m6A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization
Abstract
1. Materials and Methods
1.1. Bioinformatics of Gene Expression Database
1.2. Colorectal Clinical Tissues and CRC Cell Lines
1.3. Plasmid Construction and Cell Transfection
1.4. RNA Extraction and RT-qPCR
1.5. Immunoblotting
1.6. Cell Proliferation and Transwell Assays
1.7. Cell Apoptosis and Cell Cycle Assays
1.8. Tube Formation Assay
1.9. Dual Luciferase Reporter Assay
1.10. Tumor Xenograft Model
1.11. RNA Immunoprecipitation (RIP) Assay
1.12. RNA Pull-Down Assay
1.13. MeRIP qRT-PCR
1.14. RNA Stability Assay
1.15. Fish
1.16. Statistical Analysis
2. Background
3. Results
3.1. HNF1A-AS1 Was Upregulated in Human CRC Tissues and Correlated with Poor Prognosis
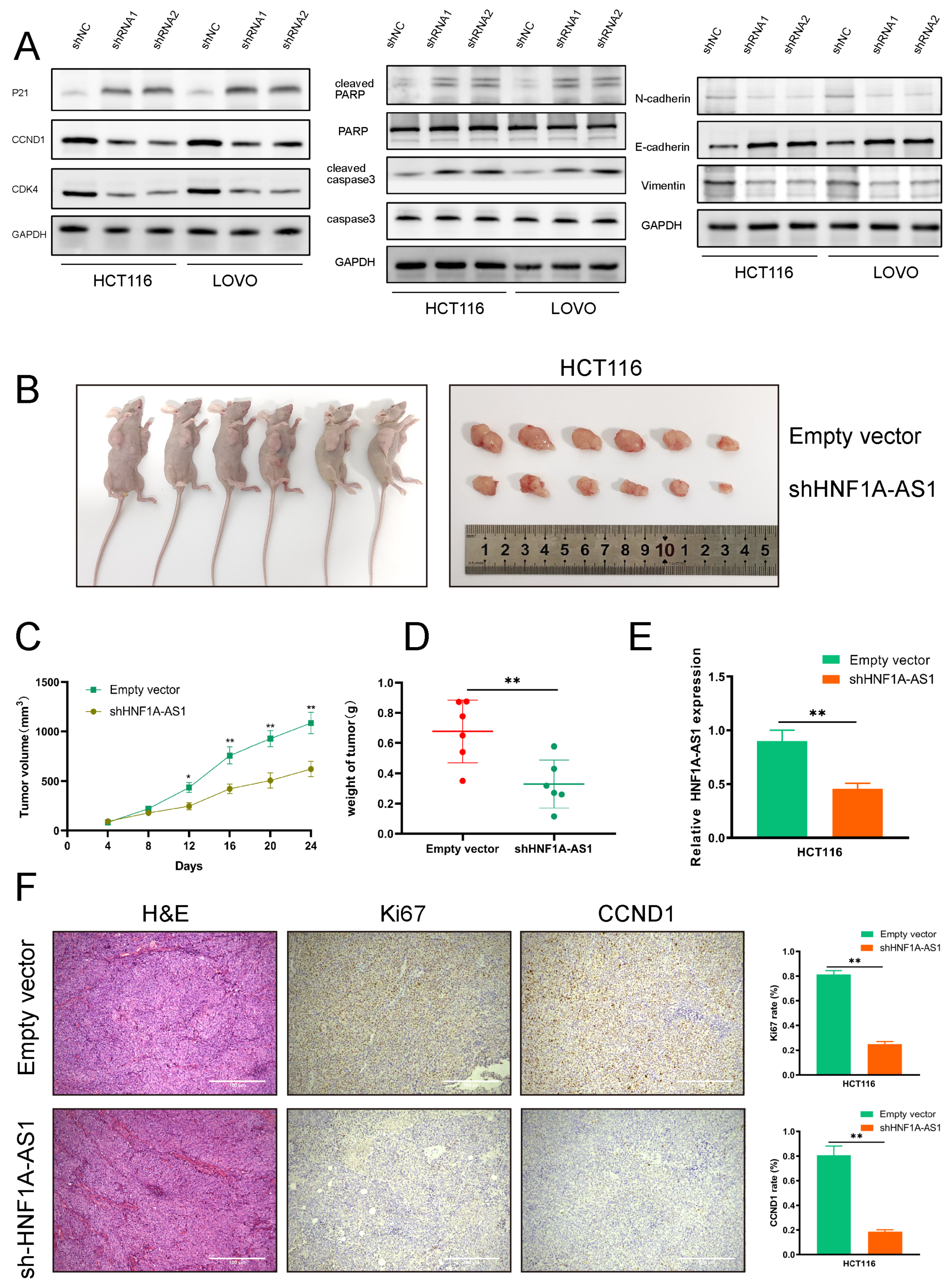
3.2. HNF1A-AS1 Promotes Progression of CRC In Vitro
3.3. HNF1A-AS1 Promotes CRC Progression In Vivo
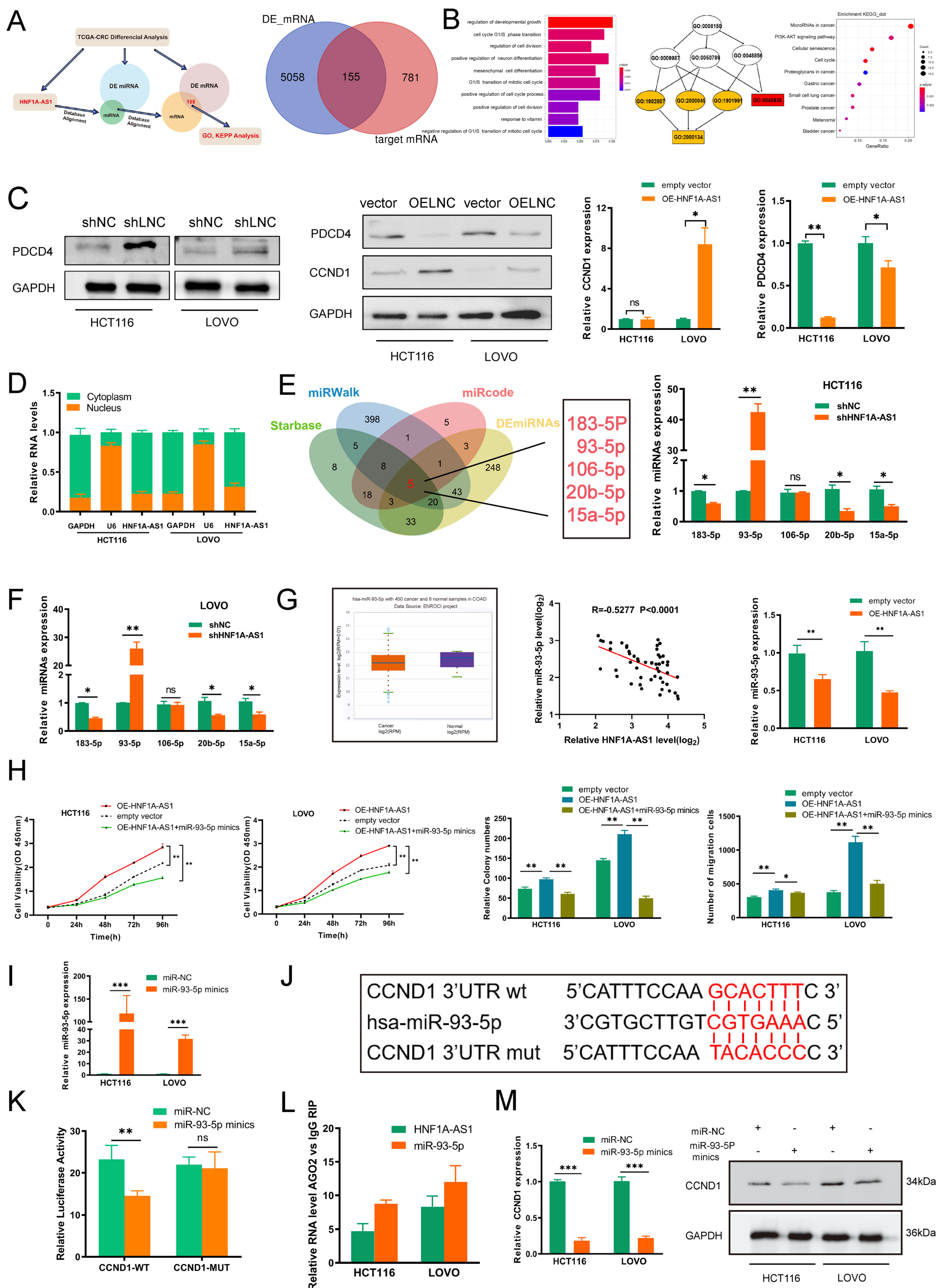
3.4. HNF1A-AS1 Affects CRC Progression through Regulating the Expression of CCND1 and PDCD4
3.5. HNF1A-AS1 Sponges miR-93-5p to Upregulate CCND1
3.6. HNF1A-AS1/IGF2BP2/CCND1 Complex Stabilizes CCND1 mRNA
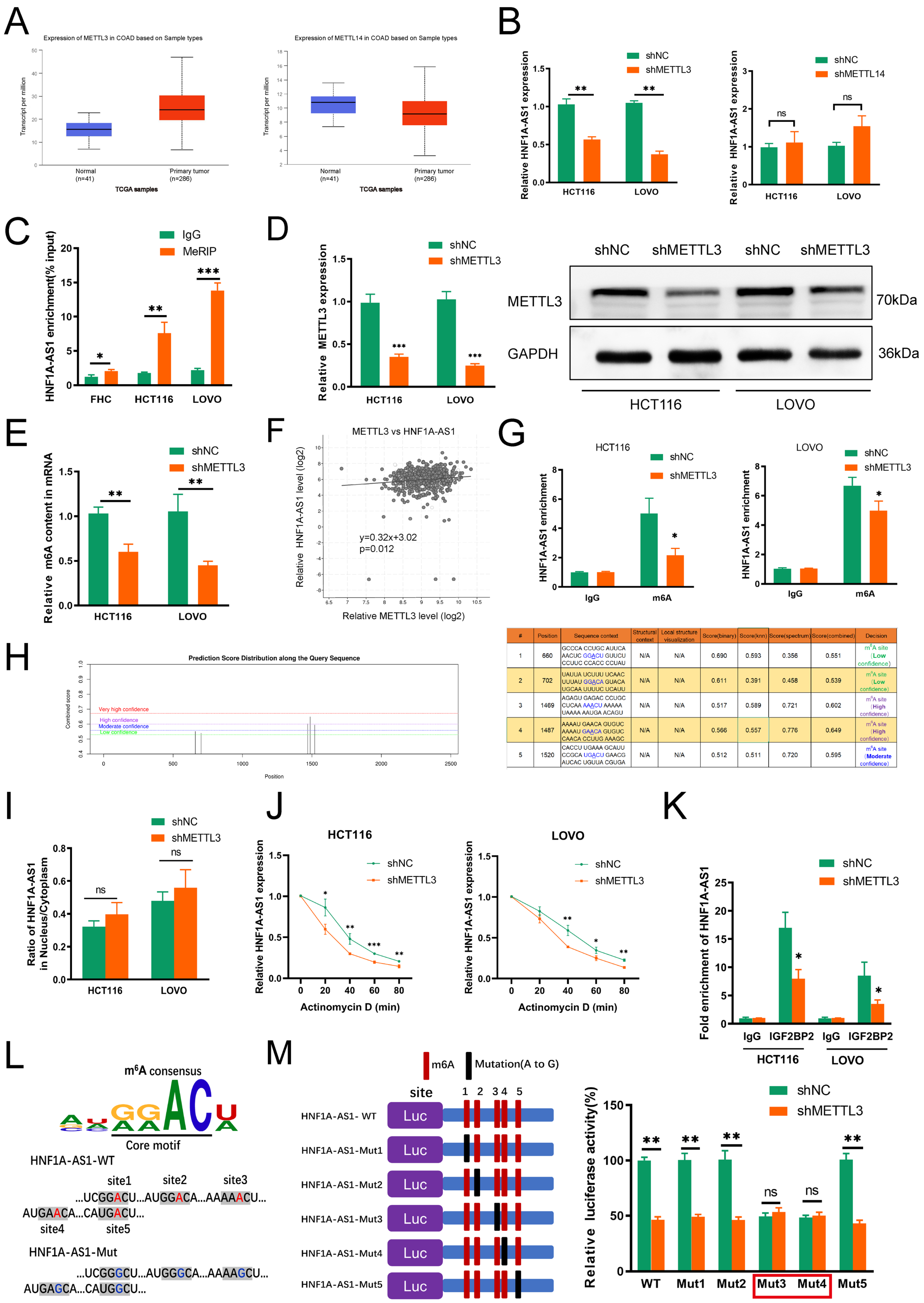
3.7. METTL3 Mediates HNF1A-AS1 m6A Modification and Contributes to Its Upregulation in CRC
3.8. m6A/HNF1A-AS1/CCND1 Axis Cooperated with PDCD4 to Regulate PI3K/AKT Pathway and Related Clinical Relationship in CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Dominissini, D.; Moshitch-Moshkovitz, S.; Salmon-Divon, M.; Amariglio, N.; Rechavi, G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013, 8, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.-G.; et al. m 6 A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Lortet-Tieulent, J.; Georges, D.; Bray, F.; Vaccarella, S. Profiling global cancer incidence and mortality by socioeconomic development. Int. J. Cancer 2020, 147, 3029–3036. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Gokulnath, M.; Partridge, N.C.; Selvamurugan, N. Runx2, a target gene for activating transcription factor-3 in human breast cancer cells. Tumor Biol. 2014, 36, 1923–1931. [Google Scholar] [CrossRef]
