A New Model Organism to Investigate Extraocular Photoreception: Opsin and Retinal Gene Expression in the Sea Urchin Paracentrotus lividus
Abstract
:1. Introduction
2. Materials & Methods
2.1. Animal Husbandry and Culture of Embryos, Larvae, and Juveniles
2.2. Rudiment Microdissection and Dissociation
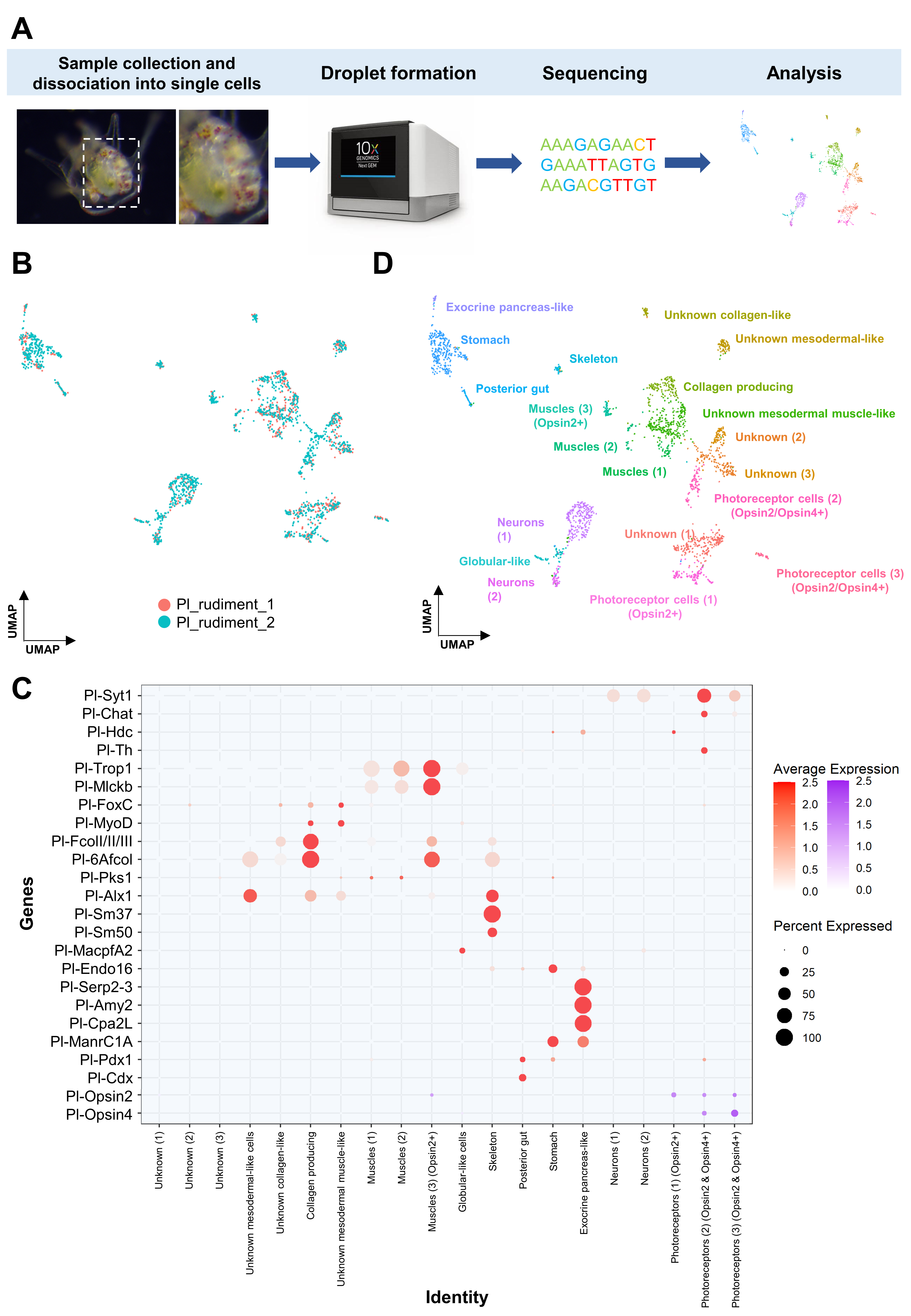
2.3. Single Cell RNA Sequencing and Computational Analysis
2.4. Whole Mount Fluorescent In Situ Hybridization (FISH)
2.5. Immunohistochemistry (IHC)
2.6. Quantitative Real Time PCR (qRT-PCR)
3. Results
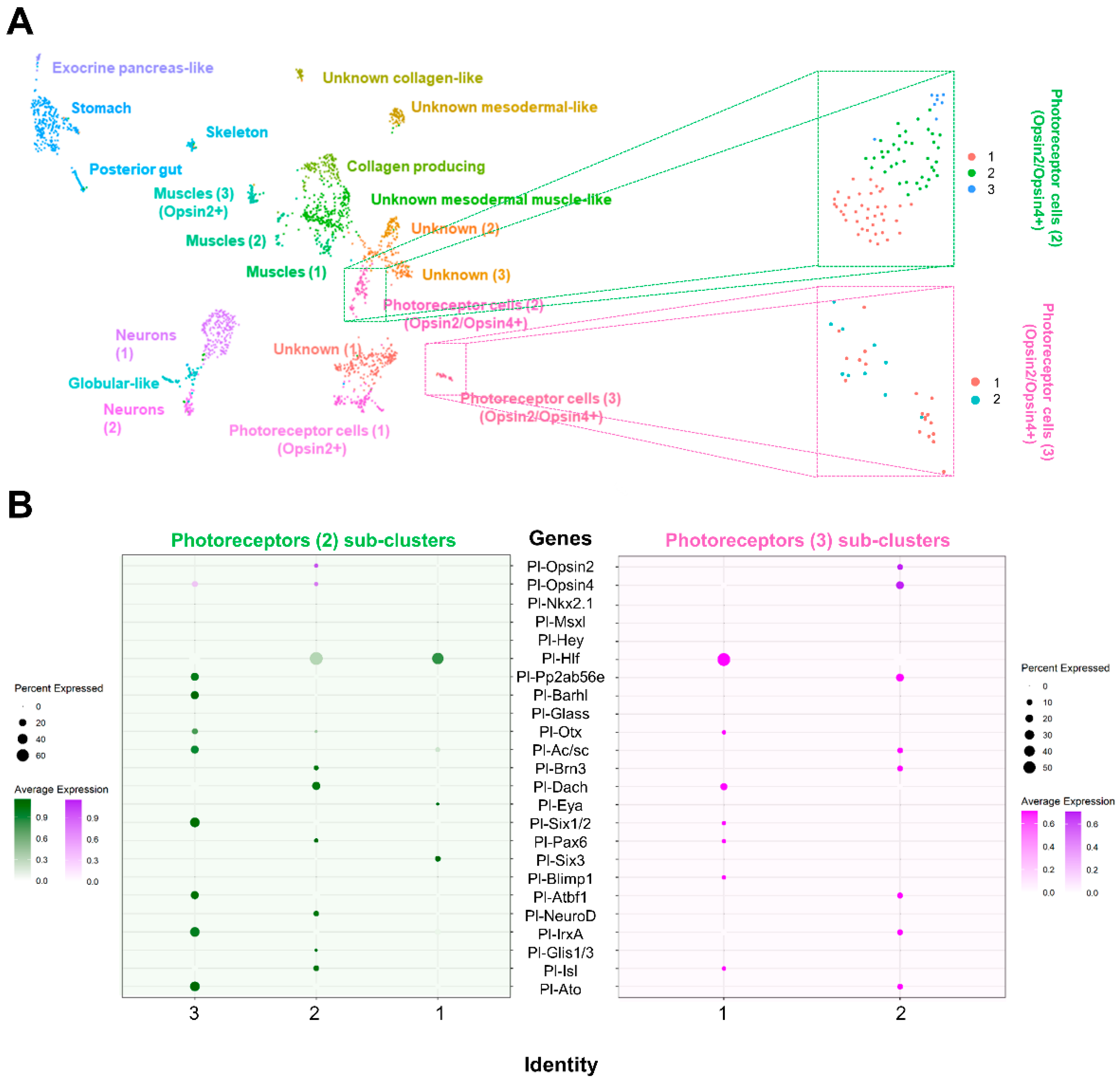
