Bio-Engineered Scaffolds Derived from Decellularized Human Esophagus for Functional Organ Reconstruction
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Sampling of Human Esophagus
2.2. Tissue Decellularization
2.3. DNA Isolation and Quantification
2.4. Histological Investigations
2.5. Immunohistochemical Study
2.6. ECM Component Quantification
2.7. Scanning Electron Microscopy (SEM)
2.8. Second-Harmonic Generation Microscopy
2.9. Mass Spectrometry Analysis
2.9.1. Sample Preparation
2.9.2. Protein Identification
2.9.3. Data Processing
2.10. Mechanical Properties
2.11. Cytotoxicity Study
2.11.1. HM1SV40 Cell Cultures
2.11.2. Preparation of Conditioned Culture Media
2.11.3. Cell Viability Assay
2.12. Statistical Analysis
3. Results
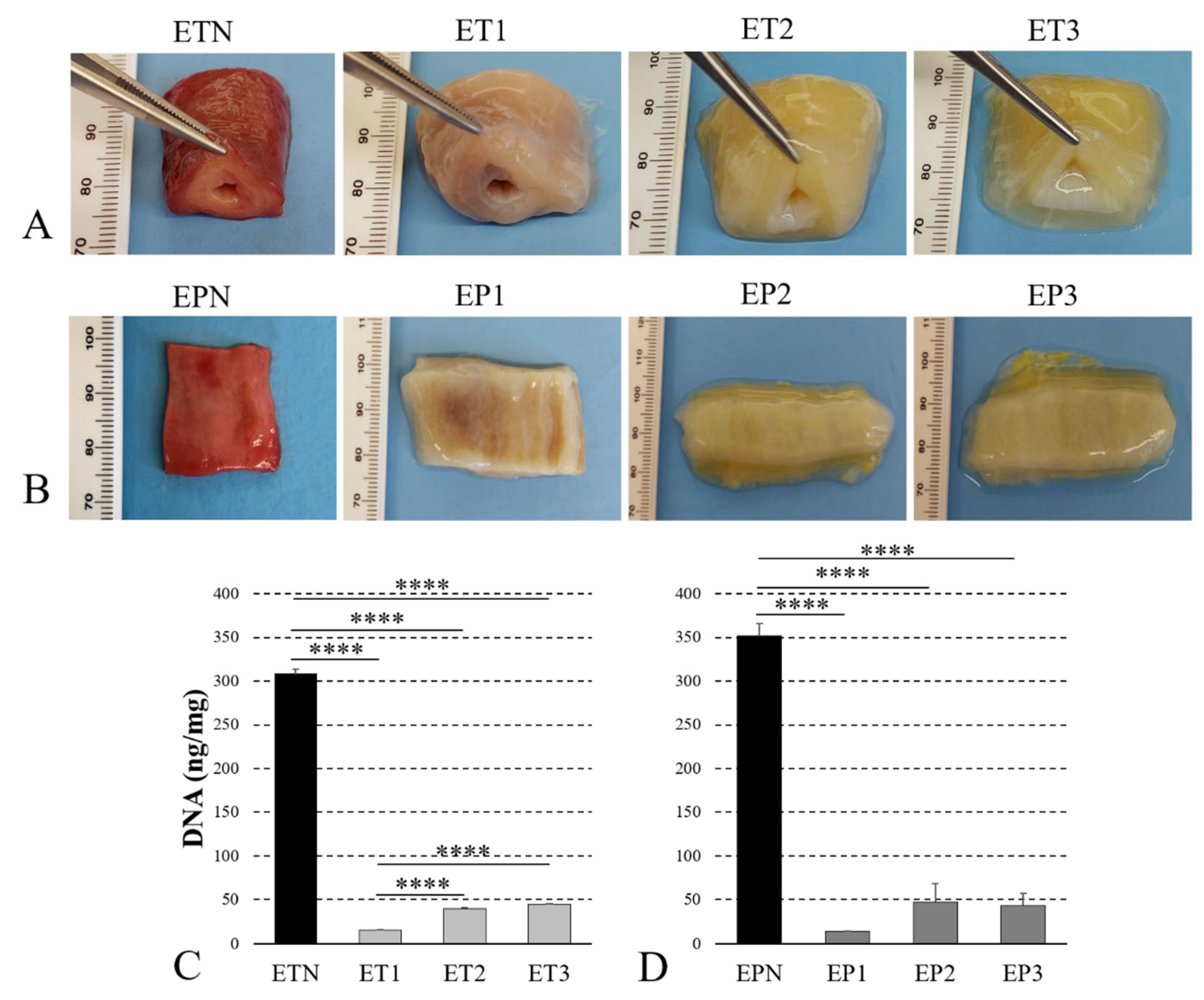
3.1. Decellularization of Human Esophagus
3.2. Histological Evaluation
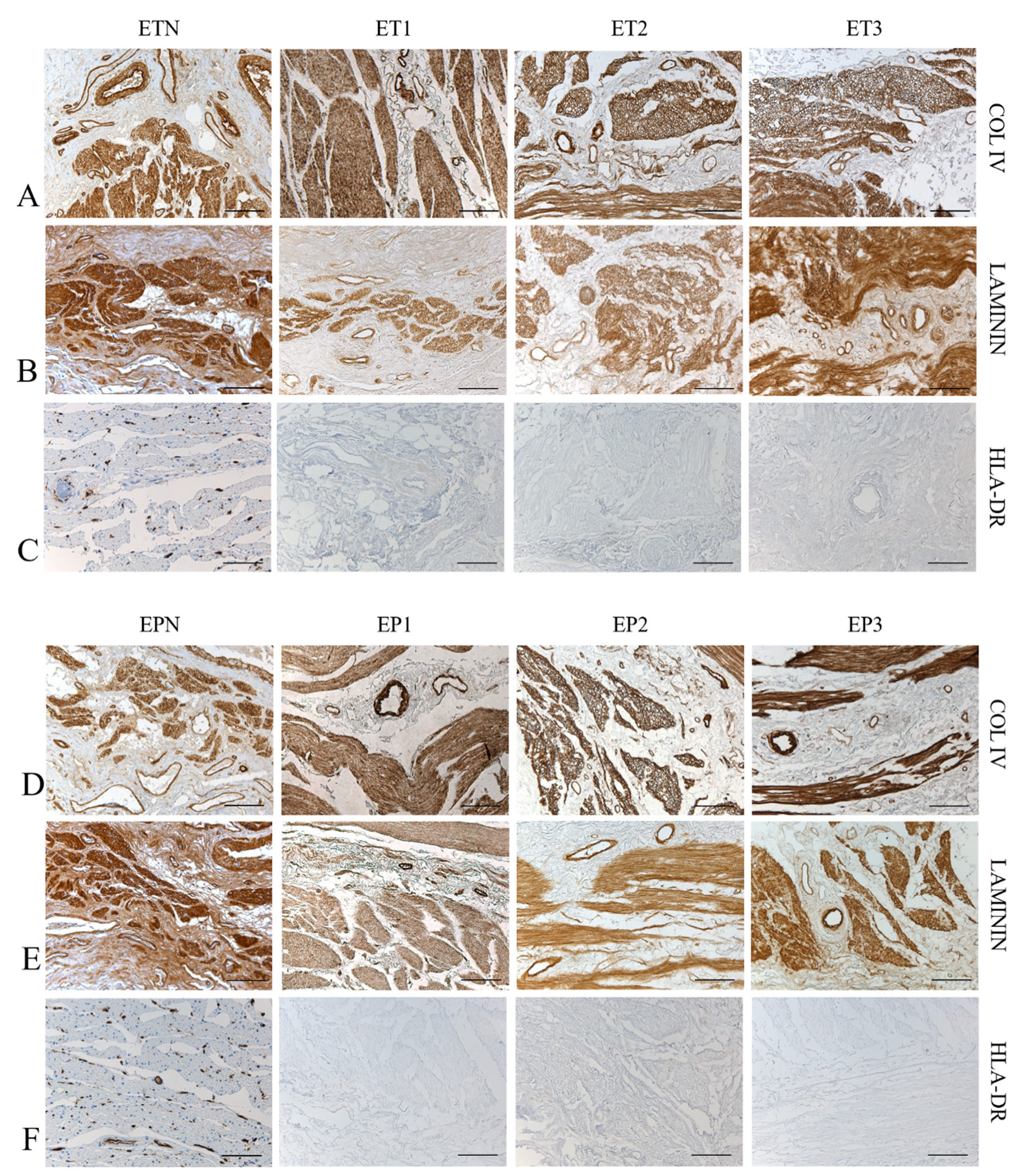
3.3. Immunohistochemical Study
3.4. Quantitative Analysis of ECM Components
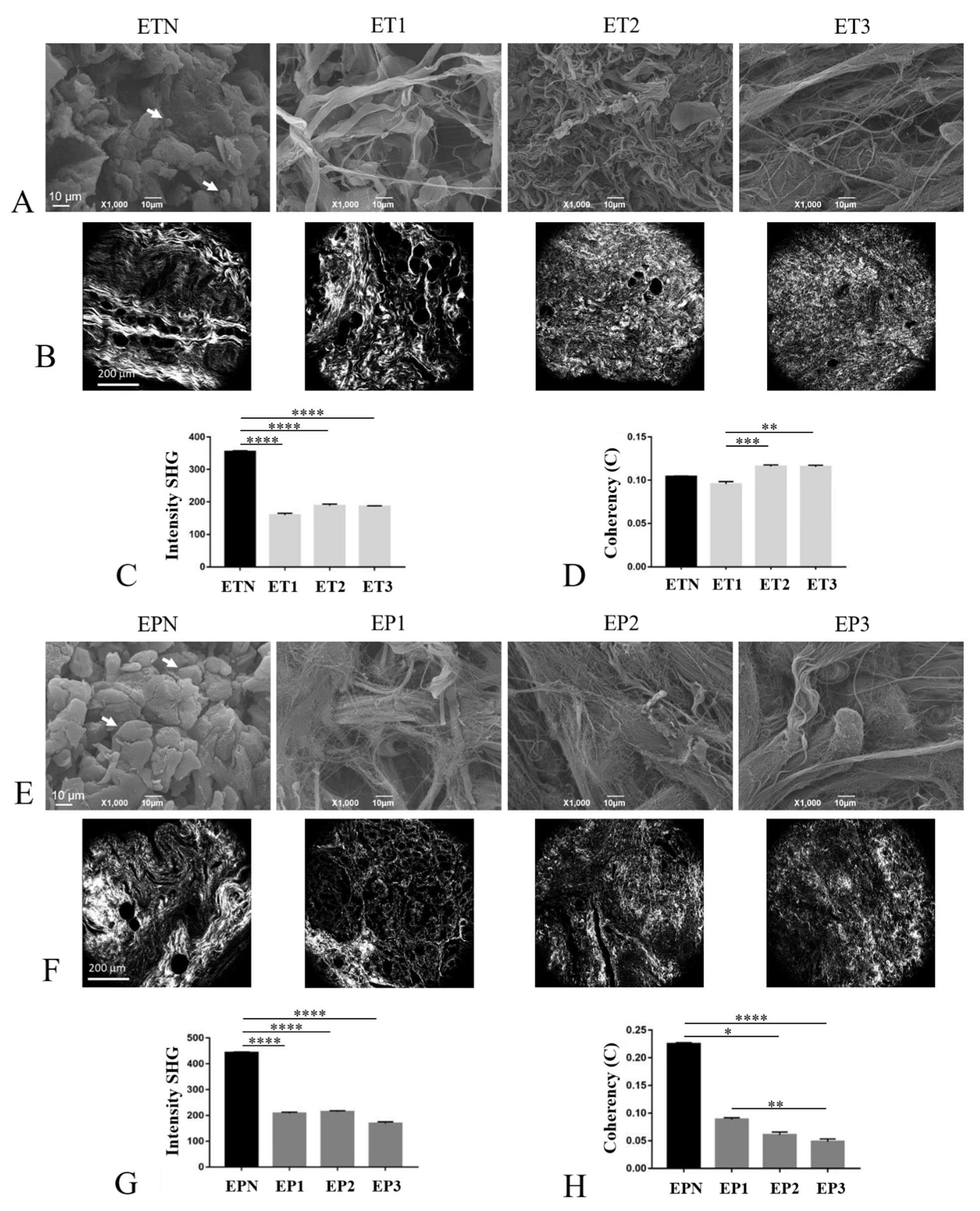
3.5. Characterization of the ECM Collagen Component
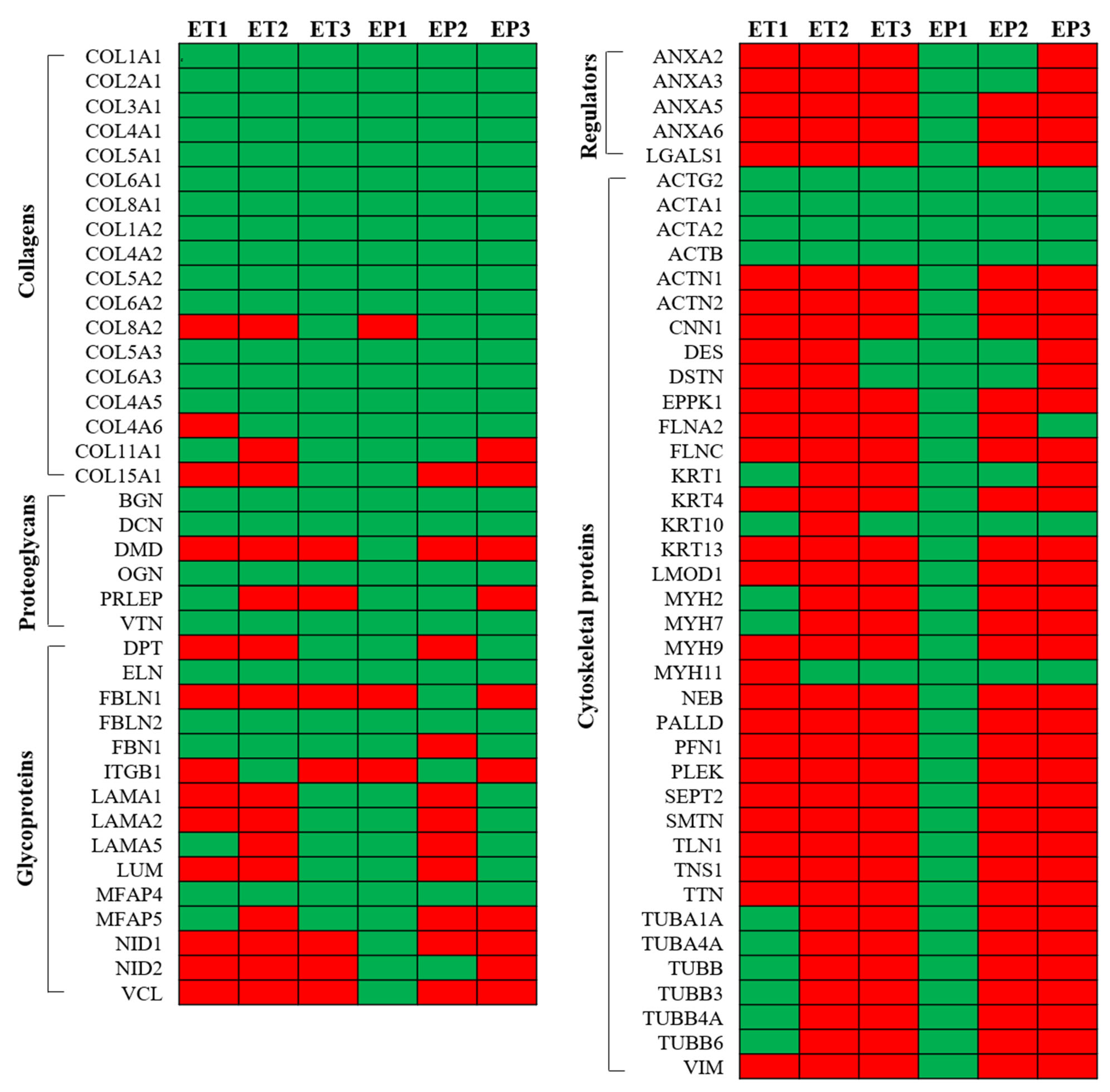
3.6. Proteomic Analysis of Decellularized Tissue Secretome
3.7. Mechanical Behavior
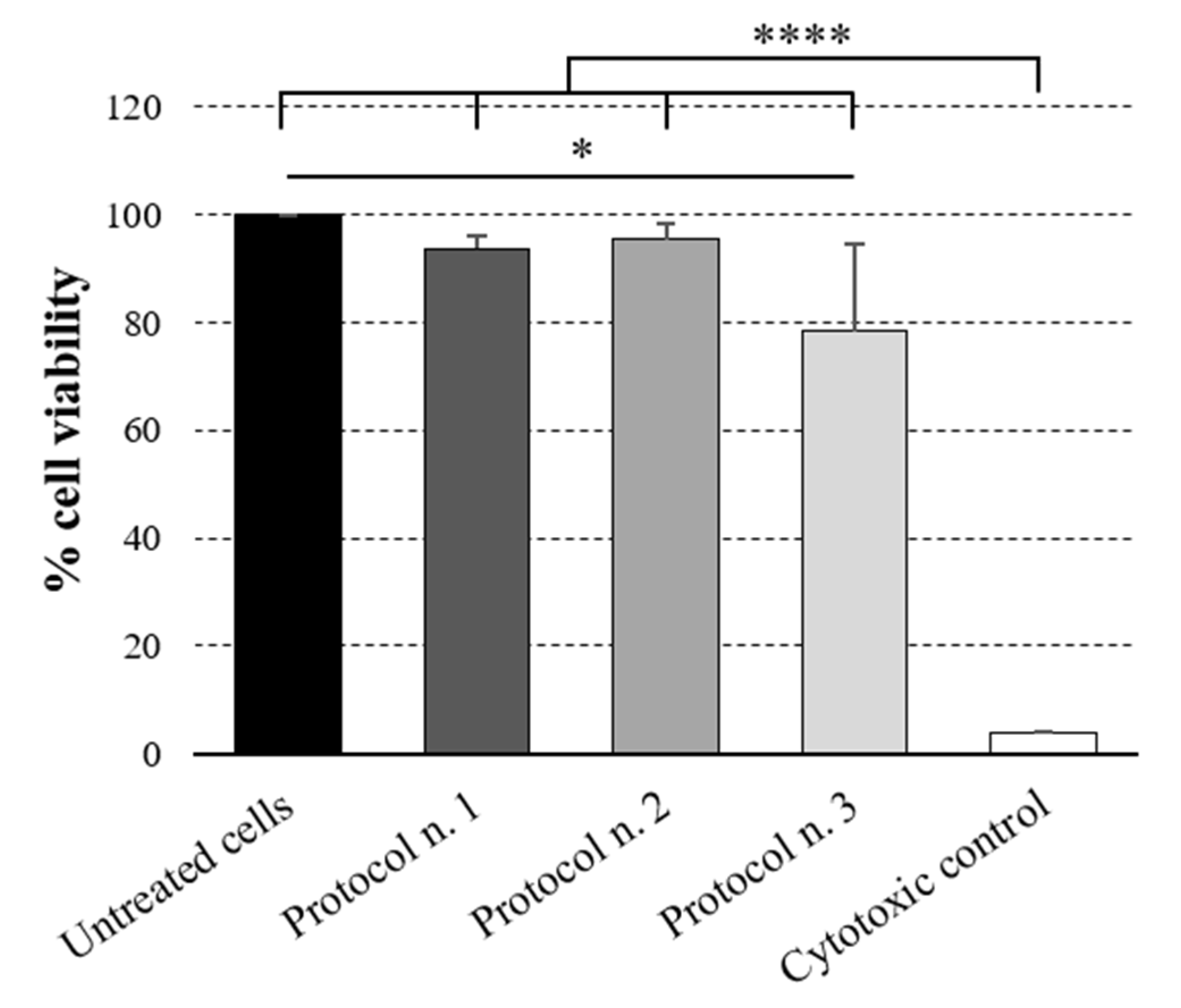
3.8. Cytotoxicity Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTA1 | Actin, Alpha, Skeletal Muscle 1 |
| ACTA2 | Actin, Alpha-2, Smooth Muscle, Aorta |
| ACTB | Actin, Beta |
| ACTG2 | Actin, Gamma-2, Smooth Muscle, Enteric |
| ACTN1 | Actinin, Alpha-1 |
| ACTN2 | Actinin, Alpha-2 |
| ANXA2 | Annexin A2 |
| ANXA3 | Annexin A3 |
| ANXA5 | Annexin A5 |
| ANXA6 | Annexin A6 |
| BGN | Biglycan |
| CNN1 | Calponin 1 |
| COL1A1 | Collagen, Type I, Alpha-1 |
| COL1A2 | Collagen, Type I, Alpha-2 |
| COL2A1 | Collagen, Type II, Alpha-1 |
| COL3A1 | Collagen, Type III, Alpha-1 |
| COL4A1 | Collagen, Type IV, Alpha-1 |
| COL4A2 | Collagen, Type IV, Alpha-2 |
| COL4A5 | Collagen, Type IV, Alpha-5 |
| COL4A2 | Collagen, Type IV, Alpha-6 |
| COL5A1 | Collagen, Type V, Alpha-1 |
| COL5A2 | Collagen, Type V, Alpha-2 |
| COL5A3 | Collagen, Type V, Alpha-3 |
| COL6A1 | Collagen, Type VI, Alpha-1 |
| COL6A2 | Collagen, Type VI, Alpha-2 |
| COL6A3 | Collagen, Type VI, Alpha-3 |
| COL8A1 | Collagen, Type VIII, Alpha-1 |
| COL8A2 | Collagen, Type VIII, Alpha-2 |
| COL11A1 | Collagen, Type XI, Alpha-1 |
| COL15A1 | Collagen, Type XV, Alpha-1 |
| DCN | Decorin |
| DES | Desmin |
| DPT | Dermatopontin |
| DSTN | Destrin |
| DMD | Dystrophin |
| ELN | Elastin |
| EPPK1 | Epiplakin 1 |
| FBN1 | Fibrillin 1 |
| FBLN1 | Fibulin 1 |
| FBLN2 | Fibulin 2 |
| FLNA2 | Filamin |
| FLNC | Filamin C |
| ITGB1 | Integrin, Beta-1 |
| KRT1 | Keratin 1, Type II |
| KRT10 | Keratin 10, Type I |
| KRT13 | Keratin 13, Type I |
| KRT4 | Keratin 4, Type II |
| LAMA1 | Laminin, Alpha-1 |
| LAMA2 | Laminin, Alpha-2 |
| LAMA5 | Laminin, Alpha-5 |
| LGALS1 | Lectin, Galactoside-Binding, Soluble, 1 |
| LMOD1 | Leiomodin 1 |
| LUM | Lumican |
| MFAP4 | Microfibrillar-Associated Protein 4 |
| MFAP5 | Microfibrillar-Associated Protein 5 |
| MYH11 | Myosin, Heavy Chain 11, Smooth Muscle |
| MYH2 | Myosin, Heavy Chain 2, Skeletal Muscle, Adult |
| MYH7 | Myosin, Heavy Chain 7, Cardiac Muscle, Beta |
| MYH9 | Myosin, Heavy Chain 9, Nonmuscle |
| NEB | Nebulin |
| NID1 | Nidogen 1 |
| NID2 | Nidogen 2 |
| OGN | Osteoglycin |
| PALLD | Palladin, Cytoskeletal-Associated Protein |
| PFN1 | Profilin 1 |
| PLEK | Pleckstrin |
| PRLEP | Prolargin |
| SEPT2 | Septin 2 |
| SMTN | Smoothelin |