- Qin, L.; Zhang, Y.; Lin, J.; Shentu, Y.; Xie, X. MicroRNA-455 regulates migration and invasion of human hepatocellular carcinoma by targeting Runx2. Oncol. Rep. 2016, 36, 3325–3332. [Google Scholar] [CrossRef]
- Berulava, T.; Buchholz, E.; Elerdashvili, V.; Pena, T.; Islam, R.; Lbik, D.; Mohamed, B.A.; Renner, A.; Von Lewinski, D.; Sacherer, M.; et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur. J. Hear. Fail. 2019, 22, 54–66. [Google Scholar] [CrossRef]
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 98. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramírez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef]
- Deng, R.; Cheng, Y.; Ye, S.; Zhang, J.; Huang, R.; Li, P.; Liu, H.; Deng, Q.; Wu, X.; Lan, P.; et al. m(6)A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019, 12, 4391–4402. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.-Q.; Li, Z.-X.; Zhou, G.-Q.; Lin, L.; Zhang, L.-L.; Lv, J.-W.; Huang, X.-D.; Liu, R.-Q.; Chen, F.; He, X.-J.; et al. Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 2019, 79, 4612–4626. [Google Scholar] [CrossRef]
- Liu, Z.; Wei, X.; Zhang, A.; Li, C.; Bai, J.; Dong, J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem. Biophys. Res. Commun. 2016, 473, 1268–1275. [Google Scholar] [CrossRef]
- Liu, H.T.; Liu, S.; Liu, L.; Ma, R.R.; Gao, P. EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Res. 2018, 78, 5877–5890. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, W.; Gao, X.; Zhang, D.; Li, Y. HNF1A?AS1 promotes growth and metastasis of esophageal squamous cell carcinoma by sponging miR?214 to upregulate the expression of SOX-4. Int. J. Oncol. 2017, 51, 657–667. