Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
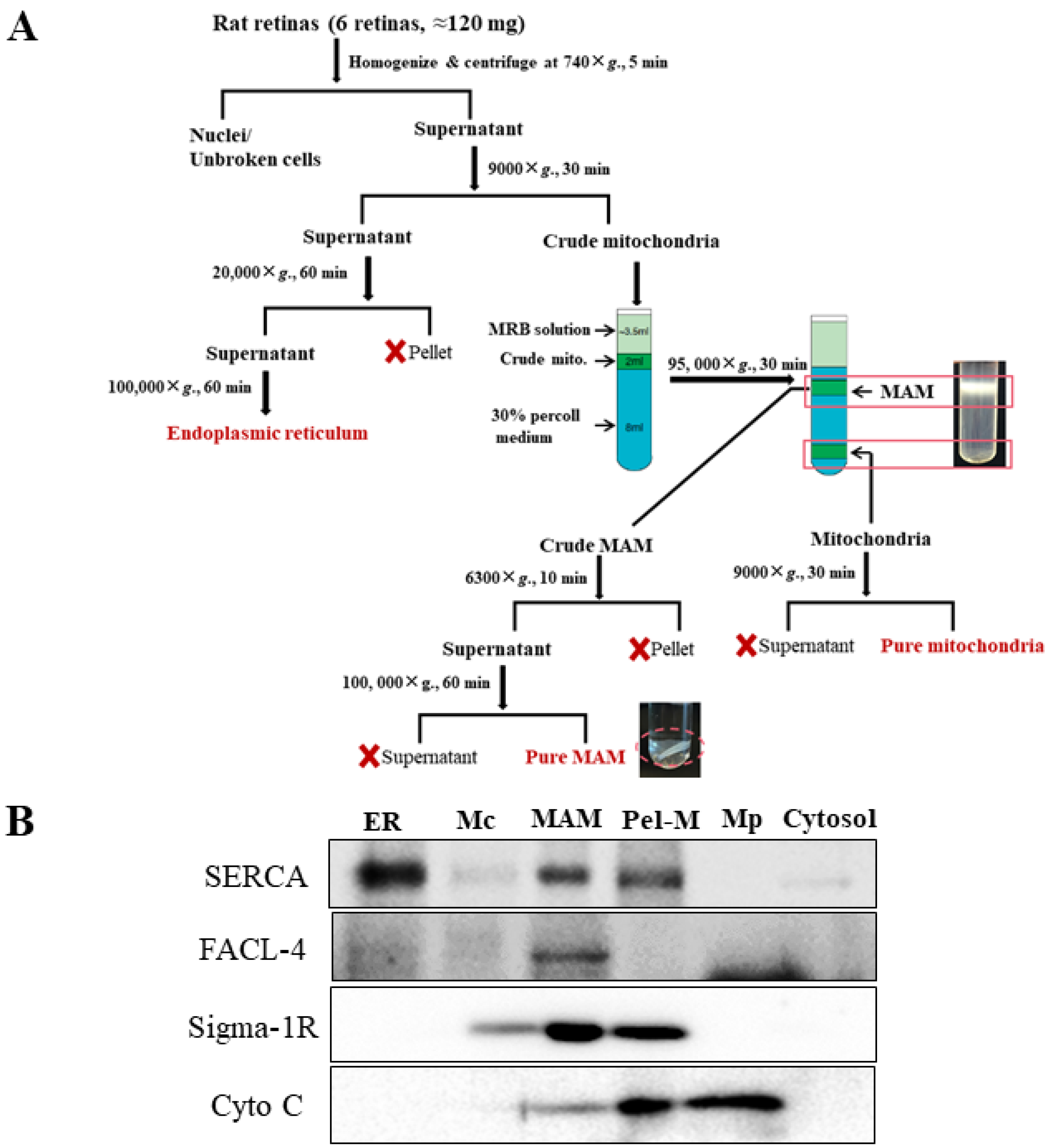
2.2. Isolation of MAM from Rat Retinas
2.3. MAM Sample Preparation
2.4. Liquid Chromatography (LC)-Mass Spectrometry (MS) Analysis
2.5. Protein Identification and Quantification
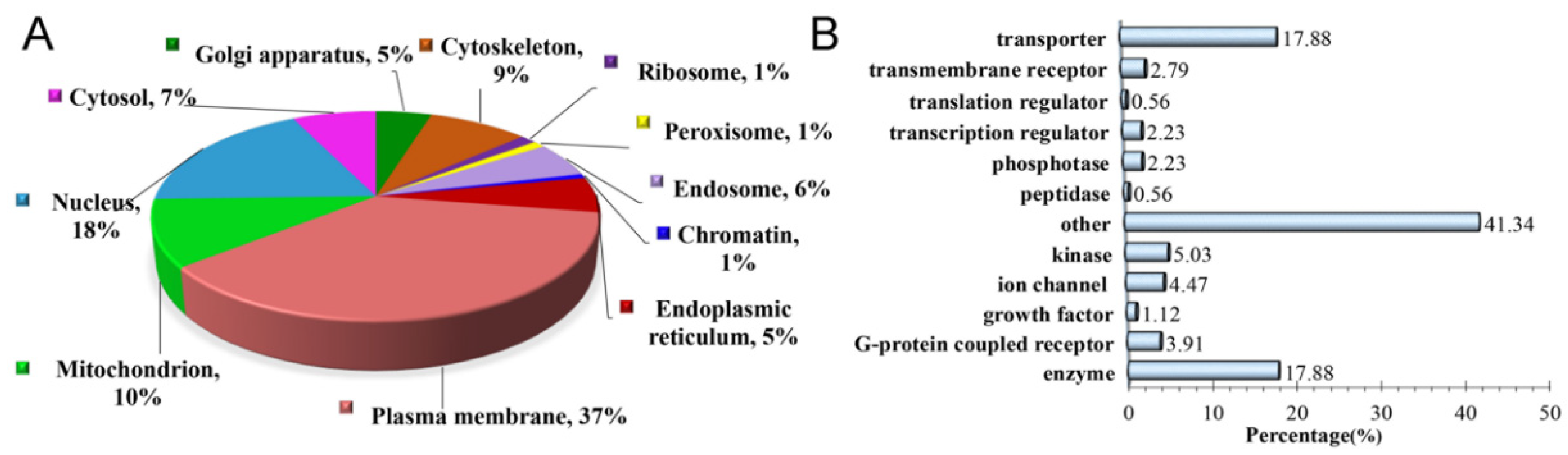
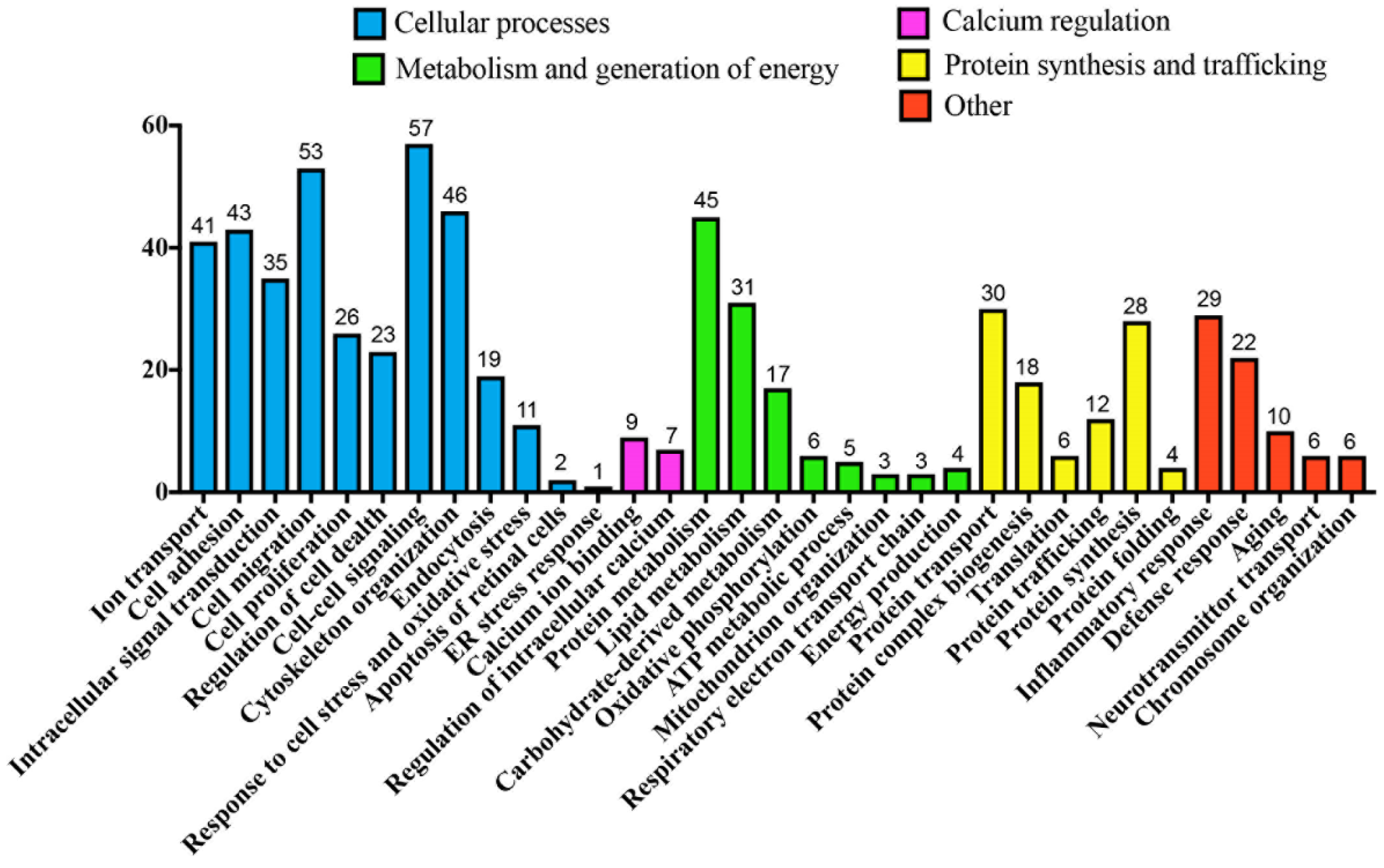
2.6. Bioinformatics Analysis
2.7. Western Blotting
2.8. Statistical Data Analysis
3. Results
3.1. Isolation of MAM from Retinas of Non-Diabetic and STZ-Induced Type 1 Diabetic Rats
3.2. Characterization of Altered MAM Proteins in STZ-Induced Diabetic Rat Retina
- (1)
- a surfactant cocktail-aided precipitation/on-pellet digestion (SEPOD) protocol for proteomics sample preparation;
- (2)
- a trapping nano LC-Orbitrap MS system for peptide separation and MS detection;
- (3)
- a data processing workflow to yield quantification results based on peptide MS1 ion currents.
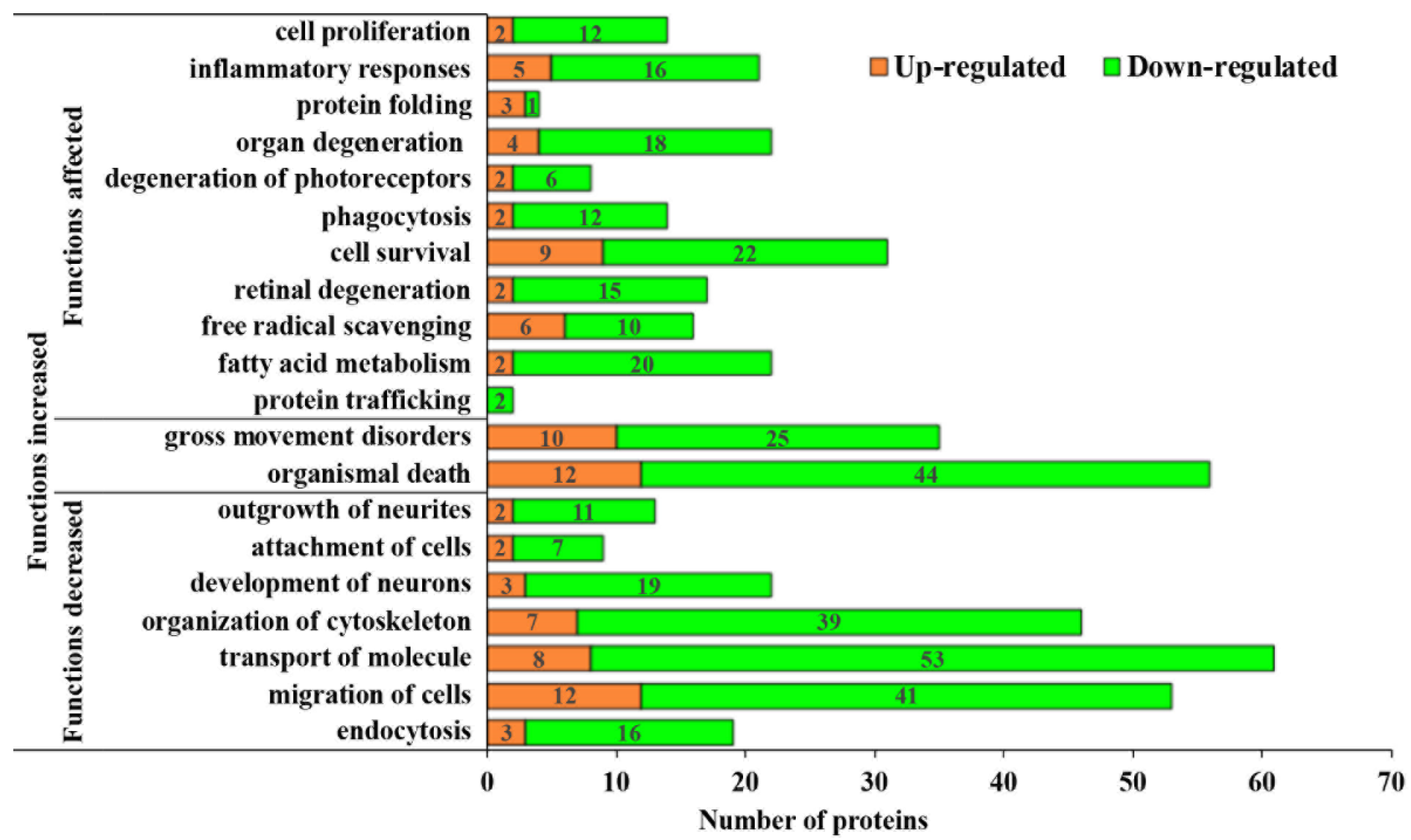
3.3. Pathological Processes Associated with Altered MAM Proteomes in STZ-Induced Diabetic Rat Retina
3.4. Bioinformatic Analyses of MAM Proteins Involved in DR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGC | Automated gain control |
| AQP4 | Aquaporin 4 |
| ATP | Adenosine triphosphate |
| CA14 | Carbonic anhydrase 14 |
| CRB1 | Crumbs cell polarity complex component 1 |
| CRYAA | Crystallin alpha A |
| CRYAA2 | Crystallin alpha A2 |
| CRYAB | Crystallin alpha B |
| DAVID | Database for annotation, visualization and integrated discovery |
| DM | Diabetes mellitus |
| DR | Diabetic retinopathy |
| DTT | Dithiothreitol |
| EDN1 | Endothelin 1 |
| EGTA | Egtazic acid |
| ER | Endoplasmic reticulum |
| FACL4 | Acyl-CoA synthetase 4 |
| FDR | False positive rate |
| FOXO1 | Forkhead box O1 |
| GFAP | Glial fibrillary acidic protein |
| GO | Gene ontology |
| GPX4 | Glutathione peroxidase 4 |
| HCD | High-energy collision dissociation |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HSPB1 | Heat shock protein beta-1 |
| IAM | Iodoacetamide |
| IL-1β | Interleukin 1 beta |
| IPA | Ingenuity pathway analysis |
| JUN | Jun proto-oncogene |
| KCNJ13 | Potassium inwardly rectifying channel subfamily J member 13 |
| LC-MS | Liquid chromatography-mass spectrometry |
| LGALS3 | Galectin 3 |
| MAF | MAF bZIP transcription factor |
| MAM | Mitochondria-associated ER membrane |
| MERTK | MER proto-oncogene, tyrosine kinase |
| MRB | Mitochondrial re-suspension buffer |
| NDM | Non-diabetes mellitus |
| NFκBIA | Nuclear factor kappa-B inhibitor alpha |
| PCDH15 | Protocadherin related 15 |
| PDGFR | Platelet-derived growth factor receptor |
| PERK | Protein kinase R (PKR)-like endoplasmic reticulum kinase |
| PMSF | Phenylmethylsulfonyl fluoride |
| PPAR | Peroxisome proliferator-activated receptor |
| PTEN | Phosphatase and tensin homolog |
| RBP3 | Retinol binding protein 3 |
| RD3 | Retinal degeneration 3 |