| TLN1 | Talin 1 |
| TNS1 | Tensin 1 |
| TTN | Titin |
| TUBA1A | Tubulin, Alpha-1a |
| TUBA4A | Tubulin, Alpha-4a |
| TUBB | Tubulin, Beta |
| TUBB3 | Tubulin, Beta-3 |
| TUBB4A | Tubulin, Beta-4a |
| TUBB6 | Tubulin, Beta-6 |
| VCL | Vinculin |
| VIM | Vimentin |
| VTN | Vitronectin |
References
- Luc, G.; Durand, M.; Collet, D.; Guillemot, F.; Bordenave, L. Esophageal tissue engineering. Expert Rev. Med. Devices 2014, 11, 225–241. [Google Scholar] [CrossRef]
- Tan, J.Y.; Chua, C.K.; Leong, K.F.; Chian, K.S.; Leong, W.S.; Tan, L.P. Esophageal tissue engineering: An in-depth review on scaffold design. Biotechnol. Bioeng. 2012, 109, 1–15. [Google Scholar] [CrossRef]
- Totonelli, G.; Maghsoudlou, P.; Fishman, J.M.; Orlando, G.; Ansari, T.; Sibbons, P.; Birchall, M.A.; Pierro, A.; Eatonz, S.; De Coppi, P. Esophageal tissue engineering: A new approach for esophageal replacement. World J. Gastroenterol. 2012, 18, 6900–6907. [Google Scholar] [CrossRef]
- Kuppan, P.; Sethuraman, S.; Krishnan, U.M. Tissue engineering interventions for esophageal disorders—Promises and challenges. Biotechnol. Adv. 2012, 30, 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Urbani, L.; Camilli, C.; Phylactopoulos, D.E.; Crowley, C.; Natarajan, D.; Scottoni, F.; Maghsoudlou, P.; McCann, C.J.; Pellegata, A.F.; Urciuolo, A.; et al. Multi-stage bioengineering of a layered oesophagus with in vitro expanded muscle and epithelial adult progenitors. Nat. Commun. 2018, 9, 4286. [Google Scholar] [CrossRef]
- Elliott, M.J.; De Coppi, P.; Speggiorin, S.; Roebuck, D.; Butler, C.R.; Samuel, E.; Crowley, C.; McLaren, C.; Fierens, A.; Vondrys, D.; et al. Stem-cell-based; tissue engineered tracheal replacement in a child: A 2-year follow-up study. Lancet 2012, 380, 994–1000. [Google Scholar] [CrossRef]
- Cheung, D.Y.; Duan, B.; Butcher, J.T. Current progress in tissue engineering of heart valves: Multiscale problems; multiscale solutions. Expert Opin. Biol. Ther. 2015, 15, 1155–1172. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, A.; Maxwell, G.P. AlloDerm RTU Integration and Clinical Outcomes When Used for Reconstructive Breast Surgery. Plast. Reconstr. Surg. Glob. Open 2018, 6, e1744. [Google Scholar] [CrossRef]
- Kuo, B.; Urma, D. Esophagus—Anatomy and development. GI Motil. Online 2006. [Google Scholar] [CrossRef]
- Totonelli, G.; Maghsoudlou, P.; Georgiades, F.; Garriboli, M.; Koshy, K.; Turmaine, M.; Ashworth, M.; Sebire, N.J.; Pierro, A.; Eaton, S.; et al. Detergent enzymatic treatment for the development of a natural acellular matrix for oesophageal regeneration. Pediatr. Surg. Int. 2013, 29, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.K. Esophagus tissue engineering: Designing and crafting the components for the “hybrid construct” approach. Eur. J. Pediatr. Surg. 2014, 24, 246–262. [Google Scholar] [CrossRef] [PubMed]
- Badylak, S.; Meurling, S.; Chen, M.; Spievack, A.; Simmons-Byrd, A. Resorbable bioscaffold for esophageal repair in a dog model. J. Pediatr. Surg. 2000, 35, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Urita, Y.; Komuro, H.; Chen, G.; Shinya, M.; Kaneko, S.; Kaneko, M.; Ushida, T. Regeneration of the esophagus using gastric acellular matrix: An experimental study in a rat model. Pediatr. Surg. Int. 2007, 23, 21–26. [Google Scholar] [CrossRef]
