrossRef]
- Takahashi, M.; Lio, C.J.; Campeau, A.; Steger, M.; Ay, F.; Mann, M.; Gonzalez, D.J.; Jain, M.; Sharma, S. The tumor suppressor kinase DAPK3 drives tumor-intrinsic immunity through the STING-IFN-beta pathway. Nat. Immunol. 2021, 22, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Xiong, Q.; Wang, N.; Chen, R.; Ren, H.; Siwko, S.; Han, H.; Liu, M.; Qian, M.; Du, B. Kisspeptin/GPR54 signaling restricts antiviral innate immune response through regulating calcineurin phosphatase activity. Sci. Adv. 2018, 4, eaas9784. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; Zhao, X.; Zhao, K.; Meng, H.; Zhao, W.; Gao, C. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J. Exp. Med. 2012, 209, 1703–1711. [Google Scholar] [CrossRef]
- Zhang, M.; Tian, Y.; Wang, R.-P.; Gao, D.; Zhang, Y.; Diao, F.-C.; Chen, D.-Y.; Zhai, Z.-H.; Shu, H.-B. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008, 18, 1096–1104. [Google Scholar] [CrossRef]
- Yu, Y.; Hayward, G.S. The Ubiquitin E3 Ligase RAUL Negatively Regulates Type I Interferon through Ubiquitination of the Transcription Factors IRF7 and IRF3. Immunity 2010, 33, 863–877. [Google Scholar] [CrossRef]
- Oke, V.; Wahren-Herlenius, M. The immunobiology of Ro52 (TRIM21) in autoimmunity: A critical review. J. Autoimmun. 2012, 39, 77–82. [Google Scholar] [CrossRef]
- Saitoh, T.; Tun-Kyi, A.; Ryo, A.; Yamamoto, M.; Finn, G.; Fujita, T.; Akira, S.; Yamamoto, N.; Lu, K.P.; Yamaoka, S. Negative regulation of interferon-regulatory factor 3–dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 2006, 7, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X. SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes. Mol. Cell 2018, 71, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.-M.; Yang, Q.; Xie, X.-Q.; Liao, C.-Y.; Lin, H.; Liu, T.-T.; Yin, L.; Shu, H.-B. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016, 45, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhong, B. Regulation of Cellular Antiviral Signaling by Modifications of Ubiquitin and Ubiquitin-like Molecules. Immune Netw. 2018, 18, e4. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Liu, T.T.; Zhou, Q.; Li, S.; Mao, A.P.; Li, Y.; Liu, L.J.; Cheng, J.K.; Shu, H.B. SENP2 negatively regulates cellular antiviral response by deSUMOy-lating IRF3 and conditioning it for ubiquitination and degradation. J. Mol. Cell Biol. 2011, 3, 283–292. [Google Scholar] [CrossRef]
- Cui, Y.; Yu, H.; Zheng, X.; Peng, R.; Wang, Q.; Zhou, Y.; Wang, R.; Wang, J.; Qu, B.; Shen, N.; et al. SENP7 Potentiates cGAS Activation by Relieving SUMO-Mediated Inhibition of Cytosolic DNA Sensing. PLoS Pathog. 2017, 13, e1006156. [Google Scholar] [CrossRef]
- Saul, V.V.; Niedenthal, R.; Pich, A.; Weber, F.; Schmitz, M.L. SUMO modification of TBK1 at the adaptor-binding C-terminal coiled-coil domain contributes to its antiviral activity. Biochim. Biophys. Acta 2015, 1853, 136–143. [Google Scholar] [CrossRef]
- Ma, T.; Chen, Y.; Zhang, F.; Yang, C.Y.; Wang, S.; Yu, X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell. 2013, 49, 897–907. [Google Scholar] [CrossRef]
- Li, C.; Zhang, L.; Qian, D.; Cheng, M.; Hu, H.; Hong, Z.; Cui, Y.; Yu, H.; Wang, Q.; Zhu, J.; et al. RNF111-facilitated neddylation potentiates cGAS-mediated anti-viral innate immune response. PLoS Pathog. 2021, 17, e1009401. [Google Scholar]