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, J.; Li, R.; Li, L.; Gu, Y.; Zhan, H.; Zhou, C.; Zhong, C. MYC-activated lncRNA HNF1A-AS1 overexpression facilitates glioma progression via cooperating with miR-32-5p/SOX4 axis. Cancer Med. 2020, 9, 6387–6398. [Google Scholar] [CrossRef] [PubMed]
- Lou, P.; Ding, T.; Zhan, X. Long Noncoding RNA HNF1A-AS1 Regulates Osteosarcoma Advancement Through Modulating the miR-32-5p/HMGB1 Axis. Cancer Biother. Radiopharm. 2020, 36, 371–381. [Google Scholar] [CrossRef]
- Fang, C.; Qiu, S.; Sun, F.; Li, W.; Wang, Z.; Yue, B.; Wu, X.; Yan, D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017, 410, 50–62. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Y.; Liu, L.; Yang, W.; Zhang, Q. HNF1A-AS1 Regulates Cell Migration, Invasion and Glycolysis via Modulating miR-124/MYO6 in Colorectal Cancer Cells. Onco Targets Ther. 2020, 13, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, X.; Yang, Y.; Yang, D. Cyclin D1 mediated by the nuclear translocation of nuclear factor kappa B exerts an oncogenic role in lung cancer. Bioengineered 2022, 13, 6866–6879. [Google Scholar] [CrossRef]
- Zhang, H.; He, C.; Guo, X.; Fang, Y.; Lai, Q.; Wang, X.; Pan, X.; Li, H.; Qin, K.; Li, Q.; et al. DDX39B contributes to the proliferation of colorectal cancer through direct binding to CDK6/CCND1. Cell Death Discov. 2022, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Tsai, C.-W.; Hsu, C.-M.; Chang, W.-S.; Shui, H.-A.; Bau, D.-T. The significant association of CCND1 genotypes with colorectal cancer in Taiwan. Tumor Biol. 2015, 36, 6533–6540. [Google Scholar] [CrossRef]
- Huang, W.; Yang, Y.; Wu, J.; Niu, Y.; Yao, Y.; Zhang, J.; Huang, X.; Liang, S.; Chen, R.; Chen, S.; et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ. 2020, 27, 1709–1727. [Google Scholar] [CrossRef]
