Small Nucleolar RNAs in Pseudoexfoliation Glaucoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Groups
- (I).
- Patients with incipient senile cataract and an advanced stage of PEXG: (a) cloudy area in the lens that causes decreased vision; (b) elevated IOP of greater than 21 mmHg, documented in medical history; (c) open-angle grade III/IV according to Schaffer’s classification; (d) glaucomatous optic nerve head damage (excavation, neuroretinal rim thinning or notching, and localized or diffuse retinal nerve fiber layer (RNFL) defect); (e) glaucomatous defect in SAP in at least two consecutive tests, with three reliability indices better than 15% (results were considered abnormal if the Glaucoma Hemifield Test result was outside normal limits and at least three contiguous points were present within the same hemifield on the pattern deviation (PD) plot at p < 1%, with at least one point at p < 0.5%); and (f) typical deposits of pathological material observed, during a slit-lamp examination, as concentric rings of grayish fringes on the peripheral lens capsule and flakes on the pupillary border of the iris.
- (II).
- Control patients with incipient senile cataract: (a) cloudy area in the lens that causes decreased vision; (b) normal IOP (from 10 mmHg to 21 mmHg); and (c) no clinical signs of glaucoma nor PEX.
2.2. Aqueous Humor
2.3. RNA Isolation
2.4. The snoRNA Profiling
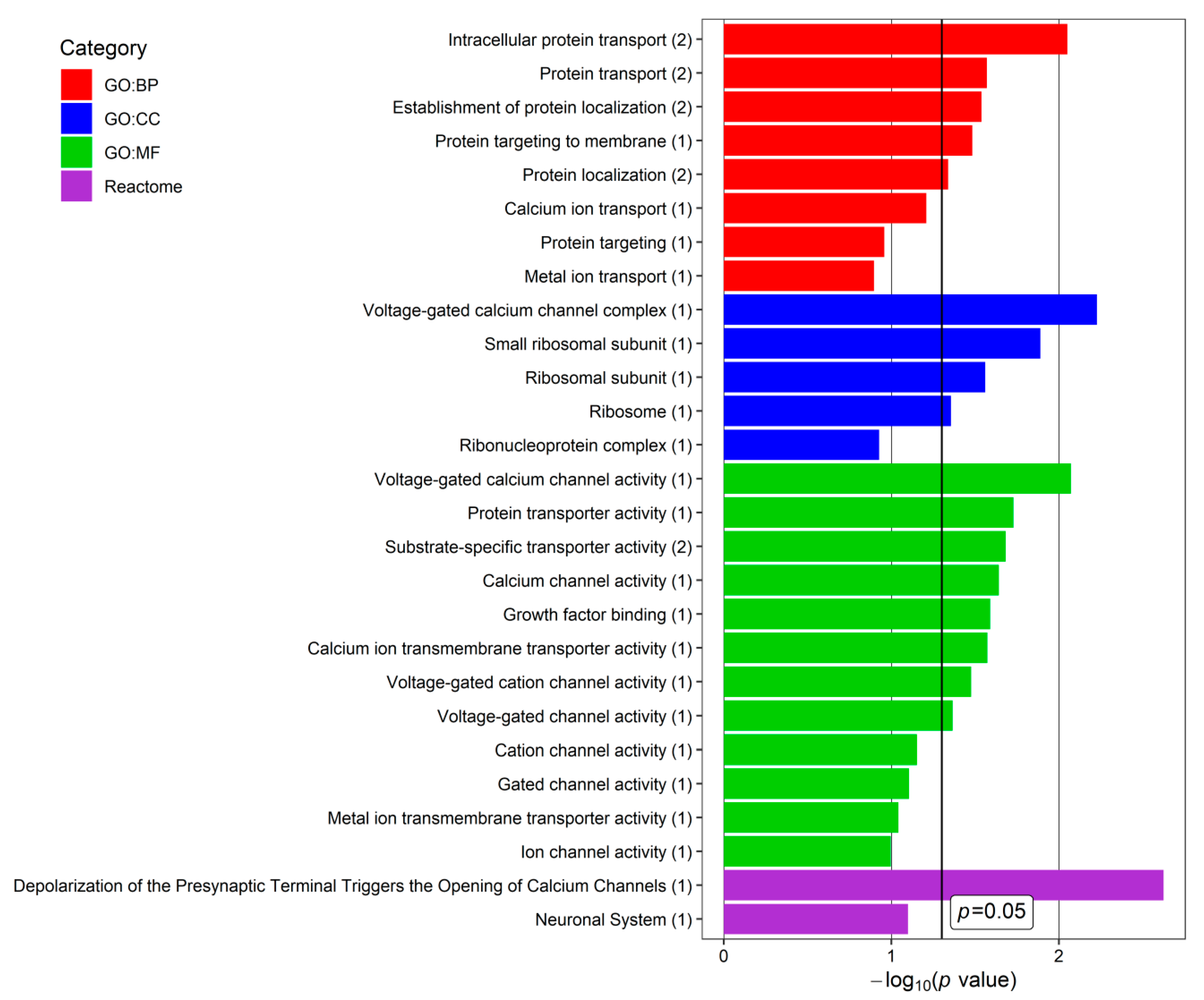
2.5. Functional and Enrichment Analysis
2.6. Statistical Analysis
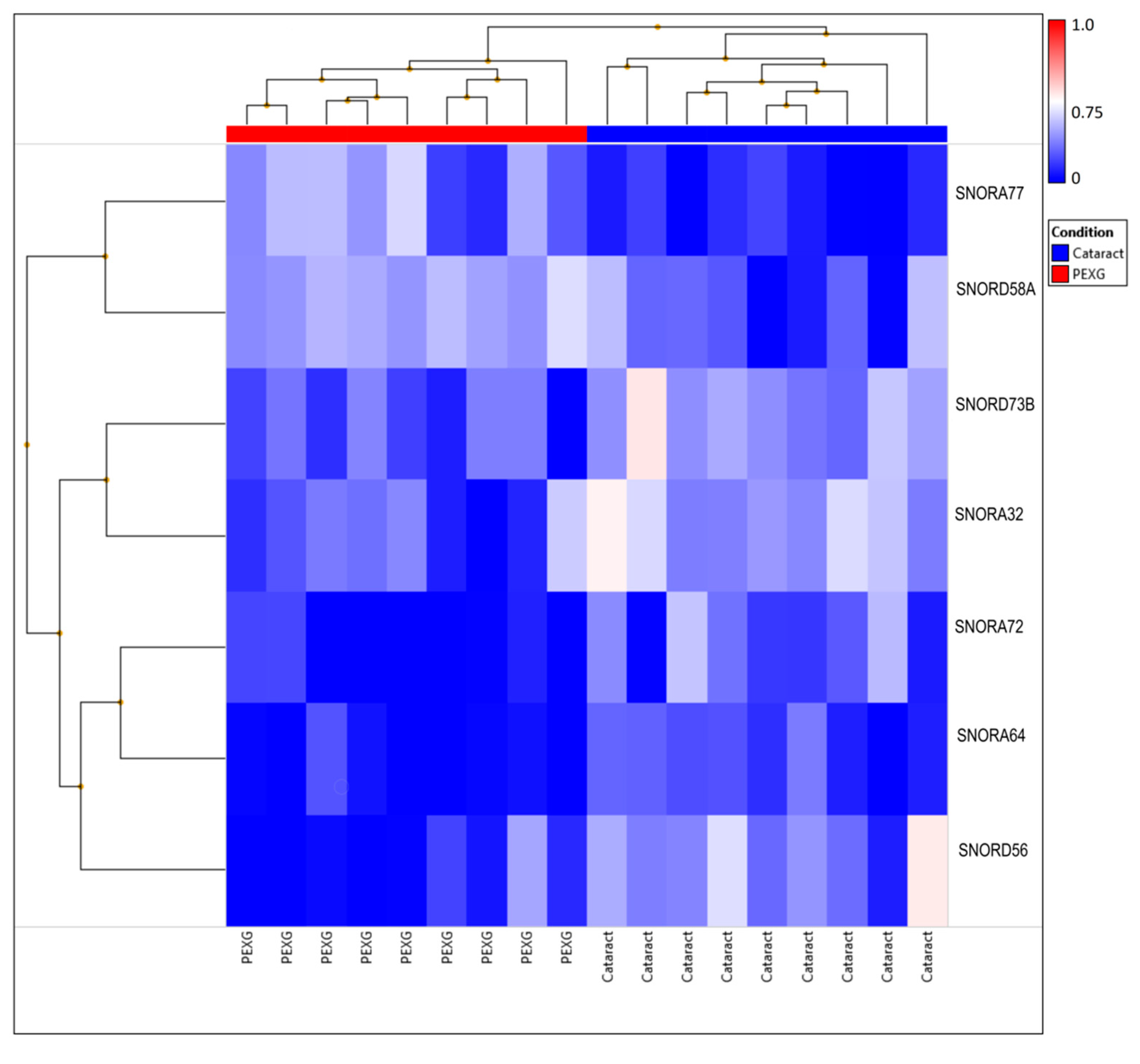
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lesiewska, H.; Łukaszewska-Smyk, A.; Odrowąż-Sypniewska, G.; Krintus, M.; Mańkowska-Cyl, A.; Malukiewicz, G. Chosen Vascular Risk Markers in Pseudoexfoliation Syndrome: An Age-Related Disorder. J. Ophthalmol. 2017, 2017, 5231095. [Google Scholar] [CrossRef]
- Schlotzer-Schrehardt, U. Genetics and Genomics of Pseudoexfoliation Syndrome/Glaucoma. Middle East Afr. J. Ophthalmol. 2011, 18, 30. [Google Scholar] [CrossRef]
- Elhawy, E.; Kamthan, G.; Dong, C.Q.; Danias, J. Pseudoexfoliation Syndrome, a Systemic Disorder with Ocular Manifestations. Hum. Genom. 2012, 6, 22. [Google Scholar] [CrossRef]
- Tuteja, S.; Chawla, H. Pseudoexfoliation Syndrome; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Andrikopoulos, G.K.; Alexopoulos, D.K.; Gartaganis, S.P. Pseudoexfoliation Syndrome and Cardiovascular Diseases. World J. Cardiol. 2014, 6, 847–854. [Google Scholar] [CrossRef]
