SUMOylation in Skeletal Development, Homeostasis, and Disease
Abstract
1. Introduction
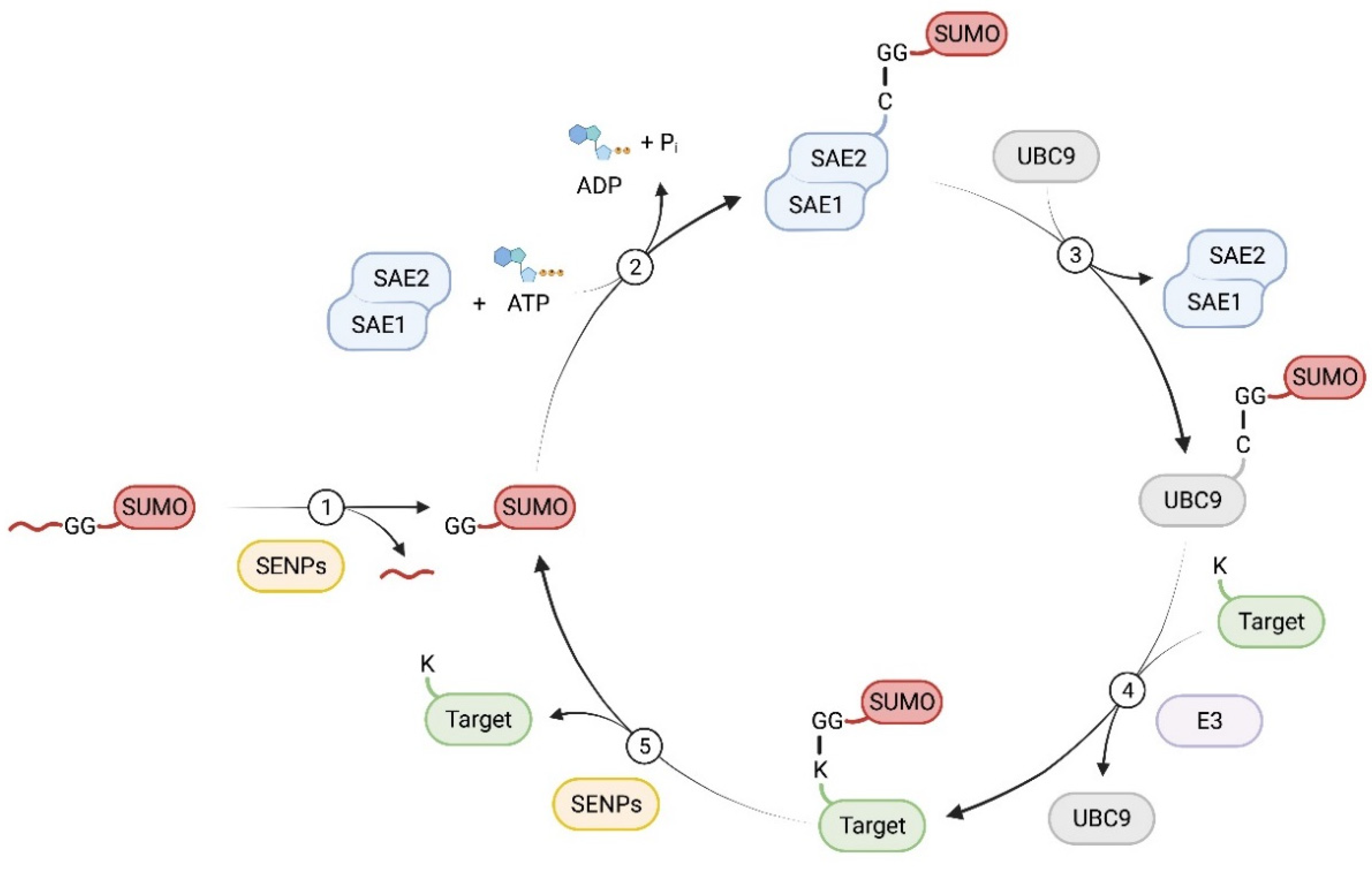
2. SUMO and SUMOylation
3. SUMOylation in Skeletal Cell Differentiation, Homeostasis, and Disease
3.1. SUMOylation in Osteogenesis, Osteoblast Homeostasis, and Bone Mass Regulation
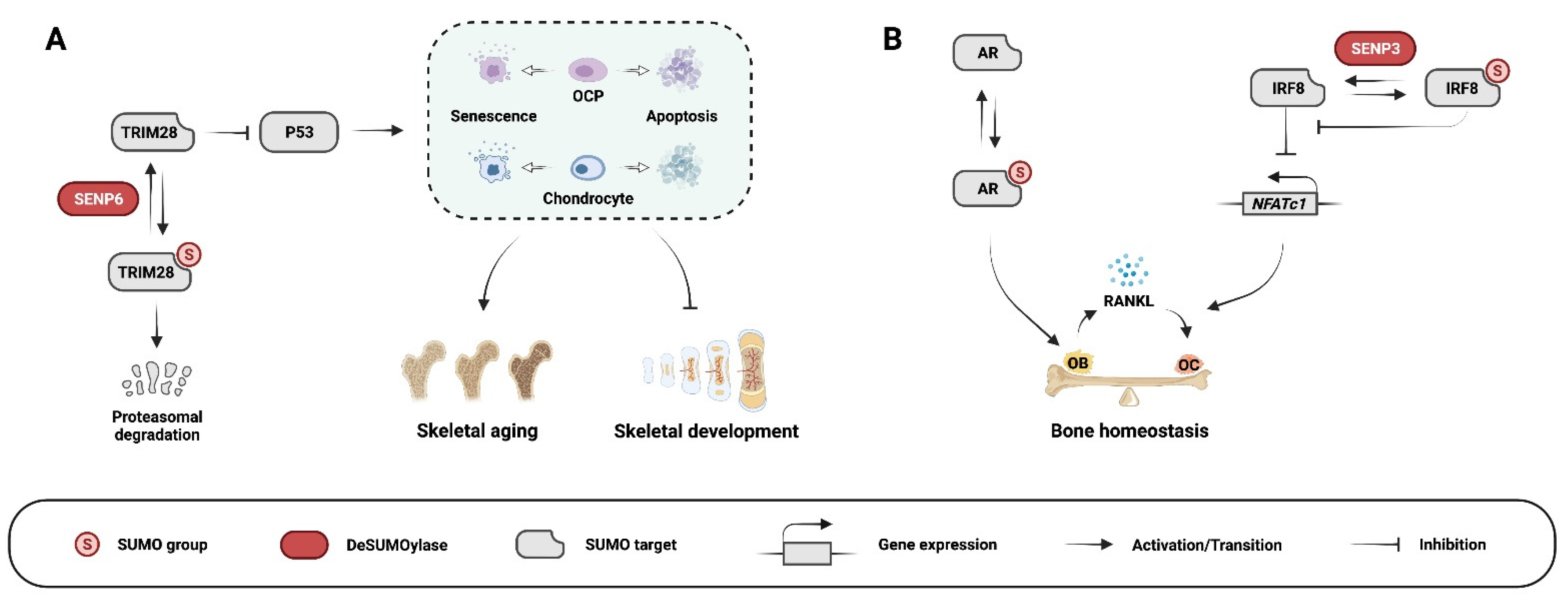
3.2. SUMOylation in Chondrogenesis, Chondrocyte Homeostasis, and Osteoarthritis
3.3. SUMOylation in Osteoclastogenesis and Osteoclast Function
3.4. SUMOylation in Developmental Diseases
3.4.1. Split Hand/Split Foot Malformation (SHFM)
3.4.2. Craniofacial Disorders
3.5. SUMOylation in rheumatoid arthritis
3.6. SUMOylation in Osteosarcoma
3.6.1. Studies Supporting a Pro-Tumorigenic Effect of SUMOylation
3.6.2. Studies Supporting an Anti-Tumorigenic Effect of SUMOylation
3.7. SUMOylation in Chondrosarcoma
4. Summary and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Percival, C.J.; Richtsmeier, J.T. Angiogenesis and intramembranous osteogenesis. Dev. Dyn. 2013, 242, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Ornitz, D.M. Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 2013, 5, a008334. [Google Scholar] [CrossRef] [PubMed]
- Ono, N.; Ono, W.; Nagasawa, T.; Kronenberg, H.M. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 2014, 16, 1157–1167. [Google Scholar] [CrossRef]
- Kronenberg, H.M. Developmental regulation of the growth plate. Nature 2003, 423, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Shao, J.; Gilbert, S.R.; Riddle, R.C.; Long, F.; Johnson, R.S.; Schipani, E.; Clemens, T.L. Role of HIF-1alpha in skeletal development. Ann. N. Y. Acad. Sci. 2010, 1192, 322–326. [Google Scholar] [CrossRef]
- Zhou, X.; von der Mark, K.; Henry, S.; Norton, W.; Adams, H.; de Crombrugghe, B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014, 10, e1004820. [Google Scholar] [CrossRef]
- Yang, L.; Tsang, K.Y.; Tang, H.C.; Chan, D.; Cheah, K.S. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. USA 2014, 111, 12097–12102. [Google Scholar] [CrossRef] [PubMed]
- Mackie, E.J.; Ahmed, Y.A.; Tatarczuch, L.; Chen, K.S.; Mirams, M. Endochondral ossification: How cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell Biol. 2008, 40, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Morini, S.; Continenza, M.A.; Ricciardi, G.; Gaudio, E.; Pannarale, L. Development of the microcirculation of the secondary ossification center in rat humeral head. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2004, 278, 419–427. [Google Scholar] [CrossRef]
- Novack, D.V.; Teitelbaum, S.L. The osteoclast: Friend or foe? Annu. Rev. Pathol. 2008, 3, 457–484. [Google Scholar] [CrossRef] [PubMed]
- Raggatt, L.J.; Partridge, N.C. Cellular and molecular mechanisms of bone remodeling. J. Biol. Chem. 2010, 285, 25103–25108. [Google Scholar] [CrossRef]
- Liu, Y.; Molchanov, V.; Yang, T. Enzymatic Machinery of Ubiquitin and Ubiquitin-Like Modification Systems in Chondrocyte Homeostasis and Osteoarthritis. Curr. Rheumatol. Rep. 2021, 23, 62. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef]
- Liang, Y.C.; Lee, C.C.; Yao, Y.L.; Lai, C.C.; Schmitz, M.L.; Yang, W.M. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci. Rep. 2016, 6, 26509. [Google Scholar] [CrossRef]
- Bohren, K.M.; Nadkarni, V.; Song, J.H.; Gabbay, K.H.; Owerbach, D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 2004, 279, 27233–27238. [Google Scholar] [CrossRef]
- Guo, D.; Li, M.; Zhang, Y.; Yang, P.; Eckenrode, S.; Hopkins, D.; Zheng, W.; Purohit, S.; Podolsky, R.H.; Muir, A.; et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 2004, 36, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef]
- Kunz, K.; Piller, T.; Muller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018, 131, jcs211904. [Google Scholar] [CrossRef]
- Matic, I.; van Hagen, M.; Schimmel, J.; Macek, B.; Ogg, S.C.; Tatham, M.H.; Hay, R.T.; Lamond, A.I.; Mann, M.; Vertegaal, A.C.O. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell Proteom. 2008, 7, 132–144. [Google Scholar] [CrossRef]