- Lopes, M.F.; Cabrita, A.; Ilharco, J.; Pessa, P.; Paiva-Carvalho, J.; Pires, A.; Patrício, J. Esophageal replacement in rat using porcine intestinal submucosa as a patch or a tube-shaped graft. Dis. Esophagus 2006, 19, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Ohki, T.; Yamato, M.; Ota, M.; Takagi, R.; Murakami, D.; Kondo, M.; Sasaki, R.; Namiki, H.; Okano, T.; Yamamoto, M. Prevention of esophageal stricture after endoscopic submucosal dissection using tissue-engineered cell sheets. Gastroenterology 2012, 143, 582–588.e2. [Google Scholar] [CrossRef] [PubMed]
- Badylak, S.F.; Vorp, D.A.; Spievack, A.R.; Simmons-Byrd, A.; Hanke, J.; Freytes, D.O.; Thapa, A.; Gilbert, T.W.; Nieponice, A. Esophageal reconstruction with ECM and muscle tissue in a dog model. J. Surg. Res. 2005, 128, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Arakelian, L.; Kanai, N.; Dua, K.; Durand, M.; Cattan, P.; Ohki, T. Esophageal tissue engineering: From bench to bedside. Ann. N. Y. Acad. Sci. 2018, 1434, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Luc, G.; Charles, G.; Gronnier, C.; Cabau, M.; Kalisky, C.; Meulle, M.; Bareille, R.; Roques, S.; Couraud, L.; Rannou, J.; et al. Decellularized and matured esophageal scaffold for circumferential esophagus replacement: Proof of concept in a pig model. Biomaterials 2018, 175, 1–18. [Google Scholar] [CrossRef]
- Spradling, A.; Drummond-Barbosa, D.; Kai, T. Stem cells find their niche. Nature 2001, 414, 98–104. [Google Scholar] [CrossRef]
- Derda, R.; Li, L.; Orner, B.P.; Lewis, R.L.; Thomson, J.A.; Kiessling, L.L. Defined substrates for human embryonic stem cell growth identified from surface arrays. ACS Chem. Biol. 2007, 2, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Muotri, A.; Gage, F.; Varki, A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat. Med. 2005, 11, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, M.; D’Angelo, E.; Crotti, S.; Sensi, F.; Urbani, L.; Maghin, E.; Burns, A.; De Coppi, P.; Fassan, M.; Rugge, M.; et al. Decellularized colorectal cancer matrix as bioactive microenvironment for in vitro 3D cancer research. J. Cell. Physiol. 2018, 233, 5937–5948. [Google Scholar] [CrossRef] [PubMed]
- Porzionato, A.; Sfriso, M.M.; Pontini, A.; Macchi, V.; Petrelli, L.; Pavan, P.G.; Natali, A.N.; Bassetto, F.; Vindigni, V.; De Caro, R. Decellularized Human Skeletal Muscle as Biologic Scaffold for Reconstructive Surgery. Int. J. Mol. Sci. 2015, 16, 14808–14831. [Google Scholar] [CrossRef] [PubMed]
- Barbon, S.; Stocco, E.; Contran, M.; Facchin, F.; Boscolo-Berto, R.; Todros, S.; Sandrin, D.; Romanato, F.; Pavan, P.; Macchi, V.; et al. Preclinical Development of Bioengineered Allografts Derived from Decellularized Human Diaphragm. Biomedicines 2022, 10, 739. [Google Scholar] [CrossRef]
- De Caro, R.; Macchi, V.; Porzionato, A. Promotion of body donation and use of cadavers in anatomical education at the University of Padova. Anat. Sci. Educ. 2009, 2, 91–92. [Google Scholar] [CrossRef]
- Macchi, V.; Porzionato, A.; Stecco, C.; Tiengo, C.; Parenti, A.; Cestrone, A.; De Caro, R. Body parts removed during surgery: A useful training source. Anat. Sci. Educ. 2011, 4, 151–156. [Google Scholar] [CrossRef]
- Porzionato, A.; Macchi, V.; Stecco, C.; Mazzi, A.; Rambaldo, A.; Sarasin, G.; Parenti, A.; Scipioni, A.; De Caro, R. Quality management of Body Donation Program at the University of Padova. Anat. Sci. Educ. 2012, 5, 264–272. [Google Scholar] [CrossRef]
- Porzionato, A.; Macchi, V.; Stecco, C.; De Caro, R. The body donation program of the University of Padua: Organizing an anatomical biobank for medical education. In New Insights on Biobanks; Caenazzo, L., Ed.; CLEUP: Padova, Italy, 2013; pp. 155–171. ISBN 9788867871216. [Google Scholar]