- Cumurcu, T.; Dorak, F.; Cumurcu, B.E.; Erbay, L.G.; Ozsoy, E. Is There Any Relation Between Pseudoexfoliation Syndrome and Alzheimer’s Type Dementia? Semin. Ophthalmol. 2013, 28, 224–229. [Google Scholar] [CrossRef]
- Nasrabady, S.E.; Rizvi, B.; Goldman, J.E.; Brickman, A.M. White Matter Changes in Alzheimer’s Disease: A Focus on Myelin and Oligodendrocytes. Acta Neuropathol. Commun. 2018, 6, 22. [Google Scholar] [CrossRef]
- Aung, T.; Ozaki, M.; Mizoguchi, T.; Allingham, R.R.; Li, Z.; Haripriya, A.; Nakano, S.; Uebe, S.; Harder, J.M.; Chan, A.S.Y.; et al. A Common Variant Mapping to CACNA1A Is Associated with Susceptibility to Exfoliation Syndrome. Nat. Genet. 2015, 47, 387–392. [Google Scholar] [CrossRef]
- Aung, T.; Ozaki, M.; Lee, M.C.; Schlötzer-Schrehardt, U.; Thorleifsson, G.; Mizoguchi, T.; Igo, R.P.; Haripriya, A.; Williams, S.E.; Astakhov, Y.S.; et al. Genetic Association Study of Exfoliation Syndrome Identifies a Protective Rare Variant at LOXL1 and Five New Susceptibility Loci. Nat. Genet. 2017, 49, 993–1004. [Google Scholar] [CrossRef]
- Hohberger, B.; Schlötzer-Schrehard, U.; Mardin, C.; Lämmer, R.; Munoz, L.; Kunze, R.; Herrmann, M.; Wallukat, G. Inhibitory and Agonistic Autoantibodies Directed Against the Β2-Adrenergic Receptor in Pseudoexfoliation Syndrome and Glaucoma. Front. Neurosci. 2021, 15, 676579. [Google Scholar] [CrossRef]
- Liu, Y.; Allingham, R.R. Molecular Genetics in Glaucoma. Exp. Eye Res. 2011, 93, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Vranka, J.A.; Kelley, M.J.; Acott, T.S.; Keller, K.E. Extracellular Matrix in the Trabecular Meshwork: Intraocular Pressure Regulation and Dysregulation in Glaucoma. Exp. Eye Res. 2015, 133, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Eivers, S.B.; Greene, A.G.; Dervan, E.; O’Brien, C.; Wallace, D. Prevalence of Pseudoexfoliation Glaucoma Risk-Associated Variants Within Lysyl Oxidase-like 1 in an Irish Population. J. Glaucoma 2020, 29, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Łukasik, U.; Kosior-Jarecka, E.; Wróbel-Dudzińska, D.; Kustra, A.; Milanowski, P.; Żarnowski, T. Clinical Features of Pseudoexfoliative Glaucoma in Treated Polish Patients. Clin. Ophthalmol. 2020, 14, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Koz, O.G.; Turkcu, M.F.; Yarangumeli, A.; Koz, C.; Kural, G. Normotensive Glaucoma and Risk Factors in Normotensive Eyes with Pseudoexfoliation Syndrome. J. Glaucoma 2009, 18, 684–688. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From SnoRNA to MiRNA: Dual Function Regulatory Non-Coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Kosior-Jarecka, E.; Czop, M.; Gasińska, K.; Wróbel-Dudzińska, D.; Zalewski, D.P.; Bogucka-Kocka, A.; Kocki, J.; Żarnowski, T. MicroRNAs in the Aqueous Humor of Patients with Different Types of Glaucoma. Graefes Arch. Clin. Exp. Ophthalmol. 2021, 259, 2337–2349. [Google Scholar] [CrossRef]