- Hendriks, I.A.; D’Souza, R.C.; Yang, B.; Verlaan-de Vries, M.; Mann, M.; Vertegaal, A.C. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Pichler, A.; Fatouros, C.; Lee, H.; Eisenhardt, N. SUMO conjugation—A mechanistic view. Biomol. Concepts 2017, 8, 13–36. [Google Scholar] [CrossRef]
- Bonne-Andrea, C.; Kahli, M.; Mechali, F.; Lemaitre, J.M.; Bossis, G.; Coux, O. SUMO2/3 modification of cyclin E contributes to the control of replication origin firing. Nat. Commun. 2013, 4, 1850. [Google Scholar] [CrossRef] [PubMed]
- Cubeñas-Potts, C.; Srikumar, T.; Lee, C.; Osula, O.; Subramonian, D.; Zhang, X.D.; Cotter, R.J.; Raught, B.; Matunis, M.J. Identification of SUMO-2/3-modified proteins associated with mitotic chromosomes. Proteomics 2015, 15, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Rogers, R.; Matunis, M.J.; Mayhew, C.N.; Goodson, M.L.; Park-Sarge, O.K.; Sarge, K.D. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 2001, 276, 40263–40267. [Google Scholar] [CrossRef] [PubMed]
- Ritho, J.; Arold, S.T.; Yeh, E.T. A Critical SUMO1 Modification of LKB1 Regulates AMPK Activity during Energy Stress. Cell Rep. 2015, 12, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Goeres, J.; Zhang, H.; Yen, T.J.; Porter, A.C.; Matunis, M.J. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 2008, 29, 729–741. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef]
- Sarge, K.D.; Park-Sarge, O.K. Sumoylation and human disease pathogenesis. Trends Biochem. Sci. 2009, 34, 200–205. [Google Scholar] [CrossRef]
- Rodriguez, M.S.; Dargemont, C.; Hay, R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001, 276, 12654–12659. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yunus, A.A.; Lima, C.D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 2009, 35, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.; Flotho, A.; Melchior, F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell 2012, 46, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Kagey, M.H.; Melhuish, T.A.; Wotton, D. The polycomb protein Pc2 is a SUMO E3. Cell 2003, 113, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fiskus, W.; Yong, B.; Atadja, P.; Takahashi, Y.; Pandita, T.K.; Wang, H.G.; Bhalla, K.N. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 6841–6846. [Google Scholar] [CrossRef] [PubMed]
- Rabellino, A.; Andreani, C.; Scaglioni, P.P. The Role of PIAS SUMO E3-Ligases in Cancer. Cancer Res. 2017, 77, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.T. SUMOylation and De-SUMOylation: Wrestling with life’s processes. J. Biol. Chem. 2009, 284, 8223–8227. [Google Scholar] [CrossRef]
- Nayak, A.; Muller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014, 15, 422. [Google Scholar] [CrossRef]
- Shin, E.J.; Shin, H.M.; Nam, E.; Kim, W.S.; Kim, J.H.; Oh, B.H.; Yun, Y. DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 2012, 13, 339–346. [Google Scholar] [CrossRef]
- Schulz, S.; Chachami, G.; Kozaczkiewicz, L.; Winter, U.; Stankovic-Valentin, N.; Haas, P.; Hofmann, K.; Urlaub, H.; Ovaa, H.; Wittbrodt, J.; et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012, 13, 930–938. [Google Scholar] [CrossRef]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Seeler, J.S.; Dejean, A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003, 4, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Davis, F.J.; Sharrocks, A.D.; Im, H.J. Emerging roles of SUMO modification in arthritis. Gene 2010, 466, 1–15. [Google Scholar] [CrossRef][Green Version]
- Chang, H.M.; Yeh, E.T.H. SUMO: From Bench to Bedside. Physiol. Rev. 2020, 100, 1599–1619. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, A.D.; Olsen, B.R. Bone development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef]
- Salhotra, A.; Shah, H.N.; Levi, B.; Longaker, M.T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020, 21, 696–711. [Google Scholar] [CrossRef]
- Li, J.; Lu, D.; Dou, H.; Liu, H.; Weaver, K.; Wang, W.; Li, J.; Yeh, E.T.H.; Williams, B.O.; Zheng, L.; et al. Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway. Nat. Commun. 2018, 9, 143. [Google Scholar] [CrossRef]
- Wan, M.; Cao, X. BMP signaling in skeletal development. Biochem. Biophys. Res. Commun. 2005, 328, 651–657. [Google Scholar] [CrossRef]