- Riederer, B.M.; Bolt, S.; Brenner, E.; Bueno-López, J.L.; Circulescu, A.R.M.; Davies, D.C.; De Caro, R.; Gerrits, P.O.; McHanwell, S.; Pais, D.; et al. The legal and ethical framework governing Body Donation in Europe—1st update on current practice. Eur. J. Anat. 2012, 16, 1–21. [Google Scholar]
- Boscolo-Berto, R.; Porzionato, A.; Stecco, C.; Macchi, V.; De Caro, R. Body donation in Italy: Lights and shadows of law No. 10/2020. Clin. Anat. 2020, 33, 950–959. [Google Scholar] [CrossRef]
- Filippi, A.; Dal Sasso, E.; Iop, L.; Armani, A.; Gintoli, M.; Sandri, M.; Gerosa, G.; Romanato, F.; Borile, G. Multimodal label-free ex vivo imaging using a dual-wavelength microscope with axial chromatic aberration compensation. J. Biomed. Opt. 2018, 23, 1–9. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, H.; Yang, H.; Zhang, X.; Li, Z.; Xu, S. Quantitative analysis on collagen morphology in aging skin based on multiphoton microscopy. J. Biomed. Opt. 2011, 16, 040502. [Google Scholar] [CrossRef] [PubMed]
- Borile, G.; Sandrin, D.; Filippi, A.; Anderson, K.I.; Romanato, F. Label-Free Multiphoton Microscopy: Much More Than Fancy Images. Int. J. Mol. Sci. 2021, 22, 2657. [Google Scholar] [CrossRef]
- Naba, A.; Clauser, K.R.; Hynes, R.O. Enrichment of Extracellular Matrix Proteins from Tissues and Digestion into Peptides for Mass Spectrometry Analysis. J. Vis. Exp. 2015, 101, e53057. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Zardin, C.; Iop, L.; Palmosi, T.; Capaldo, P.; Romanato, F.; Gerosa, G.; Bagno, A. Hybrid membranes for the production of blood contacting surfaces: Physicochemical; structural and biomechanical characterization. Biomater. Res. 2021, 25, 26. [Google Scholar] [CrossRef]
- De Angeli, S.; Di Liddo, R.; Buoro, S.; Toniolo, L.; Conconi, M.T.; Belloni, A.S.; Parnigotto, P.P.; Nussdorfer, G.G. New immortalized human stromal cell lines enhancing in vitro expansion of cord blood hematopoietic stem cells. Int. J. Mol. Med. 2004, 13, 363–371. [Google Scholar] [CrossRef]
- Barbon, S.; Stocco, E.; Dalzoppo, D.; Todros, S.; Canale, A.; Boscolo-Berto, R.; Pavan, P.; Macchi, V.; Grandi, C.; De Caro, R.; et al. Halogen-Mediated Partial Oxidation of Polyvinyl Alcohol for Tissue Engineering Purposes. Int. J. Mol. Sci. 2020, 21, 801. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef]
- Ranjit, S.; Dvornikov, A.; Stakic, M.; Hong, S.H.; Levi, M.; Evans, R.M.; Gratton, E. Imaging Fibrosis and Separating Collagens using Second Harmonic Generation and Phasor Approach to Fluorescence Lifetime Imaging. Sci. Rep. 2015, 5, 13378. [Google Scholar] [CrossRef]
- ISO 10993-5 2009; Biological Evaluation of Medical Devices—Part 5: Tests for In Vitro Cytotoxicity. International Organization for Standardization: Geneva, Switzerland, 2009. Available online: https://www.iso.org/obp/ui/#iso:std:iso:10993:-5:en (accessed on 23 March 2022).
- Stocco, E.; Barbon, S.; Dalzoppo, D.; Lora, S.; Sartore, L.; Folin, M.; Parnigotto, P.P.; Grandi, C. Tailored PVA/ECM scaffolds for cartilage regeneration. Biomed. Res. Int. 2014, 2014, 762189. [Google Scholar] [CrossRef]