| RhoGDI | Rho GDP-dissociation inhibitor |
| RIPA | Radio immune precipitation assay |
| ROS | Reactive oxygen species |
| RP2 | Retinitis pigmentosa 2 |
| RP3 | Retinitis pigmentosa 3 |
| RPE65 | Retinal pigment epithelium-specific 65 kDa |
| RPLC-MS | Reversed phase liquid chromatography-mass spectrometry |
| RS1 | Retinoschisin 1 |
| SDS | Sodium dodecyl-sulfate |
| SDS-PAGE | Sodium dodecyl-sulfate polyacrylamide gel electrophoresis |
| SERCA | Sarco/endoplasmic reticulum Ca2+-ATPase |
| STAT4 | Signal transducer and activator of transcription 4 |
| STEAP4 | Six-transmembrane epithelial antigen of the prostate 4 |
| STZ | Streptozotocin |
| SYVN1 | Synoviolin 1 |
| Tris-FA | Tris-formic acid |
| VCAM1 | Vascular cell adhesion molecule 1 |
| VEGF | Vascular endothelial growth factor |
References
- Fong, D.S.; Aiello, L.; Gardner, T.W.; King, G.L.; Blankenship, G.; Cavallerano, J.D.; Ferris, F.L., 3rd; Klein, R. American Diabetes A: Retinopathy in diabetes. Diabetes Care 2004, 27 (Suppl. 1), S84–S87. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.; Mateus, C.; Melo, P.; Figueira, J.; Cunha-Vaz, J.; Castelo-Branco, M. Pure neuroretinal dysfunction in diabetic retinopathy occurring prior to endothelial and vascular damage. Acta Ophthalmol. 2013, 91, S252. [Google Scholar] [CrossRef]
- Solomon, S.D.; Chew, E.; Duh, E.J.; Sobrin, L.; Sun, J.K.; VanderBeek, B.L.; Wykoff, C.C.; Gardner, T.W. Diabetic Retinopathy: A Position Statement by the American Diabetes Association. Diabetes Care 2017, 40, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Simo, R.; Stitt, A.W.; Gardner, T.W. Neurodegeneration in diabetic retinopathy: Does it really matter? Diabetologia 2018, 61, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.K.; Hosking, B.M.; Holeniewska, J.; Kubala, E.C.; Lundh von Leithner, P.; Gardner, P.J.; Foxton, R.H.; Shima, D.T. BTBR ob/ob mouse model of type 2 diabetes exhibits early loss of retinal function and retinal inflammation followed by late vascular changes. Diabetologia 2018, 61, 2422–2432. [Google Scholar] [CrossRef]
- Joltikov, K.A.; Sesi, C.A.; de Castro, V.M.; Davila, J.R.; Anand, R.; Khan, S.M.; Farbman, N.; Jackson, G.R.; Johnson, C.A.; Gardner, T.W. Disorganization of Retinal Inner Layers (DRIL) and Neuroretinal Dysfunction in Early Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5481–5486. [Google Scholar] [CrossRef]
- Sugimoto, M.; Sasoh, M.; Ido, M.; Wakitani, Y.; Takahashi, C.; Uji, Y. Detection of early diabetic change with optical coherence tomography in type 2 diabetes mellitus patients without retinopathy. Ophthalmologica 2005, 219, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Yonemura, D.; Aoki, T.; Tsuzuki, K. Electroretinogram in Diabetic Retinopathy. Arch. Ophthalmol. 1962, 68, 19–24. [Google Scholar] [CrossRef]
- Sokol, S.; Moskowitz, A.; Skarf, B.; Evans, R.; Molitch, M.; Senior, B. Contrast Sensitivity in Diabetics with and without Background Retinopathy. Arch. Ophthalmol. 1985, 103, 51–54. [Google Scholar] [CrossRef]