- Song, B.; Estrada, K.D.; Lyons, K.M. Smad signaling in skeletal development and regeneration. Cytokine Growth Factor Rev. 2009, 20, 379–388. [Google Scholar] [CrossRef]
- Shen, J.; Li, S.; Chen, D. TGF-beta signaling and the development of osteoarthritis. Bone Res. 2014, 2, 14002. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Akhtar, N.; Jamil, H.M.; Banik, R.S.; Asaduzzaman, S.M. TGF-beta/BMP signaling and other molecular events: Regulation of osteoblastogenesis and bone formation. Bone Res. 2015, 3, 15005. [Google Scholar] [CrossRef]
- Shimada, K.; Suzuki, N.; Ono, Y.; Tanaka, K.; Maeno, M.; Ito, K. Ubc9 promotes the stability of Smad4 and the nuclear accumulation of Smad1 in osteoblast-like Saos-2 cells. Bone 2008, 42, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Yukita, A.; Hosoya, A.; Ito, Y.; Katagiri, T.; Asashima, M.; Nakamura, H. Ubc9 negatively regulates BMP-mediated osteoblastic differentiation in cultured cells. Bone 2012, 50, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Kawano, H.; Sato, T.; Yamada, T.; Matsumoto, T.; Sekine, K.; Watanabe, T.; Nakamura, T.; Fukuda, T.; Yoshimura, K.; Yoshizawa, T.; et al. Suppressive function of androgen receptor in bone resorption. Proc. Natl. Acad. Sci. USA 2003, 100, 9416–9421. [Google Scholar] [CrossRef]
- Wu, J.; Moverare-Skrtic, S.; Zhang, F.P.; Koskela, A.; Tuukkanen, J.; Palvimo, J.J.; Sipila, P.; Poutanen, M.; Ohlsson, C. Androgen receptor SUMOylation regulates bone mass in male mice. Mol. Cell Endocrinol. 2019, 479, 117–122. [Google Scholar] [CrossRef]
- Komori, T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem. Cell Biol. 2018, 149, 313–323. [Google Scholar] [CrossRef]
- Tang, C.Y.; Chen, W.; Luo, Y.; Wu, J.; Zhang, Y.; McVicar, A.; McConnell, M.; Liu, Y.; Zhou, H.D.; Li, Y.P. Runx1 up-regulates chondrocyte to osteoblast lineage commitment and promotes bone formation by enhancing both chondrogenesis and osteogenesis. Biochem. J. 2020, 477, 2421–2438. [Google Scholar] [CrossRef]
- Tang, J.; Xie, J.; Chen, W.; Tang, C.; Wu, J.; Wang, Y.; Zhou, X.D.; Zhou, H.D.; Li, Y.P. Runt-related transcription factor 1 is required for murine osteoblast differentiation and bone formation. J. Biol. Chem. 2020, 295, 11669–11681. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, Q.; Ji, C.; Liu, X.; Li, L.; Luo, J. RUNX3 plays an important role in mediating the BMP9-induced osteogenic differentiation of mesenchymal stem cells. Int. J. Mol. Med. 2017, 40, 1991–1999. [Google Scholar] [CrossRef]
- Bauer, O.; Sharir, A.; Kimura, A.; Hantisteanu, S.; Takeda, S.; Groner, Y. Loss of osteoblast Runx3 produces severe congenital osteopenia. Mol. Cell Biol. 2015, 35, 1097–1109. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Kim, J.H.; Jang, J.W.; Lee, Y.S.; Lee, J.W.; Chi, X.Z.; Li, Y.H.; Kim, M.K.; Kim, D.M.; Choi, B.S.; Kim, J.; et al. RUNX family members are covalently modified and regulated by PIAS1-mediated sumoylation. Oncogenesis 2014, 3, e101. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Ding, Y.; Zhang, M.; Lu, Q.; Wu, S.; Zhu, H.; Song, P.; Zou, M.H. Ablation of Adenosine Monophosphate-Activated Protein Kinase alpha1 in Vascular Smooth Muscle Cells Promotes Diet-Induced Atherosclerotic Calcification In Vivo. Circ. Res. 2016, 119, 422–433. [Google Scholar] [CrossRef]
- Hosoya, A.; Yukita, A.; Ninomiya, T.; Hiraga, T.; Yoshiba, K.; Yoshiba, N.; Kasahara, E.; Nakamura, H. Localization of SUMOylation factors and Osterix in odontoblast lineage cells during dentin formation and regeneration. Histochem. Cell Biol. 2013, 140, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Yoshizawa, T.; Ishibashi, O.; Matsuda, A.; Ikegame, M.; Shimomura, J.; Mera, H.; Nakashima, K.; Kawashima, H. PIASxbeta is a key regulator of osterix transcriptional activity and matrix mineralization in osteoblasts. J. Cell Sci. 2007, 120, 2565–2573. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Li, J.; Lu, D.; Li, J.; Liu, M.; He, Y.; Williams, B.O.; Li, J.; Yang, T. Ginkgolic acid, a sumoylation inhibitor, promotes adipocyte commitment but suppresses adipocyte terminal differentiation of mouse bone marrow stromal cells. Sci. Rep. 2018, 8, 2545. [Google Scholar] [CrossRef]