- Stocco, E.; Barbon, S.; Radossi, P.; Rajendran, S.; Dalzoppo, D.; Bortolami, M.; Bagno, A.; Grandi, F.; Gamba, P.G.; Parnigotto, P.P.; et al. Autologous chondrocytes as a novel source for neo-chondrogenesis in haemophiliacs. Cell Tissue Res. 2016, 366, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Grandi, F.; Stocco, E.; Barbon, S.; Rambaldo, A.; Contran, M.; Fascetti Leon, F.; Gamba, P.; Parnigotto, P.P.; Macchi, V.; De Caro, R.; et al. Composite Scaffolds Based on Intestinal Extracellular Matrices and Oxidized Polyvinyl Alcohol: A Preliminary Study for a New Regenerative Approach in Short Bowel Syndrome. Biomed. Res. Int. 2018, 2018, 7824757. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, M.; Narita, Y.; Kagami, H.; Ohmiya, N.; Itoh, A.; Hirooka, Y.; Niwa, Y.; Ueda, M.; Goto, H. Evaluation of decellularized esophagus as a scaffold for cultured esophageal epithelial cells. J. Biomed. Mater. Res. A 2006, 79, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.A.; Brison, J.; Michel, R.; Brown, B.N.; Castner, D.G.; Badylak, S.F.; Ratner, B.D. The surface molecular functionality of decellularized extracellular matrices. Biomaterials 2011, 32, 137–143. [Google Scholar] [CrossRef]
- Mallis, P.; Chachlaki, P.; Katsimpoulas, M.; Stavropoulos-Giokas, C.; Michalopoulos, E. Optimization of Decellularization Procedure in Rat Esophagus for Possible Development of a Tissue Engineered Construct. Bioengineering 2018, 6, 3. [Google Scholar] [CrossRef]
- Saleh, T.; Ahmed, E.; Yu, L.; Kwak, H.H.; Kang, B.J.; Park, K.M.; Choi, K.Y.; Kim, B.M.; Kang, K.S.; Woo, H.M. Characterization of silver nanoparticle-modified decellularized rat esophagus for esophageal tissue engineering: Structural properties and biocompatibility. J. Biosci. Bioeng. 2019, 128, 613–621. [Google Scholar] [CrossRef]
- Melkonyan, K.; Nakokhov, R.; Rusinova, T.; Yutskevich, Y.; Bykov, I.; Redko, A.; Alekseenko, S. Serum cytokine profile during subcutaneous implantation of the decellularized esophagus matrix in rats. J. Biomater. Appl. 2020, 35, 446–455. [Google Scholar] [CrossRef]
- Eftekharzadeh, S.; Akbarzadeh, A.; Sabetkish, N.; Rostami, M.; Zabolian, A.H.; Hashemi, J.; Tavangar, S.M.; Kajbafzadeh, A.M. Esophagus tissue engineering: From decellularization to in vivo recellularization in two sites. Cell Tissue Bank 2022, 23, 301–312. [Google Scholar] [CrossRef]
- Saldin, L.T.; Klimakz, M.; Hill, R.C.; Cramer, M.C.; Huleihelx, L.; Li, X.; Quidgley-Martin, M.; Cardenas, D.; Keane, T.J.; Londono, R.; et al. The effect of normal; metaplastic; and neoplastic esophageal extracellular matrix upon macrophage activation. J. Immunol. Regen. Med. 2021, 13, 100037. [Google Scholar] [CrossRef]
- Hagen, C.K.; Maghsoudlou, P.; Totonelli, G.; Diemoz, P.C.; Endrizzi, M.; Rigon, L.; Menk, R.H.; Arfelli, F.; Dreossi, D.; Brun, E.; et al. High contrast microstructural visualization of natural acellular matrices by means of phase-based X-ray tomography. Sci. Rep. 2015, 5, 18156. [Google Scholar] [CrossRef]
- Urbani, L.; Maghsoudlou, P.; Milan, A.; Menikou, M.; Hagen, C.K.; Totonelli, G.; Camilli, C.; Eaton, S.; Burns, A.; Olivo, A.; et al. Long-term cryopreservation of decellularised oesophagi for tissue engineering clinical application. PLoS ONE 2017, 12, e0179341. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Xu, X.; Lv, D.; Lu, Y.; Li, J.; Cui, P.; Ma, R.; Luo, X.; Tang, Y.; Zheng, Y. Tissue-engineered esophagus: Recellular esophageal extracellular matrix based on perfusion-decellularized technique and mesenchymal stem cells. Biomed. Mater. 2021, 16, 055017. [Google Scholar] [CrossRef] [PubMed]