- Patra Bhattacharya, D.; Canzler, S.; Kehr, S.; Hertel, J.; Grosse, I.; Stadler, P.F. Phylogenetic Distribution of Plant SnoRNA Families. BMC Genom. 2016, 17, 969. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.; Zhang, B.; Chu, L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef]
- Kobayashi, T. A New Role of the RDNA and Nucleolus in the Nucleus—RDNA Instability Maintains Genome Integrity. BioEssays 2008, 30, 267–272. [Google Scholar] [CrossRef]
- Rimer, J.M.; Lee, J.; Holley, C.L.; Crowder, R.J.; Chen, D.L.; Hanson, P.I.; Ory, D.S.; Schaffer, J.E. Long-Range Function of Secreted Small Nucleolar RNAs That Direct 2′-O-Methylation. J. Biol. Chem. 2018, 293, 13284–13296. [Google Scholar] [CrossRef] [Green Version]
- Ellis, J.C.; Brown, D.D.; Brown, J.W. The Small Nucleolar Ribonucleoprotein (SnoRNP) Database. RNA 2010, 16, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Božič, J.; Rogelj, B. Functional Diversity of Small Nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef]
- Liu, C.-H.; Wang, Z.; Sun, Y.; SanGiovanni, J.P.; Chen, J. Retinal Expression of Small Non-Coding RNAs in a Murine Model of Proliferative Retinopathy. Sci. Rep. 2016, 6, 33947. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Zou, W. A Novel Four-snoRNA Signature for Predicting the Survival of Patients with Uveal Melanoma. Mol. Med. Rep. 2019, 19, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Contreras, A.; Ruggero, D. Small RNAs with Big Implications: New Insights into H/ACA SnoRNA Function and Their Role in Human Disease. Wiley Interdiscip. Rev. RNA 2015, 6, 173–189. [Google Scholar] [CrossRef]
- Dimitrova, D.G.; Teysset, L.; Carré, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, E117. [Google Scholar] [CrossRef]
- Muñoz-Culla, M.; Irizar, H.; Sáenz-Cuesta, M.; Castillo-Triviño, T.; Osorio-Querejeta, I.; Sepúlveda, L.; López de Munain, A.; Olascoaga, J.; Otaegui, D. SncRNA (MicroRNA &snoRNA) Opposite Expression Pattern Found in Multiple Sclerosis Relapse and Remission Is Sex Dependent. Sci. Rep. 2016, 6, 20126. [Google Scholar] [CrossRef]
- snoDB: A Database of Human snoRNA. Available online: http://scottgroup.med.usherbrooke.ca/snoDB/ (accessed on 22 June 2022).