- Sun, J.K.; Lin, M.M.; Lammer, J.; Prager, S.; Sarangi, R.; Silva, P.S.; Aiello, L.P. Disorganization of the retinal inner layers as a predictor of visual acuity in eyes with center-involved diabetic macular edema. JAMA Ophthalmol. 2014, 132, 1309–1316. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.K.; Radwan, S.H.; Soliman, A.Z.; Lammer, J.; Lin, M.M.; Prager, S.G.; Silva, P.S.; Aiello, L.B.; Aiello, L.P. Neural Retinal Disorganization as a Robust Marker of Visual Acuity in Current and Resolved Diabetic Macular Edema. Diabetes 2015, 64, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Barber, A.J.; Lieth, E.; Khin, S.A.; Antonetti, D.A.; Buchanan, A.G.; Gardner, T.W. Neural apoptosis in the retina during experimental and human diabetes. Early onset and effect of insulin. J. Clin. Investig. 1998, 102, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A. Diabetic retinopathy: Mitochondrial dysfunction and retinal capillary cell death. Antioxid. Redox Signal. 2005, 7, 1581. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.J.; Yu, Q.; Wang, M.; Zhang, S.X. Endoplasmic Reticulum Stress is implicated in Retinal Inflammation and Diabetic Retinopathy. FEBS Lett. 2009, 583, 1521–1527. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.J.; Yu, Q.; Chen, K.; Mahadev, K.; Zhang, S.X. Inhibition of reactive oxygen species by Lovastatin downregulates vascular endothelial growth factor expression and ameliorates blood-retinal barrier breakdown in db/db mice: Role of NADPH oxidase. Diabetes 2010, 59, 1528–1538. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, J.; Chen, Y.; Wang, J.J.; Ratan, R.; Zhang, S.X. Activation of Endoplasmic Reticulum Stress by Hyperglycemia Is Essential for Muller Cell-Derived Inflammatory Cytokine Production in Diabetes. Diabetes 2012, 61, 492–504. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.J.; Li, J.; Hosoya, K.I.; Ratan, R.; Townes, T.; Zhang, S.X. Activating transcription factor 4 mediates hyperglycaemia-induced endothelial inflammation and retinal vascular leakage through activation of STAT3 in a mouse model of type 1 diabetes. Diabetologia 2012, 55, 2533–2545. [Google Scholar] [CrossRef]
- Zhang, S.X.; Sanders, E.; Fliesler, S.J.; Wang, J.J. Endoplasmic reticulum stress and the unfolded protein responses in retinal degeneration. Exp. Eye Res. 2014, 125C, 30–40. [Google Scholar] [CrossRef]
- Huang, C.; Wang, J.J.; Ma, J.H.; Jin, C.; Yu, Q.; Zhang, S.X. Activation of the UPR Protects Against Cigarette Smoke-induced RPE Apoptosis through Up-Regulation of Nrf. J. Biol. Chem. 2015, 290, 5367–5380. [Google Scholar] [CrossRef]
- Huang, C.; Wang, J.J.; Jing, G.; Li, J.; Jin, C.; Yu, Q.; Falkowski, M.W.; Zhang, S.X. Erp29 Attenuates Cigarette Smoke Extract–Induced Endoplasmic Reticulum Stress and Mitigates Tight Junction Damage in Retinal Pigment Epithelial CellsProtective Role of ERp29 in RPE Cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 6196–6207. [Google Scholar] [CrossRef]