- Song, Z.-M.; Lin, H.; Yi, X.-M.; Guo, W.; Hu, M.-M.; Shu, H.-B. KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc. Natl. Acad. Sci. USA 2020, 117, 21568–21575. [Google Scholar] [CrossRef]
- Song, B.; Greco, T.M.; Lum, K.K.; Taber, C.E.; Cristea, I.M. The DNA Sensor cGAS is Decorated by Acetylation and Phosphoryla-tion Modifications in the Context of Immune Signaling. Mol. Cell. Proteom. 2020, 19, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Huang, Y.-J.; He, X.; Zhao, M.; Wang, X.; Liu, Z.-S.; Xue, W.; Cai, H.; Zhan, X.-Y.; Huang, S.-Y.; et al. Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell 2019, 176, 1447–1460.e14. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, X.; Wei, Y.; Sun, Y.; Yang, Y.; Zhang, X.; Gao, Z.; Liu, J.; Zhuang, Q. A preliminary study of KAT2A on cGAS-related immunity in in-flammation amplification of systemic lupus erythematosus. Cell Death Dis. 2021, 12, 1036. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Cheng, J.; Kong, X.; Li, S.; Li, X.; Zhang, M.; Zhang, H.; Yang, T.; Dong, Y.; Li, J.; et al. HDAC3 inhibition ameliorates ischemia/reperfusion-induced brain injury by regulating the microglial cGAS-STING pathway. Theranostics 2020, 10, 9644–9662. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Q.; Ding, Y.; Liu, Y.; Zhao, D.; Zhao, K.; Shen, Q.; Liu, X.; Zhu, X.; Li, N.; et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 2016, 17, 806–815. [Google Scholar] [CrossRef]
- Ma, D.; Yang, M.; Wang, Q.; Sun, C.; Shi, H.; Jing, W.; Bi, Y.; Shen, X.; Ma, X.; Qin, Z.; et al. Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Sci. Adv. 2021, 7, eabc1834. [Google Scholar] [CrossRef]
- Garnham, C.P.; Vemu, A.; Wilson-Kubalek, E.M.; Yu, I.; Szyk, A.; Lander, G.C.; Milligan, R.A.; Roll-Mecak, A. Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases. Cell 2015, 161, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Ye, B.; Wang, S.; Zhu, X.; Du, Y.; Xiong, Z.; Tian, Y.; Fan, Z. Glutamylation of the DNA sensor cGAS regulates its binding and syn-thase activity in antiviral immunity. Nat. Immunol. 2016, 17, 369–378. [Google Scholar] [CrossRef]
- Robinson, N.E. Protein deamidation. Proc. Natl. Acad. Sci. USA 2002, 99, 5283–5288. [Google Scholar] [CrossRef]
- Salaun, C.; Greaves, J.; Chamberlain, L.H. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 2010, 191, 1229–1238. [Google Scholar] [CrossRef]
- Haag, S.M.; Gulen, M.F.; Reymond, L.; Gibelin, A.; Abrami, L.; Decout, A.; Heymann, M.; Van Der Goot, F.G.; Turcatti, G.; Behrendt, R.; et al. Targeting STING with covalent small-molecule inhibitors. Nature 2018, 559, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Mukai, K.; Konno, H.; Akiba, T.; Uemura, T.; Waguri, T.U.S.; Kobayashi, T.; Barber, H.K.G.N.; Arai, H.; Taguchi, K.M.T.A.H.A.T. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 2016, 7, 11932. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Lemoff, A.; Wang, G.; Zarek, C.; Lowe, A.; Yan, N.; Reese, T.A. Reactive oxygen species oxidize STING and suppress interferon production. Elife 2020, 9, e57837. [Google Scholar] [CrossRef]
- Jin, L.; Lenz, L.L.; Cambier, J.C. Cellular reactive oxygen species inhibit MPYS induction of IFNbeta. PLoS ONE. 2010, 5, e15142. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Qin, D.; Zhao, C.; Chai, L.; Yu, Z.; Wang, W.; Tong, L.; Lv, L.; Wang, Y.; Rehwinkel, J.; et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 2020, 21, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Dhani, S.; Zhao, Y.; Zhivotovsky, B. A long way to go: Caspase inhibitors in clinical use. Cell Death Dis. 2021, 12, 949. [Google Scholar] [CrossRef]