- miRNet–a miRNA-Centric Network Visual Analytics Platform. Available online: https://www.mirnet.ca/miRNet/home.xhtml (accessed on 22 June 2022).
- Aw, J.G.A.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X.N.; Boon, K.-L.; Tapsin, S.; Chan, Y.-S.; Tan, C.-P.; Sim, A.Y.L.; et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of Somatic Tissue Differentiation by the Long Non-Coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, E.; Maurille, M.J. The SnoRNPs and Related Machines: Ancient Devices That Mediate Maturation of RRNA and Other RNAs. In The Nucleolus; Olson, M.O.J., Ed.; Landes Bioscience: Austin, TX, USA, 2004; pp. 225–261. [Google Scholar]
- Scott, M.S.; Avolio, F.; Ono, M.; Lamond, A.I.; Barton, G.J. Human MiRNA Precursors with Box H/ACA SnoRNA Features. PLoS Comput. Biol. 2009, 5, e1000507. [Google Scholar] [CrossRef] [PubMed]
- Soundara Pandi, S.P.; Chen, M.; Guduric-Fuchs, J.; Xu, H.; Simpson, D.A. Extremely Complex Populations of Small RNAs in the Mouse Retina and RPE/Choroid. Investig. Ophthalmol. Vis. Sci. 2013, 54, 8140. [Google Scholar] [CrossRef] [PubMed]
- Izzotti, A.; Bagnis, A.; Sacca, S. The Role of Oxidative Stress in Glaucoma. Mutat. Res. 2006, 612, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, Q.; Zhou, Z.; Fu, M.; Yang, X.; Hao, K.; Liu, Y. SNHG3 Cooperates with ELAVL2 to Modulate Cell Apoptosis and Extracellular Matrix Accumulation by Stabilizing SNAI2 in Human Trabecular Meshwork Cells under Oxidative Stress. Environ. Toxicol. 2021, 36, 1070–1079. [Google Scholar] [CrossRef]
- Rong, R.; Wang, M.; You, M.; Li, H.; Xia, X.; Ji, D. Pathogenesis and Prospects for Therapeutic Clinical Application of Noncoding RNAs in Glaucoma: Systematic Perspectives. J. Cell. Physiol. 2021, 236, 7097–7116. [Google Scholar] [CrossRef]
- Khan, S.Y.; Hackett, S.F.; Riazuddin, S.A. Non-Coding RNA Profiling of the Developing Murine Lens. Exp. Eye Res. 2016, 145, 347–351. [Google Scholar] [CrossRef]
- Dismuke, W.M.; Challa, P.; Navarro, I.; Stamer, W.D.; Liu, Y. Human Aqueous Humor Exosomes. Exp. Eye Res. 2015, 132, 73–77. [Google Scholar] [CrossRef] [PubMed]
- De Maria, M.; Iannetta, D.; Cimino, L.; Coassin, M.; Fontana, L. Measuring Anterior Chamber Inflammation After Cataract Surgery: A Review of the Literature Focusing on the Correlation with Cystoid Macular Edema. Clin. Ophthalmol. 2020, 14, 41–52. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Xie, Y.; Yan, W.; Khorram, O. Next-Generation Sequencing Reveals Differentially Expressed Small Noncoding RNAs in Uterine Leiomyoma. Fertil. Steril. 2018, 109, 919–929. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, R.; Gao, M.; Zhao, Y.; Lv, X.; Zhu, W.; Han, L.; Su, P.; Fan, Y.; Yan, Y.; et al. SNORA72 Activates the Notch1/c-Myc Pathway to Promote Stemness Transformation of Ovarian Cancer Cells. Front. Cell Dev. Biol. 2020, 8, 583087. [Google Scholar] [CrossRef]