- Ackbar, R.; Ainoedhofer, H.; Gugatschka, M.; Saxena, A.K. Decellularized ovine esophageal mucosa for esophageal tissue engineering. Technol. Health Care 2012, 20, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.P.; Gangwar, A.K.; Khangembam, S.D.; Yadav, V.K.; Kumar, R.; Verma, R.K.; Kumar, N. Decellularization of caprine esophagus using fruit pericarp extract of Sapindus mukorossi. Cell Tissue Bank 2022, 23, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.J.; Londono, R.; Carey, R.M.; Carruthers, C.A.; Reing, J.E.; Dearth, C.L.; D’Amore, A.; Medberry, C.J.; Badylak, S.F. Preparation and characterization of a biologic scaffold from esophageal mucosa. Biomaterials 2013, 34, 6729–6737. [Google Scholar] [CrossRef]
- Arakelian, L.; Caille, C.; Faivre, L.; Corté, L.; Bruneval, P.; Shamdani, S.; Flageollet, C.; Albanese, P.; Domet, T.; Jarraya, M.; et al. A clinical-grade acellular matrix for esophageal replacement. J. Tissue Eng. Regen. Med. 2019, 13, 2191–2203. [Google Scholar] [CrossRef]
- Marzaro, M.; Algeri, M.; Tomao, L.; Tedesco, S.; Caldaro, T.; Balassone, V.; Contini, A.C.; Guerra, L.; Federici D’Abriola, G.; Francalanci, P.; et al. Successful muscle regeneration by a homologous microperforated scaffold seeded with autologous mesenchymal stromal cells in a porcine esophageal substitution model. Ther. Adv. Gastroenterol. 2020, 13. [Google Scholar] [CrossRef]
- Nam, H.; Jeong, H.J.; Jo, Y.; Lee, J.Y.; Ha, D.H.; Kim, J.H.; Chung, J.H.; Cho, Y.S.; Cho, D.W.; Lee, S.J.; et al. Multi-layered Free-form 3D Cell-printed Tubular Construct with Decellularized Inner and Outer Esophageal Tissue-derived Bioinks. Sci. Rep. 2020, 10, 7255. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, J.D.; Saldin, L.T.; Sobieski, E.; Quijano, L.M.; Hill, R.C.; Chan, P.G.; Torres, C.; Dziki, J.L.; Cramer, M.C.; Lee, Y.C.; et al. Esophageal extracellular matrix hydrogel mitigates metaplastic change in a dog model of Barrett’s esophagus. Sci. Adv. 2020, 6, eaba4526. [Google Scholar] [CrossRef] [PubMed]
- Nayakawde, N.B.; Methe, K.; Banerjee, D.; Berg, M.; Premaratne, G.U.; Olausson, M. In Vitro Regeneration of Decellularized Pig Esophagus Using Human Amniotic Stem Cells. Biores. Open Access 2020, 9, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Chaitin, H.; Lu, M.L.; Wallace, M.B.; Kang, Y. Development of a Decellularized Porcine Esophageal Matrix for Potential Applications in Cancer Modeling. Cells 2021, 10, 1055. [Google Scholar] [CrossRef]
- Ha, D.H.; Chae, S.; Lee, J.Y.; Kim, J.Y.; Yoon, J.; Sen, T.; Lee, S.W.; Kim, H.J.; Cho, J.H.; Cho, D.W. Therapeutic effect of decellularized extracellular matrix-based hydrogel for radiation esophagitis by 3D printed esophageal stent. Biomaterials 2021, 266, 120477. [Google Scholar] [CrossRef] [PubMed]
- Levenson, G.; Berger, A.; Demma, J.; Perrod, G.; Domet, T.; Arakelian, L.; Bruneval, P.; Broudin, C.; Jarraya, M.; Setterblad, N.; et al. Circumferential esophageal replacement by a decellularized esophageal matrix in a porcine model. Surgery 2022, 171, 384–392. [Google Scholar] [PubMed]
- Sitthisang, S.; Leong, M.F.; Chian, K.S. Perfusion decellularization of porcine esophagus: Study of two processing factors affecting the folded mucosal structure of the esophageal scaffold. J. Biomed. Mater. Res. A 2021, 109, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Mendibil, U.; Ruiz-Hernandez, R.; Retegi-Carrion, S.; Garcia-Urquia, N.; Olalde-Graells, B.; Abarrategi, A. Tissue-Specific Decellularization Methods: Rationale and Strategies to Achieve Regenerative Compounds. Int. J. Mol. Sci. 2020, 21, 5447. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Singh, Y.P.; Mandal, B.B.; Nandi, S.K. Tissue-derived decellularized extracellular matrices toward cartilage repair and regeneration. Methods Cell. Biol. 2020, 157, 185–221. [Google Scholar]