- Wang, Y.; Ning, X.; Gao, P.; Wu, S.; Sha, M.; Lv, M.; Zhou, X.; Gao, J.; Fang, R.; Meng, G.; et al. Inflammasome Activation Triggers Caspase-1-Mediated Cleavage of cGAS to Regulate Responses to DNA Virus Infection. Immunity 2017, 46, 393–404. [Google Scholar] [CrossRef]
- Ning, X.; Wang, Y.; Jing, M.; Sha, M.; Lv, M.; Gao, P.; Zhang, R.; Huang, X.; Feng, J.-M.; Jiang, Z. Apoptotic Caspases Suppress Type I Interferon Production via the Cleavage of cGAS, MAVS, and IRF3. Mol. Cell 2019, 74, 19–31.e7. [Google Scholar] [CrossRef]
- Rongvaux, A.; Jackson, R.; Harman, C.C.; Li, T.; West, A.P.; de Zoete, M.R.; Wu, Y.; Yordy, B.; Lakhani, S.A.; Kuan, C.-Y.; et al. Apoptotic Caspases Prevent the Induction of Type I Interferons by Mitochondrial DNA. Cell 2014, 159, 1563–1577. [Google Scholar] [CrossRef]
- White, M.J.; McArthur, K.; Metcalf, D.; Lane, R.M.; Cambier, J.C.; Herold, M.J.; van Delft, M.F.; Bedoui, S.; Lessene, G.; Ritchie, M.E.; et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 2014, 159, 1549–1562. [Google Scholar] [CrossRef]
- Wang, S.; Osgood, A.O.; Chatterjee, A. Uncovering post-translational modification-associated protein–protein interactions. Curr. Opin. Struct. Biol. 2022, 74, 102352. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-S.; Cai, H.; Xue, W.; Wang, M.; Xia, T.; Li, W.-J.; Xing, J.-Q.; Zhao, M.; Huang, Y.-J.; Chen, S.; et al. G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol. 2018, 20, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Yoh, S.M.; Schneider, M.; Seifried, J.; Soonthornvacharin, S.; Akleh, R.E.; Olivieri, K.C.; De Jesus, P.D.; Ruan, C.; de Castro, E.; Ruiz, P.A.; et al. PQBP1 Is a Proximal Sensor of the cGAS-Dependent Innate Response to HIV-1. Cell 2015, 161, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Wei, J.; Zang, R.; Ye, W.; Yang, Q.; Zhang, X.-N.; Chen, Y.-D.; Fu, Y.-Z.; Hu, M.-M.; Lei, C.-Q.; et al. ZCCHC3 is a co-sensor of cGAS for dsDNA recognition in innate immune response. Nat. Commun. 2018, 9, 3349. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Li, L.; Qian, G.; Wang, Y.; Chen, Z.; Liu, J.; Fang, C.; Huang, F.; Guo, D.; et al. beta-arrestin 2 as an activator of cGAS-STING signaling and target of viral immune evasion. Nat. Commun. 2020, 11, 6000. [Google Scholar] [CrossRef]
- Liang, Q.; Seo, G.J.; Choi, Y.J.; Kwak, M.J.; Ge, J.; Rodgers, M.A.; Shi, M.; Leslie, B.J.; Hopfner, K.P.; Ha, T.; et al. Crosstalk between the cGAS DNA sensor and Beclin-1 au-tophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 2014, 15, 228–238. [Google Scholar] [CrossRef]
- Ghosh, A.; Shao, L.; Sampath, P.; Zhao, B.; Patel, N.V.; Zhu, J.; Behl, B.; Parise, R.A.; Beumer, J.H.; O’Sullivan, R.J.; et al. Oligoadenylate-Synthetase-Family Protein OASL Inhibits Activity of the DNA Sensor cGAS during DNA Virus Infection to Limit Interferon Production. Immunity 2019, 50, 51–63.e5. [Google Scholar] [CrossRef]
- Parvatiyar, K.; Zhang, Z.; Teles, R.M.; Ouyang, S.; Jiang, Y.; Iyer, S.S.; Zaver, S.A.; Schenk, M.; Zeng, S.; Zhong, W.; et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012, 13, 1155–1161. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.-J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef]