- Zhang, Y.; Shao, J.; Wang, Z.; Yang, T.; Liu, S.; Liu, Y.; Fan, X.; Ye, W. Growth differentiation factor 11 is a protective factor for osteoblastogenesis by targeting PPARgamma. Gene 2015, 557, 209–214. [Google Scholar] [CrossRef]
- Nayak, A.; Viale-Bouroncle, S.; Morsczeck, C.; Muller, S. The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation. Mol. Cell 2014, 55, 47–58. [Google Scholar] [CrossRef]
- Ali, A.; Tyagi, S. Diverse roles of WDR5-RbBP5-ASH2L-DPY30 (WRAD) complex in the functions of the SET1 histone methyltransferase family. J. Biosci. 2017, 42, 155–159. [Google Scholar] [CrossRef]
- Bochynska, A.; Luscher-Firzlaff, J.; Luscher, B. Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin. Cells 2018, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Ernst, P.; Vakoc, C.R. WRAD: Enabler of the SET1-family of H3K4 methyltransferases. Brief. Funct. Genomics 2012, 11, 217–226. [Google Scholar] [CrossRef]
- Liu, C.F.; Samsa, W.E.; Zhou, G.; Lefebvre, V. Transcriptional control of chondrocyte specification and differentiation. Semin. Cell Dev. Biol. 2017, 62, 34–49. [Google Scholar] [CrossRef]
- Decker, R.S.; Koyama, E.; Pacifici, M. Articular Cartilage: Structural and Developmental Intricacies and Questions. Curr. Osteoporos. Rep. 2015, 13, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Akkiraju, H.; Nohe, A. Role of Chondrocytes in Cartilage Formation, Progression of Osteoarthritis and Cartilage Regeneration. J. Dev. Biol. 2015, 3, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Chaboissier, M.C.; Martin, J.F.; Schedl, A.; de Crombrugghe, B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes. Dev. 2002, 16, 2813–2828. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; Behringer, R.R.; de Crombrugghe, B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthr. Cartil. 2001, 9 (Suppl. A), S69–S75. [Google Scholar] [CrossRef]
- Liu, C.F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic. Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Eberspaecher, H.; Lu, J.; Zhang, R.; Nishida, T.; Kahyo, T.; Yasuda, H.; de Crombrugghe, B. Interactions between PIAS proteins and SOX9 result in an increase in the cellular concentrations of SOX9. J. Biol. Chem. 2006, 281, 14417–14428. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.A.; Wu, M.H.; Yan, C.H.; Chau, B.K.; So, H.; Ng, A.; Chan, A.; Cheah, K.S.; Briscoe, J.; Cheung, M. Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 2882–2887. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Chen, L.; Liao, Q.M.; Wang, K.J.; Chen, S.G.; Liu, Z.J.; Zhang, Z.C. SUMO-specific protease 2 (SENP2) functions as a tumor suppressor in osteosarcoma via SOX9 degradation. Exp. Ther. Med. 2018, 16, 5359–5365. [Google Scholar] [CrossRef]
- Oh, H.J.; Kido, T.; Lau, Y.F. PIAS1 interacts with and represses SOX9 transactivation activity. Mol. Reprod. Dev. 2007, 74, 1446–1455. [Google Scholar] [CrossRef]
- Zuo, C.; Wang, L.; Kamalesh, R.M.; Bowen, M.E.; Moore, D.C.; Dooner, M.S.; Reginato, A.M.; Wu, Q.; Schorl, C.; Song, Y.; et al. SHP2 regulates skeletal cell fate by modifying SOX9 expression and transcriptional activity. Bone Res. 2018, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, K.S.; Rosario, M.; Birchmeier, C.; Birchmeier, W. The tyrosine phosphatase Shp2 in development and cancer. Adv. Cancer Res. 2010, 106, 53–89. [Google Scholar] [CrossRef] [PubMed]
- Neel, B.G.; Gu, H.; Pao, L. The ’Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003, 28, 284–293. [Google Scholar] [CrossRef]
- Fernandez-Lloris, R.; Osses, N.; Jaffray, E.; Shen, L.N.; Vaughan, O.A.; Girwood, D.; Bartrons, R.; Rosa, J.L.; Hay, R.T.; Ventura, F. Repression of SOX6 transcriptional activity by SUMO modification. FEBS Lett. 2006, 580, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Kwon, S.; Kim, D.W. A post-translational modification cascade employing HDAC9-PIASy-RNF4 axis regulates chondrocyte hypertrophy by modulating Nkx3.2 protein stability. Cell Signal 2016, 28, 1336–1348. [Google Scholar] [CrossRef]