- Ozdemir, B.H.; Aksoy, P.K.; Haberal, A.N.; Demirhan, B.; Haberal, M. Relationship of HLA-DR expression to rejection and mononuclear cell infiltration in renal allograft biopsies. Ren. Fail. 2004, 26, 247–251. [Google Scholar] [PubMed]
- Sarker, M.D.; Naghieh, S.; Sharma, N.K.; Chen, X. 3D biofabrication of vascular networks for tissue regeneration: A report on recent advances. J. Pharm. Anal. 2018, 8, 277–296. [Google Scholar] [PubMed]
- Di Liddo, R.; Aguiari, P.; Barbon, S.; Bertalot, T.; Mandoli, A.; Tasso, A.; Schrenk, S.; Iop, L.; Gandaglia, A.; Parnigotto, P.P.; et al. Nanopatterned acellular valve conduits drive the commitment of blood-derived multipotent cells. Int. J. Nanomed. 2016, 11, 5041–5055. [Google Scholar] [CrossRef]
- Zouhair, S.; Dal Sasso, E.; Tuladhar, S.R.; Fidalgo, C.; Vedovelli, L.; Filippi, A.; Borile, G.; Bagno, A.; Marchesan, M.; De Rossi, G.; et al. A comprehensive comparison of bovine and porcine decellularized pericardia: New insights for surgical applications. Biomolecules 2020, 10, 371. [Google Scholar] [CrossRef] [PubMed]
- Boso, D.; Maghin, E.; Carraro, E.; Giagante, M.; Pavan, P.; Piccoli, M. Extracellular Matrix-Derived Hydrogels as Biomaterial for Different Skeletal Muscle Tissue Replacements. Materials 2020, 13, 2483. [Google Scholar] [CrossRef] [PubMed]
- Maurer, T.; Stoffel, M.H.; Belyaev, Y.; Stiefel, N.G.; Vidondo, B.; Küker, S.; Mogel, H.; Schäfer, B.; Balmer, J. Structural characterization of four different naturally occurring porcine collagen membranes suitable for medical applications. PLoS ONE 2018, 13, e0205027. [Google Scholar] [CrossRef] [PubMed]
- Stocco, E.; Barbon, S.; Lamanna, A.; De Rose, E.; Zamuner, A.; Sandrin, D.; Marsotto, M.; Auditore, A.; Messina, G.M.L.; Licciardello, A.; et al. Bioactivated Oxidized Polyvinyl Alcohol towards Next-Generation Nerve Conduits Development. Polymers 2021, 13, 3372. [Google Scholar] [CrossRef] [PubMed]
- Merna, N.; Robertson, C.; La, A.; George, S.C. Optical imaging predicts mechanical properties during decellularization of cardiac tissue. Tissue Eng. Part C Methods 2013, 19, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Faulk, D.M.; Carruthers, C.A.; Warner, H.J.; Kramer, C.R.; Reing, J.E.; Zhang, L.; D’Amore, A.; Badylak, S.F. The effect of detergents on the basement membrane complex of a biologic scaffold material. Acta Biomater. 2014, 10, 183–193. [Google Scholar] [CrossRef]
- Hwang, J.; San, B.H.; Turner, N.J.; White, L.J.; Faulk, D.M.; Badylak, S.F.; Li, Y.; Yu, S.M. Molecular assessment of collagen denaturation in decellularized tissues using a collagen hybridizing peptide. Acta Biomater. 2017, 53, 268–278. [Google Scholar] [CrossRef]
- González-Andrades, M.; Carriel, V.; Rivera-Izquierdo, M.; Garzón, I.; González-Andrades, E.; Medialdeac, S.; Alaminos, M.; Campos, A. Effects of Detergent-Based Protocols on Decellularization of Corneas with Sclerocorneal Limbus. Evaluation of Regional Differences. Transl. Vis. Sci. Technol. 2015, 4, 13. [Google Scholar] [CrossRef]
- Emami, A.; Talaei-Khozani, T.; Vojdani, Z.; Zarei Fard, N. Comparative assessment of the efficiency of various decellularization agents for bone tissue engineering. J. Biomed. Mater. Res. B Appl. Biomater. 2021, 109, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Abass, A.; Eliasy, A.; Geraghty, B.; Elabd, M.; Hassan, A.; Elsheikh, A. Effect of freezing and thawing on the biomechanical characteristics of porcine ocular tissues. J. Biomech. 2019, 87, 93–99. [Google Scholar] [CrossRef]