- Sun, M.-S.; Zhang, J.; Jiang, L.-Q.; Pan, Y.-X.; Tan, J.-Y.; Yu, F.; Guo, L.; Yin, L.; Shen, C.; Shu, H.-B.; et al. TMED2 Potentiates Cellular IFN Responses to DNA Viruses by Reinforcing MITA Dimerization and Facilitating Its Trafficking. Cell Rep. 2018, 25, 3086–3098.e3. [Google Scholar] [CrossRef]
- Srikanth, S.; Woo, J.S.; Wu, B.; El-Sherbiny, Y.M.; Leung, J.; Chupradit, K.; Rice, L.; Seo, G.J.; Calmettes, G.; Ramakrishna, C.; et al. The Ca(2+) sensor STIM1 regulates the type I in-terferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 2019, 20, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Pokatayev, V.; Yang, K.; Tu, X.; Dobbs, N.; Wu, J.; Kalb, R.G.; Yan, N. Homeostatic regulation of STING protein at the resting state by stabilizer TOLLIP. Nat. Immunol. 2020, 21, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Almine, J.; O’Hare, C.A.J.; Dunphy, G.; Haga, I.R.; Naik, R.J.; Atrih, A.; Connolly, D.J.; Taylor, J.; Kelsall, I.R.; Bowie, A.; et al. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 2017, 8, 14392. [Google Scholar] [CrossRef] [PubMed]
- Jønsson, K.L.; Laustsen, A.; Krapp, C.; Skipper, K.; Thavachelvam, K.; Hotter, D.; Egedal, J.; Kjolby, M.; Mohammadi, P.; Prabakaran, T.; et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 2017, 8, 14391. [Google Scholar] [CrossRef]
- Guo, H.; König, R.; Deng, M.; Rieß, M.; Mo, J.; Zhang, L.; Petrucelli, A.; Yoh, S.M.; Barefoot, B.; Samo, M.; et al. NLRX1 Sequesters STING to Negatively Regulate the Interferon Response, Thereby Facilitating the Replication of HIV-1 and DNA Viruses. Cell Host Microbe 2016, 19, 515–528. [Google Scholar] [CrossRef]
- Zhang, L.; Mo, J.; Swanson, K.V.; Wen, H.; Petrucelli, A.; Gregory, S.M.; Zhang, Z.; Schneider, M.; Jiang, Y.; Fitzgerald, K.A.; et al. NLRC3, a Member of the NLR Family of Proteins, Is a Negative Regulator of Innate Immune Signaling Induced by the DNA Sensor STING. Immunity 2014, 40, 329–341. [Google Scholar] [CrossRef]
- Li, Y.; Li, C.; Xue, P.; Zhong, B.; Mao, A.-P.; Ran, Y.; Chen, H.; Wang, Y.-Y.; Yang, F.; Shu, H.-B. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. USA 2009, 106, 7945–7950. [Google Scholar] [CrossRef]
- Zhou, Q.; Lin, H.; Wang, S.; Wang, S.; Ran, Y.; Liu, Y.; Ye, W.; Xiong, X.; Zhong, B.; Shu, H.-B.; et al. The ER-Associated Protein ZDHHC1 Is a Positive Regulator of DNA Virus-Triggered, MITA/STING-Dependent Innate Immune Signaling. Cell Host Microbe 2014, 16, 450–461. [Google Scholar] [CrossRef]
- Li, Y.; James, S.J.; Wyllie, D.H.; Wynne, C.; Czibula, A.; Bukhari, A.; Pye, K.; Mustafah, S.M.B.; Fajka-Boja, R.; Szabo, E.; et al. TMEM203 is a binding partner and regulator of STING-mediated inflammatory signaling in macrophages. Proc. Natl. Acad. Sci. USA 2019, 116, 16479–16488. [Google Scholar] [CrossRef]
- Oduro, P.K.; Zheng, X.; Wei, J.; Yang, Y.; Wang, Y.; Zhang, H.; Liu, E.; Gao, X.; Du, M.; Wang, Q. The cGAS-STING signaling in cardiovascular and metabolic diseases: Future novel target option for pharmacotherapy. Acta Pharm. Sin. B 2022, 12, 50–75. [Google Scholar] [CrossRef]