- Deng, L.; Ren, R.; Liu, Z.; Song, M.; Li, J.; Wu, Z.; Ren, X.; Fu, L.; Li, W.; Zhang, W.; et al. Stabilizing heterochromatin by DGCR8 alleviates senescence and osteoarthritis. Nat. Commun. 2019, 10, 3329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Chen, C.; Huang, J.; Zhang, H.; Zhao, X.; Deng, R.; Dou, J.; Jin, H.; Chen, R.; Xu, M.; et al. SUMOylation at K707 of DGCR8 controls direct function of primary microRNA. Nucleic. Acids Res. 2015, 43, 7945–7960. [Google Scholar] [CrossRef][Green Version]
- Kobayashi, T.; Papaioannou, G.; Mirzamohammadi, F.; Kozhemyakina, E.; Zhang, M.; Blelloch, R.; Chong, M.W. Early postnatal ablation of the microRNA-processing enzyme, Drosha, causes chondrocyte death and impairs the structural integrity of the articular cartilage. Osteoarthr. Cartil. 2015, 23, 1214–1220. [Google Scholar] [CrossRef]
- Li, S.; Wang, M.; Ao, X.; Chang, A.K.; Yang, C.; Zhao, F.; Bi, H.; Liu, Y.; Xiao, L.; Wu, H. CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-α. Oncogene 2013, 32, 4883–4891. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Liu, Z.; Song, M.; Li, W.; Wu, Z.; Wang, Z.; Wang, Q.; Wang, S.; Yan, K.; Sun, L.; et al. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 2021, 31, 187–205. [Google Scholar] [CrossRef]
- Chen, D.; Shen, J.; Zhao, W.; Wang, T.; Han, L.; Hamilton, J.L.; Im, H.J. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 2017, 5, 16044. [Google Scholar] [CrossRef] [PubMed]
- arcOGEN Consortium; arcOGEN Collaborators. Identification of new susceptibility loci for osteoarthritis (arcOGEN): A genome-wide association study. Lancet 2012, 380, 815–823. [Google Scholar] [CrossRef]
- Johnson, K.; Reynard, L.N.; Loughlin, J. Functional characterisation of the osteoarthritis susceptibility locus at chromosome 6q14.1 marked by the polymorphism rs9350591. BMC Med. Genet. 2015, 16, 81. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.J.; Loeser, R.F.; Yammani, R.R. Sumoylation and nuclear translocation of S100A4 regulate IL-1beta-mediated production of matrix metalloproteinase-13. J. Biol. Chem. 2010, 285, 31517–31524. [Google Scholar] [CrossRef] [PubMed]
- Daouti, S.; Latario, B.; Nagulapalli, S.; Buxton, F.; Uziel-Fusi, S.; Chirn, G.W.; Bodian, D.; Song, C.; Labow, M.; Lotz, M.; et al. Development of comprehensive functional genomic screens to identify novel mediators of osteoarthritis. Osteoarthr. Cartil. 2005, 13, 508–518. [Google Scholar] [CrossRef]
- Sun, T.; Gao, F.; Lin, X.; Yu, R.; Zhao, Y.; Luan, J.; Li, H.; Song, M. alpha-Lipoic acid (alpha-LA) inhibits the transcriptional activity of interferon regulatory factor 1 (IRF-1) via SUMOylation. Toxicol. Vitr. 2014, 28, 1242–1248. [Google Scholar] [CrossRef]
- Im, H.J.; Sharrocks, A.D.; Lin, X.; Yan, D.; Kim, J.; van Wijnen, A.J.; Hipskind, R.A. Basic fibroblast growth factor induces matrix metalloproteinase-13 via ERK MAP kinase-altered phosphorylation and sumoylation of Elk-1 in human adult articular chondrocytes. Open. Access Rheumatol. 2009, 1, 151–161. [Google Scholar] [CrossRef][Green Version]
- Ash, P.; Loutit, J.F.; Townsend, K.M. Osteoclasts derived from haematopoietic stem cells. Nature 1980, 283, 669–670. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Teitelbaum, S.L. Osteoclasts: New Insights. Bone Res. 2013, 1, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, K.; Yang, J.; Lao, Y.; Deng, L.; Deng, G.; Yi, J.; Sun, X.; Wang, Q. SENP3 Suppresses Osteoclastogenesis by De-conjugating SUMO2/3 from IRF8 in Bone Marrow-Derived Monocytes. Cell Rep. 2020, 30, 1951–1963.e54. [Google Scholar] [CrossRef]
- Hikata, T.; Takaishi, H.; Takito, J.; Hakozaki, A.; Furukawa, M.; Uchikawa, S.; Kimura, T.; Okada, Y.; Matsumoto, M.; Yoshimura, A.; et al. PIAS3 negatively regulates RANKL-mediated osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblasts. Blood 2009, 113, 2202–2212. [Google Scholar] [CrossRef]