- Chen, X.; Chen, S.; Xiu, Y.-L.; Sun, K.-X.; Zong, Z.-H.; Zhao, Y. RhoC is a major target of microRNA-93-5P in epithelial ovarian carcinoma tumorigenesis and progression. Mol. Cancer 2015, 14, 31. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, H.Z.; Ye, F.Q.; Xu, D.C.; Fang, L.L.; Zhang, X.F.; Zhu, J.J. The MYEOV-MYC association promotes oncogenic miR-17/93-5p expression in pancreatic ductal adenocarcinoma. Cell Death Dis. 2022, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Liu, T.T.; Yu, X.N.; Balakrishnan, A.; Zhu, H.R.; Guo, H.Y.; Zhang, G.C.; Bilegsaikhan, E.; Sun, J.L.; Song, G.Q.; et al. microRNA-93-5p promotes hepatocellular carcinoma progression via a microRNA-93-5p/MAP3K2/c-Jun positive feedback circuit. Oncogene 2020, 39, 5768–5781. [Google Scholar] [CrossRef]
- Chen, X.; Liu, J.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Ge, H.; Liu, Y. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J. Exp. Clin. Cancer Res. 2020, 39, 65. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, G.; Lin, B.; Huang, J. MicroRNA-93-5p expression in tumor tissue and its tumor suppressor function via targeting programmed death ligand-1 in colorectal cancer. Cell Biol. Int. 2020, 44, 1224–1236. [Google Scholar] [CrossRef]
- Li, Q.; Yue, W.; Li, M.; Jiang, Z.; Hou, Z.; Liu, W.; Ma, N.; Gan, W.; Li, Y.; Zhou, T.; et al. Downregulating Long Non-coding RNAs CTBP1-AS2 Inhibits Colorectal Cancer Development by Modulating the miR-93-5p/TGF-beta/SMAD2/3 Pathway. Front. Oncol. 2021, 11, 626620. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chen, C.; Ji, X.; Liu, J.; Zhou, Q.; Wang, G.; Yuan, W.; Kan, Q.; Sun, Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Liu, F.; Xiao, X.; Xu, R.; Dai, L.; Zhu, M.; Xu, H.; Xu, Y.; Zhao, A.; Zhou, W.; et al. METTL3 promotes colorectal carcinoma progression by regulating the m6A–CRB3–Hippo axis. J. Exp. Clin. Cancer Res. 2022, 41, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Gao, S.; Liu, W.; Wong, C.-C.; Wu, J.; Wu, J.; Liu, D.; Gou, H.; Kang, W.; Zhai, J.; et al. RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 2021, 160, 1284–1300.e16. [Google Scholar] [CrossRef]