- Long, Z.-J.; Wang, J.-D.; Xu, J.-Q.; Lei, X.-X.; Liu, Q. cGAS/STING cross-talks with cell cycle and potentiates cancer immunotherapy. Mol. Ther. 2022, 30, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS-STING pathway as a therapeutic target in inflammatory dis-eases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.L.; Buchan, G.J.; Rühl, M.; Mukai, K.; Salvatore, S.R.; Ogawa, E.; Andersen, S.D.; Iversen, M.B.; Thielke, A.L.; Gunderstofte, C.; et al. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E7768–E7775. [Google Scholar] [CrossRef] [PubMed]
- Lara, P.N., Jr.; Douillard, J.Y.; Nakagawa, K.; von Pawel, J.; McKeage, M.J.; Albert, I.; Losonczy, G.; Reck, M.; Heo, D.S.; Fan, X.; et al. Randomized phase III place-bo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 2965–2971. [Google Scholar] [CrossRef]
- Fischer, H.; Tschachler, E.; Eckhart, L. Cytosolic DNA sensing through cGAS and STING is inactivated by gene mutations in pangolins. Apoptosis 2020, 25, 474–480. [Google Scholar] [CrossRef]
| Virus Types | Proposed Mechanism | Reference |
|---|---|---|
| DNA Viruses | ||
| HSV-1 | VC: releases extracellular vesicles; STING interaction; stabilizes STING in HEp-2 cells; enhances cGAMP levels; STING-mediated non-canonical autophagy | [27,50,51,52,53] |
| VE: dampens NF-κB activation; degrades cGAS mRNA; binds to STING and TBK1; restrains cGAS catalyze activity; the deubiquitinase (DUB) activity; mediates cGAS deamidation; inhibits cGAS-DNA phase separation; obstructs TBK1 dimerization; induces TBK1 proteasomal degradation; interacts with karyopherin | [53,54,55,56,57,58,59,60,61,62] | |
| KSHV | VC: detection by the cGAS | [16] |
| VE: inhibits cGAS activity and production of cGAMP; blocks binding of cGAS to DNA; interacts with STING; inhibits IRF3 interaction with CBP; competes with IRF3 for IFNβ promoter binding | [63,64,65,66,67] | |
| MHV68 | VC: IFN-mediated antiviral pathways | [68] |
| VE: DNA binding; DUB-dependent inhibition of STING; interacts with TBK1; blocks IRF3 and CBP interaction | [64,69,70,71] | |
| VZV | VE: restricts cGAS-DNA phase separation; interacts with IRF3; prevents IRF3 phosphorylation at S396; inhibits IκBa ubiquitination | [60,72,73,74] |
| MDV | VE: hampers the combination of IRF7 and TBK1 with STING; impedes IRF7 phosphorylation and nuclear translocation | [75,76] |
| EBV | VE: regulation of LMP1; restricts cGAS-DNA phase separation | [60,77] |
| HPV | VC: binds to DNA virus | [78] |
| VE: vesicular trafficking; degradation of STING; translation inhibition; STING interaction | [79,80,81,82] | |
| Adenovirus | VC: detection by the cGAS | [83] |
| VE: STING interaction | [82] | |
| HBV | VC: detection by the cGAS and STING; | [84] |
| VE: bounds to STING and attenuates K63-linked polyubiquitination of STING | [85] | |
| HCMV | VC: induces cGAS; binds to IFI16 and relocalizes IFI16 to the cytoplasm | [86,87] |
| VE: removes K63 ubiquitination of STING; inhibits the translocation of STING and impairs the recruitment of TBK1 and IRF3; induces STING degradation and inhibits cGAMP-mediated IFN-β induction; cGAS interaction; disrupts STING oligomerization and STING-TBK1 association; protein S-nitrosylation; hinders STAT1 phosphorylation | [88,89,90,91,92,93,94] | |
| VACV | VC: activation of NF-κB and IRF3 | [95,96] |
| VE: selective 2’3’cGAMP degradation; interaction of STING with sulfated glycosaminoglycans; suppresses STING phosphorylation and dimerization; mTOR-dependent cGAS degradation; preventes cGAMP spread; blocks the activation of TBK1 and IKKε; binds to the Ku70-Ku80 complex and blocks DNA sensing by DNA-PK in fibroblasts | [97,98,99,100,101,102] | |