- Rusiñol, A.E.; Cui, Z.; Chen, M.H.; Vance, J.E. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994, 269, 27494–27502. [Google Scholar] [CrossRef]
- Betz, C.; Stracka, D.; Prescianotto-Baschong, C.; Frieden, M.; Demaurex, N.; Hall, M.N. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA 2013, 110, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Bononi, A.; Bonora, M.; Marchi, S.; Missiroli, S.; Poletti, F.; Giorgi, C.; Pandolfi, P.P.; Pinton, P. Identification of PTEN at the ER and MAMs and its regulation of Ca(2+) signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ. 2013, 20, 1631–1643. [Google Scholar] [CrossRef] [PubMed]
- Verfaillie, T.; Rubio, N.; Garg, A.D.; Bultynck, G.; Rizzuto, R.; Decuypere, J.P.; Piette, J.; Linehan, C.; Gupta, S.; Samali, A.; et al. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012, 19, 1880–1891. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.H.; Shen, S.; Wang, J.J.; He, Z.; Poon, A.; Li, J.; Qu, J.; Zhang, S.X. Comparative Proteomic Analysis of the Mitochondria-associated ER Membrane (MAM) in a Long-term Type 2 Diabetic Rodent Model. Sci. Rep. 2017, 7, 2062. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Shen, S.; Li, J.; Hu, Q.; Nie, L.; Tu, C.; Wang, X.; Poulsen, D.J.; Orsburn, B.C.; Wang, J.; et al. IonStar enables high-precision, low-missing-data proteomics quantification in large biological cohorts. Proc. Natl. Acad. Sci. USA 2018, 115, E4767–E4776. [Google Scholar] [CrossRef]
- Wang, X.; Jin, L.; Hu, C.; Shen, S.; Qian, S.; Ma, M.; Zhu, X.; Li, F.; Wang, J.; Tian, Y.; et al. Ultra-High-Resolution IonStar Strategy Enhancing Accuracy and Precision of MS1-Based Proteomics and an Extensive Comparison with State-of-the-Art SWATH-MS in Large-Cohort Quantification. Anal. Chem. 2021, 93, 4884–4893. [Google Scholar] [CrossRef]
- Gastinger, M.J.; Singh, R.S.J.; Barber, A.J. Loss of Cholinergic and Dopaminergic Amacrine Cells in Streptozotocin-Diabetic Rat and Ins2Akita-Diabetic Mouse Retinas. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3143–3150. [Google Scholar] [CrossRef]
- Zhang, S.X.; Wang, J.J.; Gao, G.; Shao, C.; Mott, R.; Ma, J.-X. Pigment epithelium-derived factor (PEDF) is an endogenous anti-inflammatory factor. FASEB J. 2006, 20, 323–325. [Google Scholar] [CrossRef]
- Pardue, M.T.; Barnes, C.S.; Kim, M.K.; Aung, M.H.; Amarnath, R.; Olson, D.E.; Thulé, P.M. Rodent Hyperglycemia-Induced Inner Retinal Deficits are Mirrored in Human Diabetes. Transl. Vis. Sci. Technol. 2014, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Obrosova, I.G.; Drel, V.R.; Kumagai, A.K.; Szábo, C.; Pacher, P.; Stevens, M.J. Early diabetes-induced biochemical changes in the retina: Comparison of rat and mouse models. Diabetologia 2006, 49, 2525–2533. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, M.R.; Giorgi, C.; Lebiedzinska, M.; Duszynski, J.; Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009, 4, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Poston, C.N.; Krishnan, S.C.; Bazemore-Walker, C.R. In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J. Proteom. 2013, 79, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Horner, S.M.; Liu, H.M.; Park, H.S.; Briley, J.; Gale, M. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. USA 2011, 108, 14590–14595. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.; Tsai, S.Y.; Su, T.P. Detection of Isolated Mitochondria-Associated ER Membranes Using the Sigma-1 Receptor. Methods Mol. Biol. 2016, 1376, 133–140. [Google Scholar] [PubMed]
- Xiao, F.; Zhang, J.; Zhang, C.; An, W. Hepatic stimulator substance inhibits calcium overflow through the mitochondria-associated membrane compartment during nonalcoholic steatohepatitis. Lab. Investig. 2017, 97, 289–301. [Google Scholar] [CrossRef]
- Sala-Vila, A.; Navarro-Lérida, I.; Sánchez-Alvarez, M.; Bosch, M.; Calvo, C.; López, J.A.; Calvo, E.; Ferguson, C.; Giacomello, M.; Serafini, A.; et al. Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 2016, 6, 27351. [Google Scholar] [CrossRef]
- Kolesnikov, A.V.; Tang, P.H.; Kefalov, V.J. Examining the Role of Cone-expressed RPE65 in Mouse Cone Function. Sci. Rep. 2018, 8, 14201. [Google Scholar] [CrossRef]
- Flaxel, C.J.; Jay, M.; Thiselton, D.L.; Nayudu, M.; Hardcastle, A.J.; Wright, A.; Bird, A.C. Difference between RP2 and RP3 phenotypes in X linked retinitis pigmentosa. Br. J. Ophthalmol. 1999, 83, 1144–1148. [Google Scholar] [CrossRef]
- Al-Khersan, H.; Shah, K.P.; Jung, S.C.; Rodriguez, A.; Madduri, R.K.; Grassi, M.A. A novel MERTK mutation causing retinitis pigmentosa. Graefes Arch. Clin. Exp. Ophthalmol. 2017, 255, 1613–1619. [Google Scholar] [CrossRef]
- Bujakowska, K.; Audo, I.; Mohand-Said, S.; Lancelot, M.E.; Antonio, A.; Germain, A.; Leveillard, T.; Letexier, M.; Saraiva, J.P.; Lonjou, C.; et al. CRB1 mutations in inherited retinal dystrophies. Hum. Mutat. 2012, 33, 306–315. [Google Scholar] [CrossRef]
- Preising, M.N.; Hausotter-Will, N.; Solbach, M.C.; Friedburg, C.; Ruschendorf, F.; Lorenz, B. Mutations in RD3 are associated with an extremely rare and severe form of early onset retinal dystrophy. Investig. Ophthalmol. Vis. Sci. 2012, 53, 3463–3472. [Google Scholar] [CrossRef] [PubMed]
- Pimenides, D.; George, N.D.; Yates, J.R.; Bradshaw, K.; Roberts, S.A.; Moore, A.T.; Trump, D. X-linked retinoschisis: Clinical phenotype and RS1 genotype in 86 UK patients. J. Med. Genet 2005, 42, e35. [Google Scholar] [CrossRef]
- Akasaki, Y.; Alvarez-Garcia, O.; Saito, M.; Carames, B.; Iwamoto, Y.; Lotz, M.K. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014, 66, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; He, H.; Ma, Q.S.; Zhang, Y.; Zhu, Y.; Wan, X.; Wang, F.W.; Wang, S.S.; Liu, L.; Li, B. Experimental study of the protective effects of SYVN1 against diabetic retinopathy. Sci. Rep. 2015, 5, 14036. [Google Scholar] [CrossRef] [PubMed]