- Christel, C.; Lee, A. Ca2+-Dependent Modulation of Voltage-Gated Ca2+ Channels. Biochim. Biophys. Acta 2012, 1820, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Petkova, T.D.; Laritsky, E.; Kessler, N.; Baker, M.S.; Zhu, S.; Waterland, R.A. Early Postnatal Overnutrition Accelerates Aging-Associated Epigenetic Drift in Pancreatic Islets. Environ. Epigenet. 2019, 5, dvz015. [Google Scholar] [CrossRef] [PubMed]
- Vendel, A.C.; Rithner, C.D.; Lyons, B.A.; Horne, W.A. Solution Structure of the N-Terminal A Domain of the Human Voltage-Gated Ca2+ channel Beta4a Subunit. Protein Sci. 2006, 15, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Buraei, Z.; Yang, J. The ß Subunit of Voltage-Gated Ca2+ Channels. Physiol. Rev. 2010, 90, 1461–1506. [Google Scholar] [CrossRef]
- Cork, R.J.; Namkung, Y.; Shin, H.-S.; Mize, R.R. Development of the Visual Pathway Is Disrupted in Mice with a Targeted Disruption of the Calcium Channel Beta(3)-Subunit Gene. J. Comp. Neurol. 2001, 440, 177–191. [Google Scholar] [CrossRef]
- Jin, K.; Mao, X.O.; Eshoo, M.W.; Nagayama, T.; Minami, M.; Simon, R.P.; Greenberg, D.A. Microarray Analysis of Hippocampal Gene Expression in Global Cerebral Ischemia. Ann. Neurol. 2001, 50, 93–103. [Google Scholar] [CrossRef]
- Bavamian, S.; Mellios, N.; Lalonde, J.; Fass, D.M.; Wang, J.; Sheridan, S.D.; Madison, J.M.; Zhou, F.; Rueckert, E.H.; Barker, D.; et al. Dysregulation of MiR-34a Links Neuronal Development to Genetic Risk Factors for Bipolar Disorder. Mol. Psychiatry 2015, 20, 573–584. [Google Scholar] [CrossRef]
- Qin, Y.; Sun, W.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, H. Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine-Related Cancers. OTT 2020, 13, 7699–7717. [Google Scholar] [CrossRef]
- Mostmans, Y.; Cutolo, M.; Giddelo, C.; Decuman, S.; Melsens, K.; Declercq, H.; Vandecasteele, E.; De Keyser, F.; Distler, O.; Gutermuth, J.; et al. The Role of Endothelial Cells in the Vasculopathy of Systemic Sclerosis: A Systematic Review. Autoimmun. Rev. 2017, 16, 774–786. [Google Scholar] [CrossRef]
- Errasti-Murugarren, E.; Bartoccioni, P.; Palacín, M. Membrane Protein Stabilization Strategies for Structural and Functional Studies. Membranes 2021, 11, 155. [Google Scholar] [CrossRef]