- Wang, H.; Wei, W.; Zhang, Z.-Y.; Liu, Y.; Bin Shi, B.; Zhong, W.; Zhang, H.-S.; Fang, X.; Sun, C.-L.; Wang, J.-B.; et al. TCF4 and HuR mediated-METTL14 suppresses dissemination of colorectal cancer via N6-methyladenosine-dependent silencing of ARRDC4. Cell Death Dis. 2021, 13, 3. [Google Scholar] [CrossRef]
- Lang, C.; Yin, C.; Lin, K.; Li, Y.; Yang, Q.; Wu, Z.; Du, H.; Ren, D.; Dai, Y.; Peng, X. m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 2021, 11, e426. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.-H.; Wu, Q.-N.; Jin, Y.; Wang, D.-S.; Chen, Y.-X.; Liu, J.; Luo, X.-J.; Meng, Q.; Pu, H.-Y.; et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 2019, 18, 174. [Google Scholar] [CrossRef]
- Li, T.; Hu, P.-S.; Zuo, Z.; Lin, J.-F.; Li, X.; Wu, Q.-N.; Chen, Z.-H.; Zeng, Z.-L.; Wang, F.; Zheng, J.; et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 2019, 18, 112. [Google Scholar] [CrossRef]
- Zhen, Y.; Fang, W.; Zhao, M.; Luo, R.; Liu, Y.; Fu, Q.; Chen, Y.; Cheng, C.; Zhang, Y.; Liu, Z. miR-374a-CCND1-pPI3K/AKT-c-JUN feedback loop modulated by PDCD4 suppresses cell growth, metastasis, and sensitizes nasopharyngeal carcinoma to cisplatin. Oncogene 2016, 36, 275–285. [Google Scholar] [CrossRef]
- Yang, X.; Song, J.H.; Cheng, Y.L.; Wu, W.J.; Bhagat, T.; Yu, Y.T.; Abraham, J.M.; Ibrahim, S.; Ravich, W.; Roland, B.C.; et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 2014, 63, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Liu, L.; Du, Y.K.; Mi, Y.; Wang, L. The HNF1A-AS1/miR-92a-3p axis affects the radiosensitivity of non-small cell lung cancer by competitively regulating the JNK pathway. Cell Biol. Toxicol. 