- Zervou, M.I.; Mamoulakis, D.; Panierakis, C.; Boumpas, D.T.; Goulielmos, G.N. STAT4: A risk factor for type 1 diabetes? Hum. Immunol. 2008, 69, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Rajan, M.R.; Nyman, E.; Brannmark, C.; Olofsson, C.S.; Stralfors, P. Inhibition of FOXO1 transcription factor in primary human adipocytes mimics the insulin-resistant state of type 2 diabetes. Biochem. J. 2018, 475, 1807–1820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guo, Z.M. Multiple functions of Maf in the regulation of cellular development and differentiation. Diabetes Metab. Res. Rev. 2015, 31, 773–778. [Google Scholar] [CrossRef]
- Mendonca, H.R.; Carvalho, J.N.A.; Abreu, C.A.; Mariano de Souza Aguiar Dos Santos, D.; Carvalho, J.R.; Marques, S.A.; da Costa Calaza, K.; Martinez, A.M.B. Lack of Galectin-3 attenuates neuroinflammation and protects the retina and optic nerve of diabetic mice. Brain Res. 2018, 1700, 126–137. [Google Scholar] [CrossRef]
- Collin, G.B.; Gogna, N.; Chang, B.; Damkham, N.; Pinkney, J.; Hyde, L.F.; Stone, L.; Naggert, J.K.; Nishina, P.M.; Krebs, M.P. Mouse Models of Inherited Retinal Degeneration with Photoreceptor Cell Loss. Cells 2020, 9, 931. [Google Scholar] [CrossRef] [Green Version]
- Canning, P.; Glenn, J.V.; Hsu, D.K.; Liu, F.T.; Gardiner, T.A.; Stitt, A.W. Inhibition of advanced glycation and absence of galectin-3 prevent blood-retinal barrier dysfunction during short-term diabetes. Exp. Diabetes Res. 2007, 2007, 51837. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Xu, G.; Fan, J.; Witt, R.E.; Da, C. High-salt loading exacerbates increased retinal content of aquaporins AQP1 and AQP4 in rats with diabetic retinopathy. Exp. Eye Res. 2009, 89, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Oosuka, S.; Kida, T.; Oku, H.; Horie, T.; Morishita, S.; Fukumoto, M.; Sato, T.; Ikeda, T. Effects of an Aquaporin 4 Inhibitor, TGN-020, on Murine Diabetic Retina. Int. J. Mol. Sci. 2020, 21, 2324. [Google Scholar] [CrossRef] [PubMed]
- Da, T.; Verkman, A.S. Aquaporin-4 gene disruption in mice protects against impaired retinal function and cell death after ischemia. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4477–4483. [Google Scholar] [CrossRef]
- Kase, S.; Ishida, S.; Rao, N.A. Increased expression of alphaA-crystallin in human diabetic eye. Int. J. Mol. Med. 2011, 28, 505–511. [Google Scholar]
- Fort, P.E.; Freeman, W.M.; Losiewicz, M.K.; Singh, R.S.; Gardner, T.W. The retinal proteome in experimental diabetic retinopathy: Up-regulation of crystallins and reversal by systemic and periocular insulin. Mol. Cell Proteom. 2009, 8, 767–779. [Google Scholar] [CrossRef]