- Zenkel, M.; Schlotzer-Schrehardt, U.; Berner, D.; Kruse, F.E.; Reis, A.; Khor, C.C.; Aung, T.; Pasutto, F. Dysregulated Expression of POMP and TMEM136 May Contribute to Impaired Proteasome Function and Endothelial Dysfunction in Eyes with Pseudoexfoliation Syndrome/Glaucoma. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4902. [Google Scholar]
- Pinelli, M.; Carissimo, A.; Cutillo, L.; Lai, C.-H.; Mutarelli, M.; Moretti, M.N.; Singh, M.V.; Karali, M.; Carrella, D.; Pizzo, M.; et al. An Atlas of Gene Expression and Gene Co-Regulation in the Human Retina. Nucleic Acids Res. 2016, 44, 5773–5784. [Google Scholar] [CrossRef] [PubMed]
- Orphal, M.; Gillespie, A.; Böhme, K.; Subrova, J.; Eisenreich, A.; Kreutz, R. TMEM63C, a Potential Novel Target for Albuminuria Development, Is Regulated by MicroRNA-564 and Transforming Growth Factor Beta in Human Renal Cells. Kidney Blood Press. Res. 2020, 45, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Ye, C.; Wu, D.; Zang, Y.-Y.; Zhang, L.; Chen, C.; He, X.-Y.; Yang, J.-J.; Hu, P.; Xu, Z.; et al. The Cation Channel TMEM63B Is an Osmosensor Required for Hearing. Cell Rep. 2020, 31, 107596. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.N.; Chagot, B.; Chazin, W.J. Calcium-Dependent Regulation of Ion Channels. Calcium Bind. Proteins 2006, 1, 203–212. [Google Scholar]
- Ma, L.; Liu, X.; Liu, Q.; Jin, S.; Chang, H.; Liu, H. The Roles of Transient Receptor Potential Ion Channels in Pathologies of Glaucoma. Front. Physiol. 2022, 13, 806786. [Google Scholar] [CrossRef]
- Jiang, Y.; Chiu, C.-Y.; Yan, Q.; Chen, W.; Gorin, M.B.; Conley, Y.P.; Lakhal-Chaieb, M.L.; Cook, R.J.; Amos, C.I.; Wilson, A.F.; et al. Gene-Based Association Testing of Dichotomous Traits with Generalized Functional Linear Mixed Models Using Extended Pedigrees: Applications to Age-Related Macular Degeneration. J. Am. Stat. Assoc. 2021, 116, 531–545. [Google Scholar] [CrossRef]
- Schulz, A.; Müller, N.V.; van de Lest, N.A.; Eisenreich, A.; Schmidbauer, M.; Barysenka, A.; Purfürst, B.; Sporbert, A.; Lorenzen, T.; Meyer, A.M.; et al. Analysis of the Genomic Architecture of a Complex Trait Locus in Hypertensive Rat Models Links Tmem63c to Kidney Damage. Elife 2019, 8, e42068. [Google Scholar] [CrossRef]
- Lim, B.J.; Yang, J.W.; Do, W.S.; Fogo, A.B. Pathogenesis of Focal Segmental Glomerulosclerosis. J. Pathol. Transl. Med. 2016, 50, 405–410. [Google Scholar] [CrossRef]
- Villarreal, G.; Oh, D.-J.; Kang, M.H.; Rhee, D.J. Coordinated Regulation of Extracellular Matrix Synthesis by the MicroRNA-29 Family in the Trabecular Meshwork. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3391. [Google Scholar] [CrossRef]