- Lin, C.H.; Kao, Y.C.; Ma, H.; Tsay, R.Y. An investigation on the correlation between the mechanical property change and the alterations in composition and microstructure of a porcine vascular tissue underwent trypsin-based decellularization treatment. J. Mech. Behav. Biomed. Mater. 2018, 86, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Grauss, R.W.; Hazekamp, M.G.; Oppenhuizen, F.; van Munsteren, C.J.; Gittenberger-de Groot, A.C.; DeRuiter, M.C. Histological evaluation of decellularised porcine aortic valves: Matrix changes due to different decellularisation methods. Eur. J. Cardio-Thorac. Surg. 2005, 27, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhou, J.; Xu, Y. Study of the in vitro cytotoxicity testing of medical devices. Biomed. Rep. 2015, 3, 617–620. [Google Scholar] [CrossRef] [PubMed]



| Protocol No. | 1 | 2 | 3 |
|---|---|---|---|
| Method | dH2O (overnight at 4 °C) 4% SDC (4 h at RT, under agitation) 2000 kU DNase I in 1 M NaCl (3 h at RT, under agitation) dH2O (overnight at 4 °C) After the last cycle: PBS 1X + 3% Pen/strep (72 h at 4 °C, under agitation) Peracetic acid 0.1 M (1 h at RT, under agitation) | dH2O (24 h at 4 °C) 0.05% Trypsin + 0.02% EDTA in PBS (1 h at 37 °C) 0.002% SDS + 0.8% NH4OH in PBS (72 h at 4 °C, under agitation) dH2O (72 h at 4 °C) | dH2O (24 h at 4 °C) 0.05% Trypsin + 0.02% EDTA in PBS (1 h at 37 °C) 2% TergitolTM + 0.8% NH4OH in PBS (72 h at 4 °C, under agitation) dH2O (72 h at 4 °C) |
| Sample | Weight (g) | Width (cm) | Length (cm) |
|---|---|---|---|
| ETN | 4.27 ± 0.12 | 1.8 ± 0.01 | 2.1 ± 0.14 |
| ET1 | 4.73 ± 0.24 | 2.4 ± 0.05 | 2.1 ± 0.05 |
| ET2 | 4.81 ± 0.11 | 1.9 ± 0.01 | 2.4 ± 0.14 |
| ET3 | 4.95 ± 0.09 | 2.0 ± 0.03 | 2.0 ± 0.07 |
| EPN | 2.06 ± 0.09 | 2.0 ± 0.04 | 2.4 ± 0.06 |
| EP1 | 2.45 ± 0.52 | 1.9 ± 0.01 | 2.9 ± 0.02 |
| EP2 | 2.53 ± 0.34 | 2.1 ± 0.12 | 4.5 ± 0.10 |
| EP3 | 2.69 ± 0.20 | 2.3 ± 0.12 | 4.1 ± 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbon, S.; Biccari, A.; Stocco, E.; Capovilla, G.; D’Angelo, E.; Todesco, M.; Sandrin, D.; Bagno, A.; Romanato, F.; Macchi, V.; et al. Bio-Engineered Scaffolds Derived from Decellularized Human Esophagus for Functional Organ Reconstruction. Cells 2022, 11, 2945. https://doi.org/10.3390/cells11192945
Barbon S, Biccari A, Stocco E, Capovilla G, D’Angelo E, Todesco M, Sandrin D, Bagno A, Romanato F, Macchi V, et al. Bio-Engineered Scaffolds Derived from Decellularized Human Esophagus for Functional Organ Reconstruction. Cells. 2022; 11(19):2945. https://doi.org/10.3390/cells11192945
Chicago/Turabian StyleBarbon, Silvia, Andrea Biccari, Elena Stocco, Giovanni Capovilla, Edoardo D’Angelo, Martina Todesco, Deborah Sandrin, Andrea Bagno, Filippo Romanato, Veronica Macchi, and et al. 2022. "Bio-Engineered Scaffolds Derived from Decellularized Human Esophagus for Functional Organ Reconstruction" Cells 11, no. 19: 2945. https://doi.org/10.3390/cells11192945
APA StyleBarbon, S., Biccari, A., Stocco, E., Capovilla, G., D’Angelo, E., Todesco, M., Sandrin, D., Bagno, A., Romanato, F., Macchi, V., De Caro, R., Agostini, M., Merigliano, S., Valmasoni, M., & Porzionato, A. (2022). Bio-Engineered Scaffolds Derived from Decellularized Human Esophagus for Functional Organ Reconstruction. Cells, 11(19), 2945. https://doi.org/10.3390/cells11192945