| ECTV | VC: detection by the cGAS-STING pathway; induces the Phosphorylation of TBK1 and IRF3 | [95] |
| VE: suppresses activation of STING and IRF3 | [99] | |
| VE: activation of IRF7 and NF-κB signaling | [103] | |
| ASFV | VC: induces STING phosphorylation and trafficking | [104] |
| VE: suppresses TBK1 phosphorylation and IKKβ; impairs STING activation; IKKε interaction; the autophagy-mediated lysosomal degradation of TBK1; suppression of NF-κB and IRF3 activation | [104,105,106,107,108] | |
| Lentiviruses | ||
| HIV | VC: cGAS interaction; recognizes cDNA (ssDNA) reverse-transcribed from HIV-1 virus; intercellular transfer of cGAMP; detects/disrupts nuclear viral capsid | [47,109,110,111,112] |
| VE: dampens IRF3 and NF-kB nuclear translocation | [62,113] | |
| RNA viruses | ||
| DENV | VC: modulates the overall refractoriness of cells; induces mtDNA release; interacts with RIG-I and MAVS | [28,68,114] |
| VE: binds to and cleaves STING; targets cGAS for degradation | [115,116] | |
| HCV | VC: addition of cGAMP or STING inhibits viral replication | [117] |
| VE: interacts with STING to disrupt the interaction of STING with TBK1 or MAVS and downstream IFN signalling; suppresses STING accumulation | [117,118,119] | |
| SARS-CoV-2 | VC: activates of STING; cell fusion; triggers the cGAS/STING axis | [120,121,122,123] |
| VE: cleaves ubiquitin and ISG15; STING interaction; disrupts dimerization and K63-linked polyubiquitination of STING; interacts with TBK1 and impedes the phosphorylation and nuclear translocation of IRF3; interaction with the STING-TRAF3-TBK1 complex; the deubiquitinating activity | [120,124,125,126,127] | |
| PEDV | VE: interacts with STING and represses K63-linked polyubiquitination of STING | [128] |
| TGEV | VE: deubiquitination of STING | [129] |
| HCoV-NL63 | VE: binds to STING to disrupt its dimerisation/ubiquitination and downstream IFN production; DUB activity | [127,130] |
| CHIKV | VC: restricts CHIKV replication | [131,132] |
| VE: induces cGAS degradation | [131] | |
| WNV | VC: modulates T cell responses and T cell frequencies; restricts WNV infection | [68,133] |
| VE: cleaves hSTING; inhibits the phosphorylation of TBK1 and IRF3 | [134,135] | |
| MNV | VC: limits MNV infection; drives cytosolic DNA accumulation and cGAS/STING activation | [136] |
| ZIKV | VC: STING interaction; induces antiviral autophagy | [137,138,139] |
| VE: promotes the cleavage of cGAS; cleaves STING | [134,140] | |
| MeV | VC: induces phosphorylation and ubiquitination of STING; induces mtDNA release | [141,142] |
| NiV | VC: induces phosphorylation and ubiquitination of STING | [141] |
| SeV | VC: translation inhibition; anti-SeV activity; induces STING expression | [143,144,145,146] |
| VSV | VC: translation promotion | [143] |
| VE: reduces the ratio of full-length wt hSTING/truncated STING isoforms | [147] | |
| SINV | VC: translation inhibition | [143] |
| EMCV | VC: stimulates mtDNA release and consequent cGAS activation; restricts EMCV replication | [144,148] |
| IAV | VC: membrane fusion and STING interaction; monoubiquitination of cGAS; triggers mtDNA release and binds to mtDNA; inhibits IAV replication | [146,148,149,150] |
| PRRSV | VC: suppress PRRSV replication | [150] |