- Li, H.; Raghunathan, V.; Stamer, W.D.; Ganapathy, P.S.; Herberg, S. Extracellular Matrix Stiffness and TGFβ2 Regulate YAP/TAZ Activity in Human Trabecular Meshwork Cells. Front. Cell Dev. Biol. 2022, 10, 844342. [Google Scholar] [CrossRef] [PubMed]
- Fuchshofer, R.; Stephan, D.A.; Russell, P.; Tamm, E.R. Gene Expression Profiling of TGFbeta2- and/or BMP7-Treated Trabecular Meshwork Cells: Identification of Smad7 as a Critical Inhibitor of TGF-Beta2 Signaling. Exp. Eye Res. 2009, 88, 1020–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydın Yaz, Y.; Yıldırım, N.; Yaz, Y.; Tekin, N.; İnal, M.; Şahin, F.M. Role of Oxidative Stress in Pseudoexfoliation Syndrome and Pseudoexfoliation Glaucoma. Turk. J. Ophthalmol. 2019, 49, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, P.P.; Maddala, R.; Rao, P.V. Regulation of Plasticity and Fibrogenic Activity of Trabecular Meshwork Cells by Rho GTPase Signaling. J. Cell. Physiol. 2014, 229, 927–942. [Google Scholar] [CrossRef]
- Tanna, A.P.; Johnson, M. Rho Kinase Inhibitors as a Novel Treatment for Glaucoma and Ocular Hypertension. Ophthalmology 2018, 125, 1741–1756. [Google Scholar] [CrossRef]
- Filla, M.S.; Faralli, J.A.; Peotter, J.L.; Peters, D.M. The Role of Integrins in Glaucoma. Exp. Eye Res. 2017, 158, 124–136. [Google Scholar] [CrossRef]
- Tezel, G.; Wax, M.B. Increased Production of Tumor Necrosis Factor-Alpha by Glial Cells Exposed to Simulated Ischemia or Elevated Hydrostatic Pressure Induces Apoptosis in Cocultured Retinal Ganglion Cells. J. Neurosci. 2000, 20, 8693–8700. [Google Scholar] [CrossRef]
| PEXG N = 9 | Cataract N = 9 | p-Value | ||
| Age (mean ± SD) | 77.56 ± 6.54 | 76.00 ± 6.95 | 0.6314 ^ | |
| Gender (N, %) | Male | 8 (88.89%) | 8 (88.89%) | 0.7647 # |
| Female | 1 (11.11%) | 1 (11.11%) | ||
| BCVA | 0.19 ± 0.17 | 0.42 ± 0.12 | 0.0047 ^ | |
| MAX IOP | 30.44 ± 13.1 | 15.67 ± 2.12 | 0.0042 ^ | |
| C/D | 0.91 ± 0.07 | 0.29 ± 0.15 | <0.0001 ^ | |
| RNFL | 61.22 ± 5.12 | 93.8 ± 5.61 | 0.0005 ^ | |
| MD | −22.55 ± 4.77 | −0.03 ± 0.98 | <0.0001 ^ | |
| Symbol | Box Type | Host Genes | Target Genes | Target snoRNAs |
|---|---|---|---|---|
| SNORD73B | C/D | RPS3A | - | - |
| SNORD58A | C/D | RPL17 | - | - |
| SNORD56 | C/D | EDEM2 | - | - |
| LINC02463 | - | - | ||
| AC012501.2 | - | - | ||
| ITGB8 | - | - | ||
| NOP56 | - | SNORA7B | ||
| SGSM1 | - | SNORA65, SNORD50A | ||
| - | FAM120AOS | - | ||
| SNORA77 | H/ACA | RERE | - | - |
| ATP2B4 OR ATP2B2 | ||||