- Palvimo, J.J. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 2007, 35, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Elliott, A.M.; Evans, J.A.; Chudley, A.E. Split hand foot malformation (SHFM). Clin. Genet. 2005, 68, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Ianakiev, P.; Kilpatrick, M.W.; Toudjarska, I.; Basel, D.; Beighton, P.; Tsipouras, P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 2000, 67, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Van Bokhoven, H.; Hamel, B.C.; Bamshad, M.; Sangiorgi, E.; Gurrieri, F.; Duijf, P.H.; Vanmolkot, K.R.; van Beusekom, E.; van Beersum, S.E.; Celli, J.; et al. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am. J. Hum. Genet. 2001, 69, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Ghioni, P.; D’Alessandra, Y.; Mansueto, G.; Jaffray, E.; Hay, R.T.; La Mantia, G.; Guerrini, L. The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle 2005, 4, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.P.; Wu, G.; Guo, Z.; Osada, M.; Fomenkov, T.; Park, H.L.; Trink, B.; Sidransky, D.; Fomenkov, A.; Ratovitski, E.A. Altered sumoylation of p63alpha contributes to the split-hand/foot malformation phenotype. Cell Cycle 2004, 3, 1587–1596. [Google Scholar] [CrossRef]
- Ranieri, M.; Vivo, M.; De Simone, M.; Guerrini, L.; Pollice, A.; La Mantia, G.; Calabro, V. Sumoylation and ubiquitylation crosstalk in the control of DeltaNp63alpha protein stability. Gene 2018, 645, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef]
- Alkuraya, F.S.; Saadi, I.; Lund, J.J.; Turbe-Doan, A.; Morton, C.C.; Maas, R.L. SUMO1 haploinsufficiency leads to cleft lip and palate. Science 2006, 313, 1751. [Google Scholar] [CrossRef]
- Song, T.; Li, G.; Jing, G.; Jiao, X.; Shi, J.; Zhang, B.; Wang, L.; Ye, X.; Cao, F. SUMO1 polymorphisms are associated with non-syndromic cleft lip with or without cleft palate. Biochem. Biophys. Res. Commun. 2008, 377, 1265–1268. [Google Scholar] [CrossRef] [PubMed]
- Mostowska, A.; Hozyasz, K.K.; Wojcicki, P.; Biedziak, B.; Paradowska, P.; Jagodzinski, P.P. Association between genetic variants of reported candidate genes or regions and risk of cleft lip with or without cleft palate in the polish population. Birth. Defects Res. A Clin. Mol. Teratol. 2010, 88, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Carter, T.C.; Molloy, A.M.; Pangilinan, F.; Troendle, J.F.; Kirke, P.N.; Conley, M.R.; Orr, D.J.; Earley, M.; McKiernan, E.; Lynn, E.C.; et al. Testing reported associations of genetic risk factors for oral clefts in a large Irish study population. Birth Defects Res. A Clin. Mol. Teratol. 2010, 88, 84–93. [Google Scholar] [CrossRef]
- Jia, Z.L.; Li, Y.; Meng, T.; Shi, B. Association between polymorphisms at small ubiquitin-like modifier 1 and nonsyndromic orofacial clefts in Western China. DNA Cell Biol. 2010, 29, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Pauws, E.; Stanier, P. FGF signalling and SUMO modification: New players in the aetiology of cleft lip and/or palate. Trends Genet. 2007, 23, 631–640. [Google Scholar] [CrossRef]
- Dobreva, G.; Dambacher, J.; Grosschedl, R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes. Dev. 2003, 17, 3048–3061. [Google Scholar] [CrossRef] [PubMed]
- Antonio Urrutia, G.; Ramachandran, H.; Cauchy, P.; Boo, K.; Ramamoorthy, S.; Boller, S.; Dogan, E.; Clapes, T.; Trompouki, E.; Torres-Padilla, M.E.; et al. ZFP451-mediated SUMOylation of SATB2 drives embryonic stem cell differentiation. Genes. Dev. 2021, 35, 1142–1160. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Bei, M. Modification of Msx1 by SUMO-1. Biochem. Biophys. Res. Commun. 2006, 345, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.J.; Lee, H. PIAS1 negatively regulates ubiquitination of Msx1 homeoprotein independent of its SUMO ligase activity. Mol. Cells 2011, 32, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.M.; Pauws, E.; Jones, M.C.; Singh, M.K.; Bussen, M.; Doudney, K.; Moore, G.E.; Kispert, A.; Brosens, J.J.; Stanier, P. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. Am. J. Hum. Genet. 2007, 81, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, F.J.; Ludecke, H.J.; Weger, S. SUMOylation modulates transcriptional repression by TRPS1. Biol. Chem. 2007, 388, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wang, Y.; Xu, D.; Nossent, J.; Pavlos, N.J.; Xu, J. Rheumatoid arthritis: Pathological mechanisms and modern pharmacologic therapies. Bone Res. 2018, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Rheumatoid arthritis. Nat. Rev. Dis. Primers 2018, 4, 18002. [CrossRef] [PubMed]