| SFTSV | VC: Cytosolic SAFA senses viral RNA and activates STING antiviral signal | [151] |
| T3D | VC: translation inhibition | [143] |
| Target Protein | Factors | Functions | PTM Effects | Reference |
|---|---|---|---|---|
| cGAS | G3BP1 | Promoting the formation of large cGAS complexes | Efficient activation of cGAS | [282] |
| cGAS | PQBP1 | Increasing DNA-binding affinity | Initiates an IRF3-dependent innate response | [283] |
| cGAS | ZCCHC3 | Enhancing the binding of cGAS to dsDNA | Acts as a general co-sensor of cGAS | [284] |
| cGAS | β-arrestin 2 | Enhancing the DNA-binding ability of cGAS | Promotes IFN-β signaling | [285] |
| cGAS | Beclin-1 | Regulating cGAMP production and autophagy | Balances anti-microbial immune responses | [286] |
| cGAS | OASL | Binding to cGAS and inhibiting cGAMP synthesis | Inhibits cGAS-mediated IFN induction | [287] |
| STING | SAR1A and SEC24C | Blocking STING puncta formation induced by cGAMP | Affects STING trafficking and signalling | [27] |
| STING | DDX41 | Interacting with DNA and STING | Activates protein kinases, TBK1, NF-κB and IRF3 | [288,289] |
| STING | TMED2 | Reinforcing STING dimerization and facilitating its trafficking | Potentiates cellular IFN responses to DNA viruses | [290] |
| STING | iRhom2 | Recruiting the translocon-associated protein TRAPβ | Facilitates the trafficking of STING | [240] |
| STING | STIM1 | Retaining the signaling adaptor STING at the ER | Regulates the type I interferon response | [291] |
| STING | TOLLIP | Interacting with STING N-terminus | Stabilizes resting-state STING protein levels | [292] |
| STING/TBK1 | IFI16 | Promoting production and function of cGAMP | Regulates the activation of STING and the recruitment of TBK1 | [293,294] |
| STING/TBK1 | NLRX1 | Associating with STING to disrupt STING-TBK1 interaction | Inhibits innate immunity and facilitates viral spread | [295] |
| STING/TBK1 | NLRC3 | Impeding the interaction between STING and TBK1 | Negatively regulates the STING-mediated DNA-sensing pathway | [296] |
| STING/TBK1 | ISG56 | Disrupting the interactions between STING and TBK1, | Inhibits antiviral signaling | [297] |
| STING/TBK1 | ZDHHC1 | Mediating dimerization/aggregation of STING and recruitment of TBK1 and IRF3 | Positively regulates the innate immune response against DNA viruses | [298] |
| STING/TBK1 | TMEM203 | Forming a functional signaling complex with STING | Promotes the TBK1-IRF3-IFN activation | [299] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Liu, J.; Liu, C.; Liu, R.; Liu, L.; Yu, Z.; Zhuang, J.; Sun, C. Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation. Cells 2022, 11, 3043. https://doi.org/10.3390/cells11193043
Yu Y, Liu J, Liu C, Liu R, Liu L, Yu Z, Zhuang J, Sun C. Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation. Cells. 2022; 11(19):3043. https://doi.org/10.3390/cells11193043
Chicago/Turabian StyleYu, Yang, Jingyang Liu, Cun Liu, Ruijuan Liu, Lijuan Liu, Zhenhai Yu, Jing Zhuang, and Changgang Sun. 2022. "Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation" Cells 11, no. 19: 3043. https://doi.org/10.3390/cells11193043
APA StyleYu, Y., Liu, J., Liu, C., Liu, R., Liu, L., Yu, Z., Zhuang, J., & Sun, C. (2022). Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation. Cells, 11(19), 3043. https://doi.org/10.3390/cells11193043