2021, 37, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.-H.; Yin, C.; Chen, S.-J.; Wen, L.-Z.; Ding, K.; Lei, S.-J.; Liu, J.-P.; Wang, J.; Chen, K.-X.; Jiang, H.-L.; et al. The HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer 2018, 17, 63. [Google Scholar] [CrossRef] [PubMed]




| Variables | Total Patients | Low Expression of HNF1A-AS1 | High Expression of HNF1A-AS1 | p Value Chi-Squared Test |
|---|---|---|---|---|
| All cases | 52 | 18 | 34 | |
| Age ≤60 >60 | 20 32 | 8 10 | 12 22 | 0.5188 |
| Sex male female | 30 22 | 11 7 | 19 15 | 0.7165 |
| TNM stage I and II III and IV | 27 25 | 13 5 | 14 20 | 0.0330 * |
| Tumor size ≤4 cm >4 cm | 24 28 | 13 5 | 11 23 | 0.0061 ** |
| Differentiation High/moderate Poor | 44 8 | 15 3 | 29 5 | 0.8278 |
| Lymph node N0 N1/2/3 | 27 25 | 13 5 | 14 20 | 0.0330 * |
| Distant metastasis Negative Positive | 37 15 | 16 2 | 21 13 | 0.0400 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, Y.; Wang, Y.; Xu, S.; Gao, Z.; Li, C.; Fan, Z.; Ding, J.; Wang, K. m6A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization. Cells 2022, 11, 3008. https://doi.org/10.3390/cells11193008
Bian Y, Wang Y, Xu S, Gao Z, Li C, Fan Z, Ding J, Wang K. m6A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization. Cells. 2022; 11(19):3008. https://doi.org/10.3390/cells11193008
Chicago/Turabian StyleBian, Yibo, Yang Wang, Shufen Xu, Zhishuang Gao, Chao Li, Zongyao Fan, Jie Ding, and Keming Wang. 2022. "m6A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization" Cells 11, no. 19: 3008. https://doi.org/10.3390/cells11193008
APA StyleBian, Y., Wang, Y., Xu, S., Gao, Z., Li, C., Fan, Z., Ding, J., & Wang, K. (2022). m6A Modification of Long Non-Coding RNA HNF1A-AS1 Facilitates Cell Cycle Progression in Colorectal Cancer via IGF2BP2-Mediated CCND1 mRNA Stabilization. Cells, 11(19), 3008. https://doi.org/10.3390/cells11193008