| SNORA72 | H/ACA | RPL30 | - | - |
| NUCKS1 | - | - | ||
| ECT2 | - | - | ||
| DEGS1 | - | - | ||
| SNORA64 | H/ACA | MYRIP | - | - |
| RPS2 | RPS2, TNPO2 | - | ||
| PLS3-AS1 | - | - | ||
| - | CACNB3 | - | ||
| SNORA32 | H/ACA | TAF1D | - | - |
| TMEM63C | - | - |
| Symbol | Chromosome | Description |
| CACNB3 | 12q13.12 | Calcium voltage-gated channel auxiliary subunit beta 3 |
| FAM120AOS | 9q22.31 | Family with sequence similarity 120A opposite strand |
| RPS2 | 16p13.3 | Ribosomal protein S2 |
| TNPO2 | 19p13.13 | Transportin 2 |
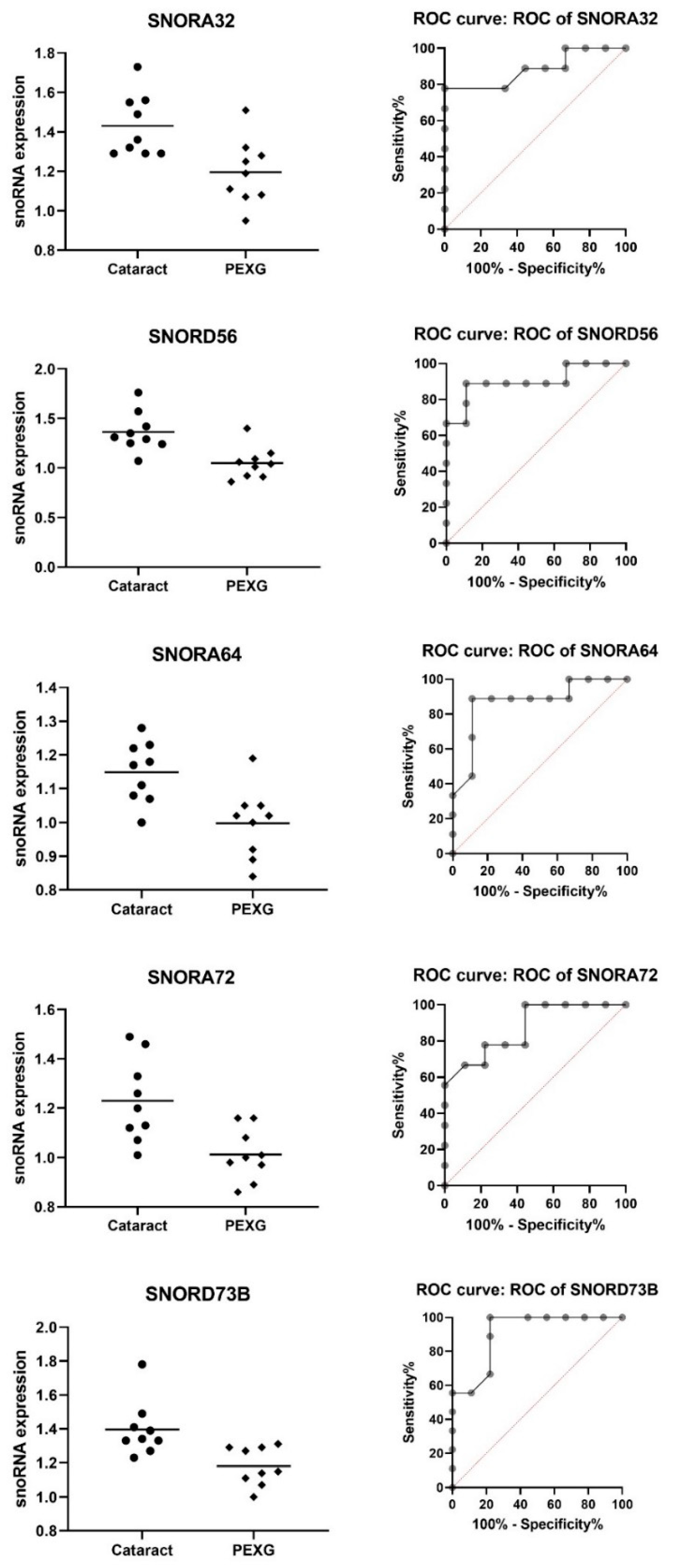
| snoRNA | Group | N | Mean | SD | Fold Change | p-Value | AUC | AUC p-Value |
|---|---|---|---|---|---|---|---|---|
| SNORA32 | Cataract | 9 | 1.431 | 0.158 | −1.14 | 0.0072 | 0.883 | 0.0062 |
| PEXG | 9 | 1.196 | 0.166 | |||||
| SNORA72 | Cataract | 9 | 1.230 | 0.169 | −1.15 | 0.0047 | 0.870 | 0.0081 |
| PEXG | 9 | 1.012 | 0.106 | |||||
| SNORD56 | Cataract | 9 | 1.362 | 0.202 | −1.21 | 0.0022 | 0.901 | 0.0041 |
| PEXG | 9 | 1.049 | 0.161 | |||||
| SNORA64 | Cataract | 9 | 1.149 | 0.090 | −1.10 | 0.0045 | 0.870 | 0.0081 |
| PEXG | 9 | 0.998 | 0.104 | |||||
| SNORD73b | Cataract | 9 | 1.397 | 0.163 | −1.12 | 0.0048 | 0.907 | 0.0036 |
| PEXG | 9 | 1.181 | 0.112 |
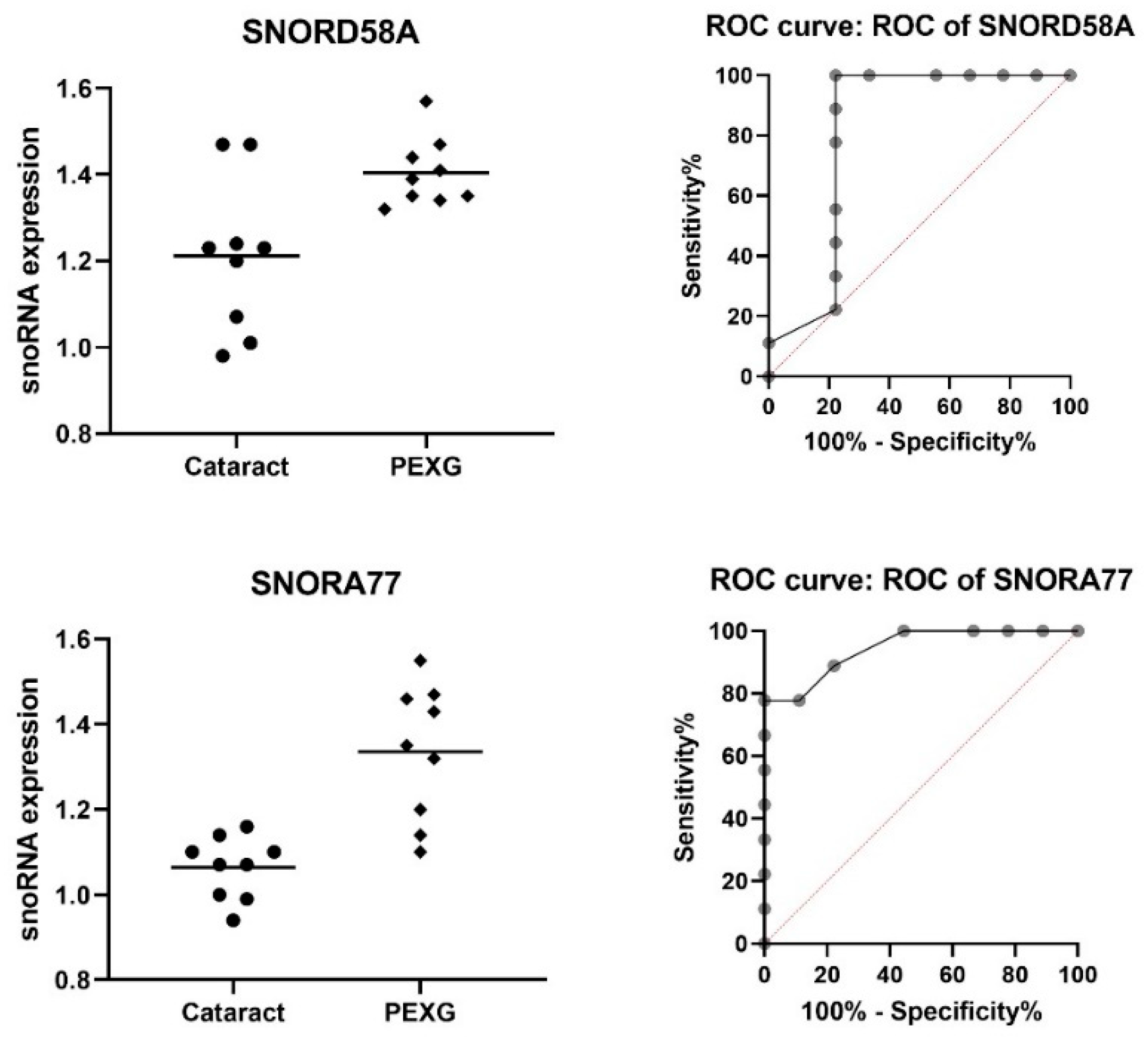
| snoRNA | Group | N | Mean | SD | Fold Change | p-Value | AUC | AUC p-Value |
|---|---|---|---|---|---|---|---|---|
| SNORD58A | Cataract | 9 | 1.211 | 0.177 | 1.13 | 0.0086 | 0.815 | 0.0243 |
| PEXG | 9 | 1.404 | 0.079 | |||||
| SNORA77 | Cataract | 9 | 1.063 | 0.073 | 1.21 | 0.0003 | 0.944 | 0.0015 |
| PEXG | 9 | 1.336 | 0.159 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasińska, K.; Czop, M.; Kosior-Jarecka, E.; Wróbel-Dudzińska, D.; Kocki, J.; Żarnowski, T. Small Nucleolar RNAs in Pseudoexfoliation Glaucoma. Cells 2022, 11, 2738. https://doi.org/10.3390/cells11172738
Gasińska K, Czop M, Kosior-Jarecka E, Wróbel-Dudzińska D, Kocki J, Żarnowski T. Small Nucleolar RNAs in Pseudoexfoliation Glaucoma. Cells. 2022; 11(17):2738. https://doi.org/10.3390/cells11172738
Chicago/Turabian StyleGasińska, Karolina, Marcin Czop, Ewa Kosior-Jarecka, Dominika Wróbel-Dudzińska, Janusz Kocki, and Tomasz Żarnowski. 2022. "Small Nucleolar RNAs in Pseudoexfoliation Glaucoma" Cells 11, no. 17: 2738. https://doi.org/10.3390/cells11172738
APA StyleGasińska, K., Czop, M., Kosior-Jarecka, E., Wróbel-Dudzińska, D., Kocki, J., & Żarnowski, T. (2022). Small Nucleolar RNAs in Pseudoexfoliation Glaucoma. Cells, 11(17), 2738. https://doi.org/10.3390/cells11172738