- Franz, J.K.; Pap, T.; Hummel, K.M.; Nawrath, M.; Aicher, W.K.; Shigeyama, Y.; Muller-Ladner, U.; Gay, R.E.; Gay, S. Expression of sentrin, a novel antiapoptotic molecule, at sites of synovial invasion in rheumatoid arthritis. Arthritis Rheum. 2000, 43, 599–607. [Google Scholar] [CrossRef]
- Lao, M.; Shi, M.; Zou, Y.; Huang, M.; Ye, Y.; Qiu, Q.; Xiao, Y.; Zeng, S.; Liang, L.; Yang, X.; et al. Protein Inhibitor of Activated STAT3 Regulates Migration, Invasion, and Activation of Fibroblast-like Synoviocytes in Rheumatoid Arthritis. J. Immunol. 2016, 196, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Lao, M.; Zhan, Z.; Li, N.; Xu, S.; Shi, M.; Zou, Y.; Huang, M.; Zeng, S.; Liang, L.; Xu, H. Role of small ubiquitin-like modifier proteins-1 (SUMO-1) in regulating migration and invasion of fibroblast-like synoviocytes from patients with rheumatoid arthritis. Exp. Cell Res. 2019, 375, 52–61. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Y.; Lao, M.; Wang, J.; Xu, S.; Li, R.; Xu, X.; Kuang, Y.; Shi, M.; Zou, Y.; et al. Increased SUMO-activating enzyme SAE1/UBA2 promotes glycolysis and pathogenic behavior of rheumatoid fibroblast-like synoviocytes. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Meinecke, I.; Cinski, A.; Baier, A.; Peters, M.A.; Dankbar, B.; Wille, A.; Drynda, A.; Mendoza, H.; Gay, R.E.; Hay, R.T.; et al. Modification of nuclear PML protein by SUMO-1 regulates Fas-induced apoptosis in rheumatoid arthritis synovial fibroblasts. Proc. Natl. Acad. Sci. USA 2007, 104, 5073–5078. [Google Scholar] [CrossRef]
- Maciejewska-Rodrigues, H.; Karouzakis, E.; Strietholt, S.; Hemmatazad, H.; Neidhart, M.; Ospelt, C.; Gay, R.E.; Michel, B.A.; Pap, T.; Gay, S.; et al. Epigenetics and rheumatoid arthritis: The role of SENP1 in the regulation of MMP-1 expression. J. Autoimmun. 2010, 35, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, X.; Kou, L.; Li, Y.; Meng, F.; Ma, F. SUMO-conjugating enzyme UBC9 promotes proliferation and migration of fibroblast-like synoviocytes in rheumatoid arthritis. Inflammation 2014, 37, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.; Peters, M.A.; Wehmeyer, C.; Strietholt, S.; Koers-Wunrau, C.; Bertrand, J.; Heitzmann, M.; Hillmann, A.; Sherwood, J.; Seyfert, C.; et al. Regulation of matrixmetalloproteinase-3 and matrixmetalloproteinase-13 by SUMO-2/3 through the transcription factor NF-kappaB. Ann. Rheum. Dis. 2013, 72, 1874–1881. [Google Scholar] [CrossRef]
- Vatsyayan, J.; Qing, G.; Xiao, G.; Hu, J. SUMO1 modification of NF-kappaB2/p100 is essential for stimuli-induced p100 phosphorylation and processing. EMBO Rep. 2008, 9, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sha, M.; Wang, Q.; Ma, Y.; Geng, X.; Gao, Y.; Feng, L.; Shen, Y.; Shen, Y. Small ubiquitin-related modifier 2/3 interacts with p65 and stabilizes it in the cytoplasm in HBV-associated hepatocellular carcinoma. BMC Cancer 2015, 15, 675. [Google Scholar] [CrossRef] [PubMed]
- Leidner, J.; Voogdt, C.; Niedenthal, R.; Moller, P.; Marienfeld, U.; Marienfeld, R.B. SUMOylation attenuates the transcriptional activity of the NF-kappaB subunit RelB. J. Cell Biochem. 2014, 115, 1430–1440. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Z.; Han, D.; Wei, C.; Liang, Y.; Jiang, T.; Chen, L.; Sha, M.; Cao, Y.; Huang, F.; et al. Mesencephalic Astrocyte-Derived Neurotrophic Factor Inhibits Liver Cancer Through Small Ubiquitin-Related Modifier (SUMO)ylation-Related Suppression of NF-kappaB/Snail Signaling Pathway and Epithelial-Mesenchymal Transition. Hepatology 2020, 71, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Whelan, J.S.; Davis, L.E. Osteosarcoma, Chondrosarcoma, and Chordoma. J. Clin. Oncol. 2018, 36, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Ritter, J.; Bielack, S.S. Osteosarcoma. Ann. Oncol. 2010, 21 (Suppl. 7), vii320–vii325. [Google Scholar] [CrossRef]
- Seeler, J.S.; Dejean, A. SUMO and the robustness of cancer. Nat. Rev. Cancer 2017, 17, 184–197. [Google Scholar] [CrossRef]