- Jadeja, S.; Mort, R.L.; Keighren, M.; Hart, A.W.; Joynson, R.; Wells, S.; Potter, P.K.; Jackson, I.J. A CNS-specific hypomorphic Pdgfr-beta mutant model of diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3569–3578. [Google Scholar] [CrossRef]
- Arboleda-Velasquez, J.F.; Valdez, C.N.; Marko, C.K.; D’Amore, P.A. From pathobiology to the targeting of pericytes for the treatment of diabetic retinopathy. Curr. Diab. Rep. 2015, 15, 573. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, Q.; Wang, H.; Chen, C.; Luo, D. TRIM46 contributes to high glucose-induced ferroptosis and cell growth inhibition in human retinal capillary endothelial cells by facilitating GPX4 ubiquitination. Exp. Cell Res. 2021, 407, 112800. [Google Scholar] [CrossRef]
- Perez, R.V.; Machado, C.G.; Santos-Bezerra, D.P.; Admoni, S.N.; Patente, T.A.; Monteiro, M.B.; Cavaleiro, A.M.; Queiroz, M.S.; Nery, M.; Correa-Giannella, M.L. Allelic variations in genes belonging to glutathione system increase proliferative retinopathy risk in type 1 diabetes individuals. Gene 2019, 703, 120–124. [Google Scholar] [CrossRef]
- VanGuilder, H.D.; Bixler, G.V.; Kutzler, L.; Brucklacher, R.M.; Bronson, S.K.; Kimball, S.R.; Freeman, W.M. Multi-modal proteomic analysis of retinal protein expression alterations in a rat model of diabetic retinopathy. PLoS ONE 2011, 6, e16271. [Google Scholar] [CrossRef] [PubMed]
- Ruebsam, A.; Dulle, J.E.; Myers, A.M.; Sakrikar, D.; Green, K.M.; Khan, N.W.; Schey, K.; Fort, P.E. A specific phosphorylation regulates the protective role of alphaA-crystallin in diabetes. JCI Insight 2018, 3, e97919. [Google Scholar] [CrossRef] [PubMed]
- Nath, M.; Shan, Y.; Myers, A.M.; Fort, P.E. HspB4/alphaA-Crystallin Modulates Neuroinflammation in the Retina via the Stress-Specific Inflammatory Pathways. J. Clin. Med. 2021, 10, 2384. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Sureda, A.; Jaña, F.; Urra, H.; Durand, S.; Mortenson, D.E.; Sagredo, A.; Bustos, G.; Hazari, Y.; Ramos-Fernández, E.; Sassano, M.L.; et al. Non-canonical function of IRE1α determines mitochondria-associated endoplasmic reticulum composition to control calcium transfer and bioenergetics. Nat. Cell Biol. 2019, 21, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Mignard, V.; Dubois, N.; Lanoé, D.; Joalland, M.P.; Oliver, L.; Pecqueur, C.; Heymann, D.; Paris, F.; Vallette, F.M.; Lalier, L. Sphingolipid distribution at mitochondria-associated membranes (MAMs) upon induction of apoptosis. J. Lipid Res. 2020, 61, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
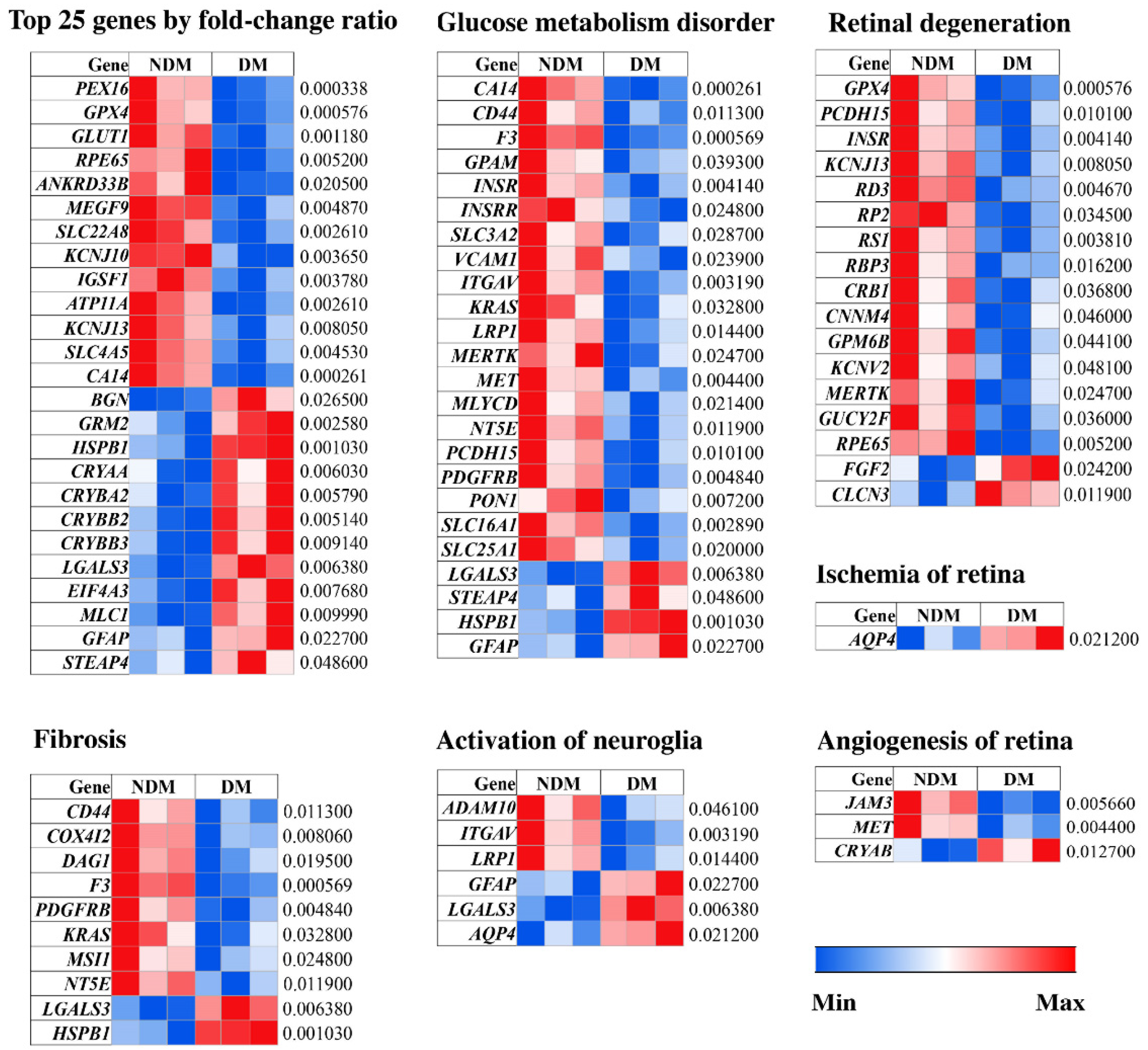
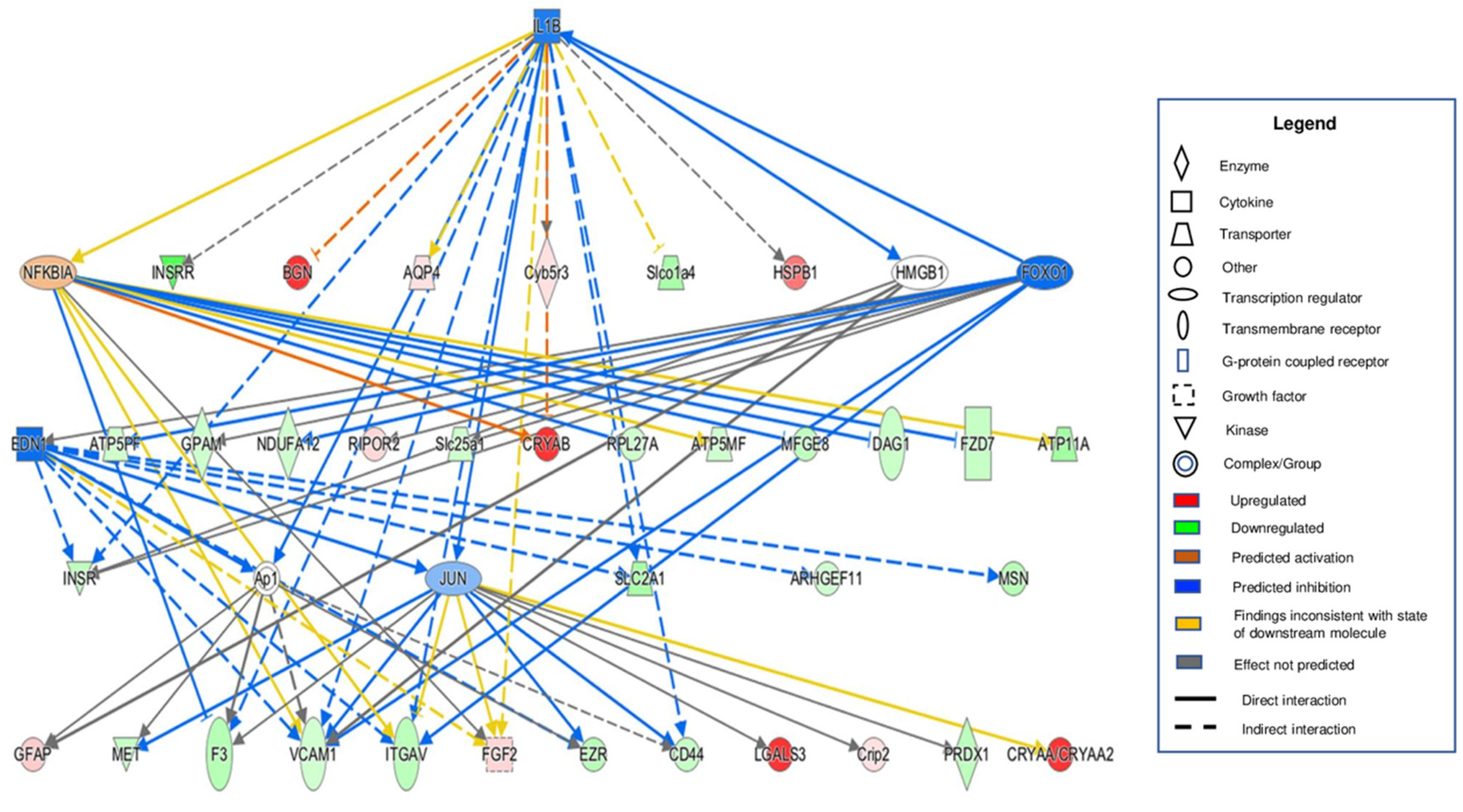
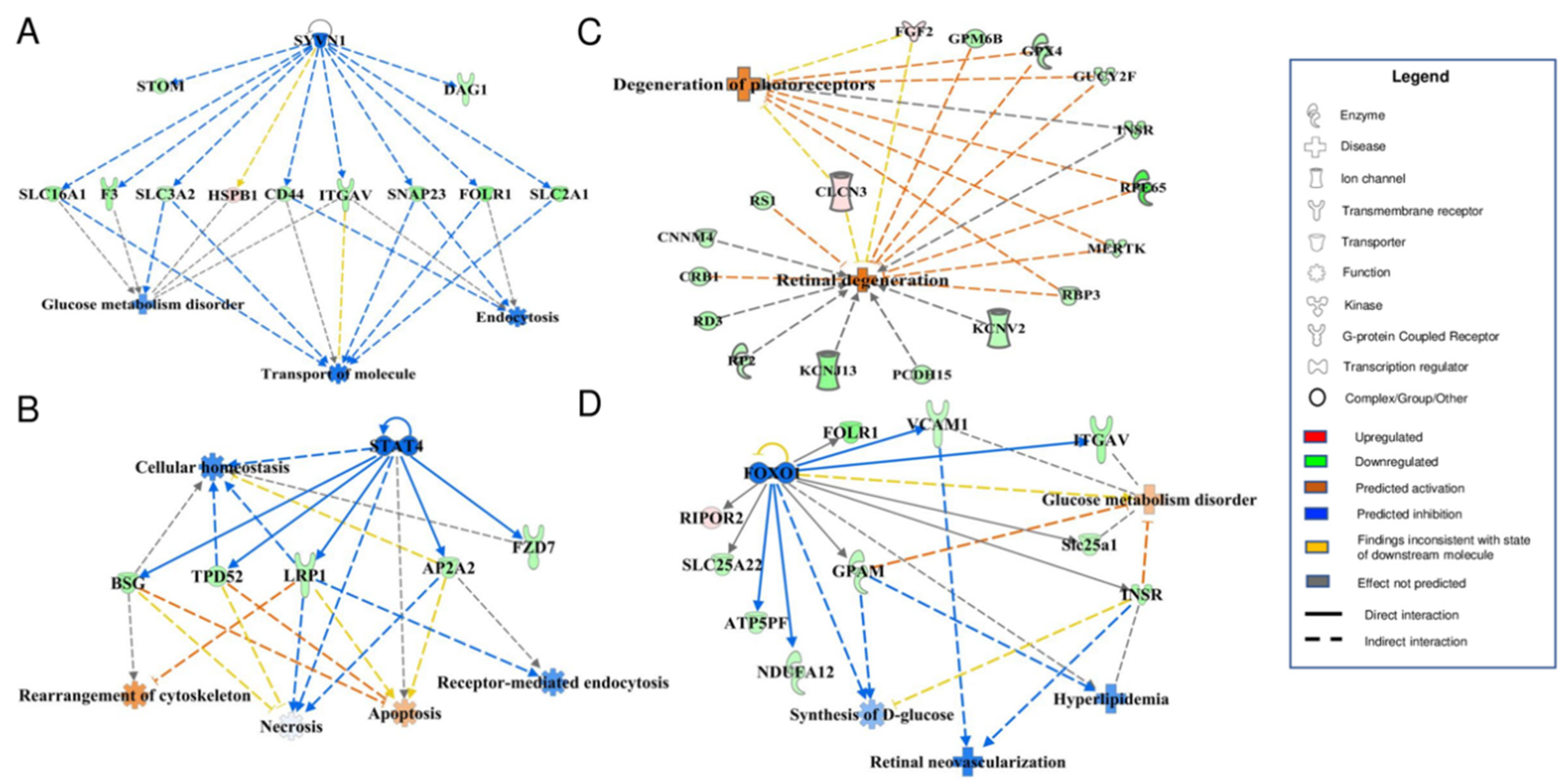
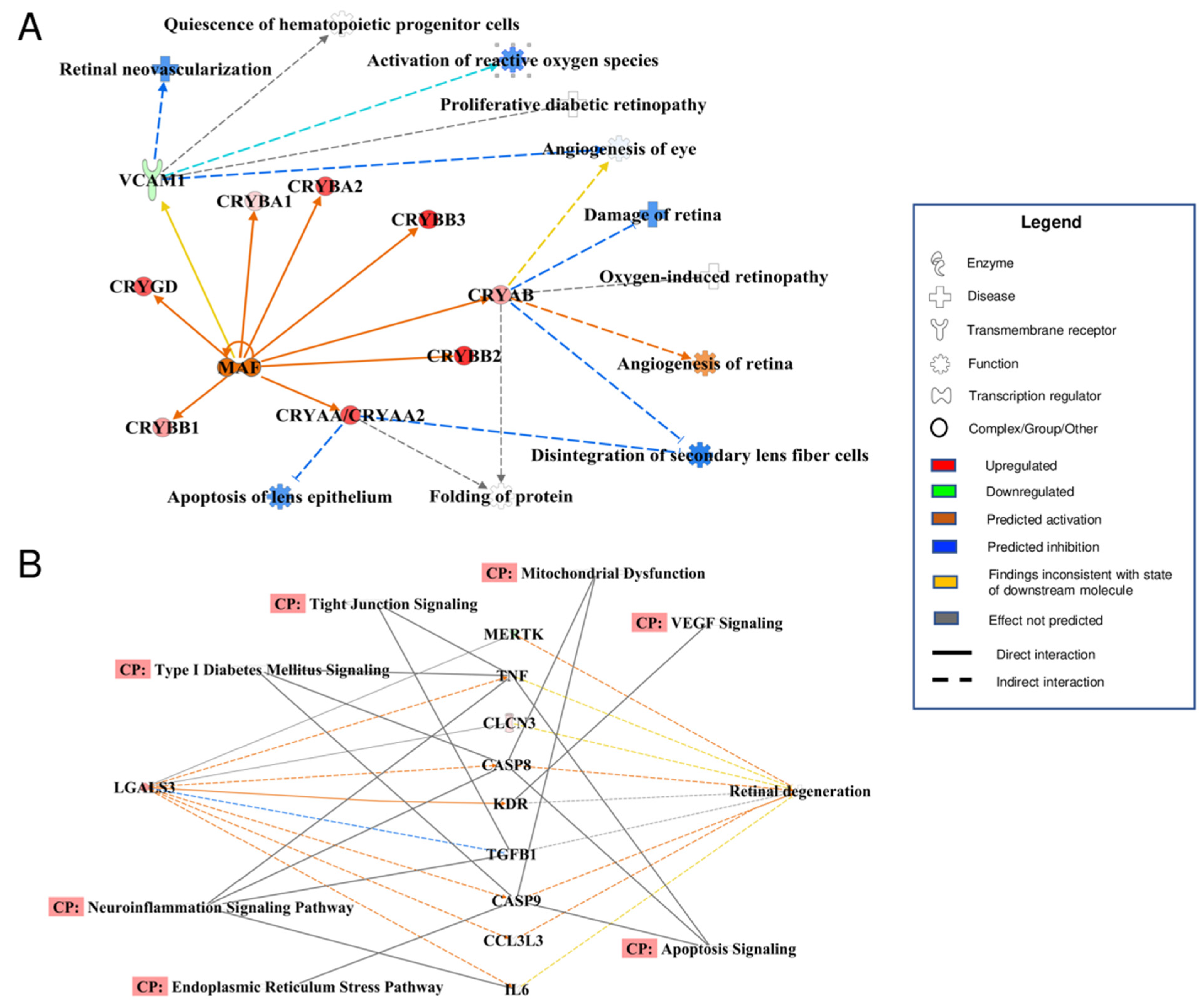
| NDM | DM | p-Value | |
|---|---|---|---|
| n | 9 | 9 | |
| Body weight (g) | 879.4 ± 47.6 | 411.3 ± 14.7 | 6.54 × 10−8 |
| Blood glucose (mg/dL) | 111.0 ± 4.2 | 377.3 ± 36.4 | 1.9 × 10−6 |
| Biological Processes | Differentially Expressed MAM Proteins Observed Only in Mouse Brain with T2DM | Common Proteins | Differentially Expressed MAM Proteins Observed Only in Rat Retina with T1DM |