- Eifler, K.; Vertegaal, A.C.O. SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem. Sci. 2015, 40, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yu, K.; Yang, Z.; Li, Y.; Ma, X.; Bian, X.; Liu, F.; Li, L.; Liu, X.; Wu, W. Silencing Ubc9 expression suppresses osteosarcoma tumorigenesis and enhances chemosensitivity to HSV-TK/GCV by regulating connexin 43 SUMOylation. Int. J.Oncol. 2018, 53, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, L.; Li, Y.; Ma, X.; Bian, X.; Liu, X.; Wang, G.; Zhang, D. Overexpression of SENP1 reduces the stemness capacity of osteosarcoma stem cells and increases their sensitivity to HSVtk/GCV. Int. J. Oncol. 2018, 53, 2010–2020. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Barker, D.; Gibbins, J.M.; Dash, P.R. Talin is a substrate for SUMOylation in migrating cancer cells. Exp. Cell Res. 2018, 370, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, X.C. Inhibition of SENP5 suppresses cell growth and promotes apoptosis in osteosarcoma cells. Exp. Ther. Med. 2014, 7, 1691–1695. [Google Scholar] [CrossRef]
- Wang, J.; Ni, J.; Yi, S.; Song, D.; Ding, M. Protein inhibitor of activated STAT xalpha depresses cyclin D and cyclin D kinase, and contributes to the inhibition of osteosarcoma cell progression. Mol. Med. Rep. 2016, 13, 1645–1652. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, P.; Yang, X.; Ying, M.; Chaudhry, P.; Wang, A.; Shimada, H.; May, W.A.; Adams, G.B.; Mock, D.; Triche, T.J.; et al. Retinoid-suppressed phosphorylation of RARalpha mediates the differentiation pathway of osteosarcoma cells. Oncogene 2010, 29, 2772–2783. [Google Scholar] [CrossRef][Green Version]
- Zhou, Q.; Zhang, L.; Chen, Z.; Zhao, P.; Ma, Y.; Yang, B.; He, Q.; Ying, M. Small ubiquitin-related modifier-1 modification regulates all-trans-retinoic acid-induced differentiation via stabilization of retinoic acid receptor alpha. FEBS J. 2014, 281, 3032–3047. [Google Scholar] [CrossRef]
- Wang, X.; Liang, X.; Liang, H.; Wang, B. SENP1/HIF-1alpha feedback loop modulates hypoxia-induced cell proliferation, invasion, and EMT in human osteosarcoma cells. J. Cell Biochem. 2018, 119, 1819–1826. [Google Scholar] [CrossRef]
- Wang, L.; Wu, J.; Song, S.; Chen, H.; Hu, Y.; Xu, B.; Liu, J. Plasma Exosome-Derived Sentrin SUMO-Specific Protease 1: A Prognostic Biomarker in Patients With Osteosarcoma. Front. Oncol. 2021, 11, 625109. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Plow, E.F.; Qin, J. Initiation of focal adhesion assembly by talin and kindlin: A dynamic view. Protein Sci. 2021, 30, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Kroonen, J.S.; Kruisselbrink, A.B.; Briaire-de Bruijn, I.H.; Olaofe, O.O.; Bovee, J.; Vertegaal, A.C.O. SUMOylation Is Associated with Aggressive Behavior in Chondrosarcoma of Bone. Cancers 2021, 13, 3823. [Google Scholar] [CrossRef] [PubMed]
- Brackett, C.M.; Blagg, B.S.J. Current Status of SUMOylation Inhibitors. Curr. Med. Chem. 2021, 28, 3892–3912. [Google Scholar] [CrossRef] [PubMed]
- Kukkula, A.; Ojala, V.K.; Mendez, L.M.; Sistonen, L.; Elenius, K.; Sundvall, M. Therapeutic Potential of Targeting the SUMO Pathway in Cancer. Cancers 2021, 13, 4402. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Craig, S.E.L.; Molchanov, V.; Floramo, J.S.; Zhao, Y.; Yang, T. SUMOylation in Skeletal Development, Homeostasis, and Disease. Cells 2022, 11, 2710. https://doi.org/10.3390/cells11172710
Liu H, Craig SEL, Molchanov V, Floramo JS, Zhao Y, Yang T. SUMOylation in Skeletal Development, Homeostasis, and Disease. Cells. 2022; 11(17):2710. https://doi.org/10.3390/cells11172710
Chicago/Turabian StyleLiu, Huadie, Sonya E. L. Craig, Vladimir Molchanov, Joseph S. Floramo, Yaguang Zhao, and Tao Yang. 2022. "SUMOylation in Skeletal Development, Homeostasis, and Disease" Cells 11, no. 17: 2710. https://doi.org/10.3390/cells11172710
APA StyleLiu, H., Craig, S. E. L., Molchanov, V., Floramo, J. S., Zhao, Y., & Yang, T. (2022). SUMOylation in Skeletal Development, Homeostasis, and Disease. Cells, 11(17), 2710. https://doi.org/10.3390/cells11172710