|---|---|---|---|
| Retinal degeneration | ABCA4, ATP1B2, GNAT1, GNGT1, GRK1, GUCY2D, PDE6B, PRPH2, RHO, SLC1A3, SLC6A6 | GUCY2F, RS1 | CLCN3, CNNM4, GPM6B, RP2, INSR, FGF2, RPE65, RD3, GPX4, MERTK, PCDH15, RBP3, KCNV2, CRB1, KCNJ13 |
| Transport of amino acids | ARL6IP5, SLC17A7, SLC1A2, SLC1A3, SLC38A3, SLC6A9, SLC7A5, SLC32A1, | SLC3A2, KCNJ10, SLC7A8 | SLC6A20, SLC7A10, INSR, SLCO1C1, FGF2, Slco1a4, SLC6A13, SLC25A22 |
| Ingenuity Canonical Pathways | −Log (p-Value) | Z-Score | Down-Regulated | Up-Regulated |
|---|---|---|---|---|
| Actin Cytoskeleton Signaling | 1.94 | −2.236 | 59/227 (26%) | 17/227 (7%) |
| Integrin Signaling | 1.45 | −2.236 | 55/219 (25%) | 18/219 (8%) |
| Leukocyte Extravasation Signaling | 2.09 | −2.236 | 40/211 (19%) | 9/211 (4%) |
| EIF2 Signaling | 1.99 | −2 | 87/221 (39%) | 9/221 (4%) |
| Glioblastoma Multiform Signaling | 1.36 | −2 | 33/160 (21%) | 7/160 (4%) |
| Glioma Invasiveness Signaling | 2.57 | −2 | 17/70 (24%) | 2/70 (3%) |
| IL-8 Signaling | 2.23 | −2 | 54/197 (27%) | 9/197 (5%) |
| NF-κB Signaling | 1.2 | −2 | 25/181 (14%) | 3/181 (2%) |
| RhoA Signaling | 1.71 | −2 | 40/124 (32%) | 8/124 (6%) |
| STAT3 Pathway | 2.49 | −2 | 23/74 (31%) | 3/74 (4%) |
| Gαi Signaling | 5.16 | −1.633 | 44/120 (37%) | 8/120 (7%) |
| Signaling by Rho Family GTPases | 2.34 | −1.342 | 78/248 (31%) | 17/248 (7%) |
| Synaptic Long Term Depression | 2.13 | −1.342 | 50/147 (34%) | 10/147 (7%) |
| CREB Signaling in Neurons | 2.36 | −1 | 64/185 (35%) | 14/185 (8%) |
| mTOR Signaling | 2.19 | −1 | 57/201 (28%) | 11/201 (5%) |
| Synaptic Long Term Potentiation | 1.75 | −1 | 41/121 (34%) | 13/121(11%) |
| ERK/MAPK Signaling | 1.08 | 0 | 38/199 (19%) | 9/199 (5%) |
| PPARα/RXRα Activation | 1.21 | 1 | 46/180 (26%) | 9/180 (5%) |
| PPAR Signaling | 2.1 | 2 | 19/95 (20%) | 1/95 (1%) |
| PTEN Signaling | 1.77 | 2 | 27/119 (23%) | 6/119 (5%) |
| RhoGDI Signaling21 | 3.23 | 2.236 | 61/173 (35%) | 13/173 (8%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.J.; Park, K.S.; Dhimal, N.; Shen, S.; Tang, X.; Qu, J.; Zhang, S.X. Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes. Cells 2022, 11, 2819. https://doi.org/10.3390/cells11182819
Wang JJ, Park KS, Dhimal N, Shen S, Tang X, Qu J, Zhang SX. Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes. Cells. 2022; 11(18):2819. https://doi.org/10.3390/cells11182819
Chicago/Turabian StyleWang, Joshua J., Karen Sophia Park, Narayan Dhimal, Shichen Shen, Xixiang Tang, Jun Qu, and Sarah X. Zhang. 2022. "Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes" Cells 11, no. 18: 2819. https://doi.org/10.3390/cells11182819
APA StyleWang, J. J., Park, K. S., Dhimal, N., Shen, S., Tang, X., Qu, J., & Zhang, S. X. (2022). Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes. Cells, 11(18), 2819. https://doi.org/10.3390/cells11182819