3.1. Generating a Cell Type Atlas of the Mature Rudiment Stage with Single-Cell Transcriptomics
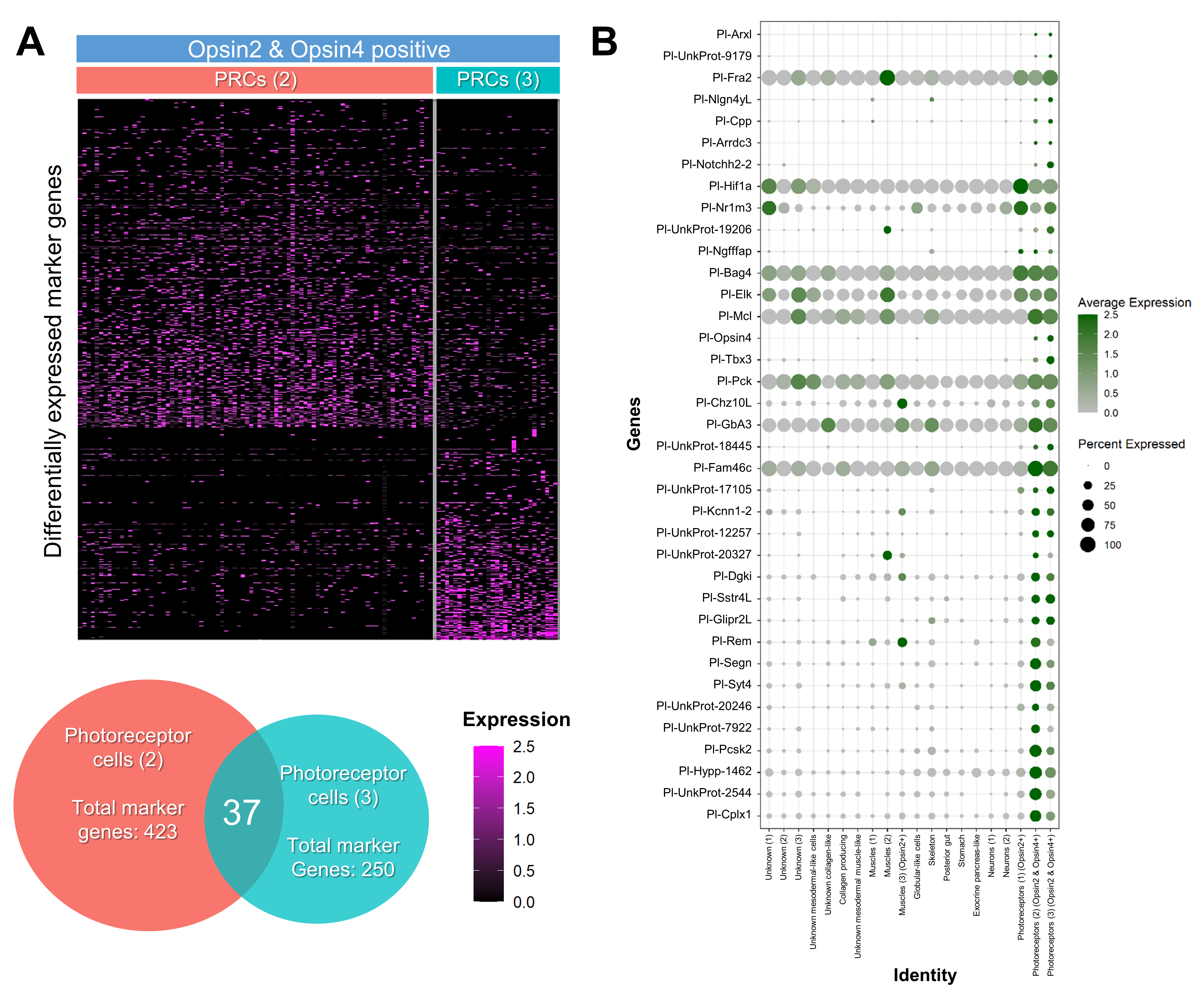
3.2. Investigating the Molecular Fingerprint of Prcs in Mature Rudiment
3.3. Spatial and Temporal Expression of Opsin4, Pax6, Neurod and Six3 during Juvenile Development
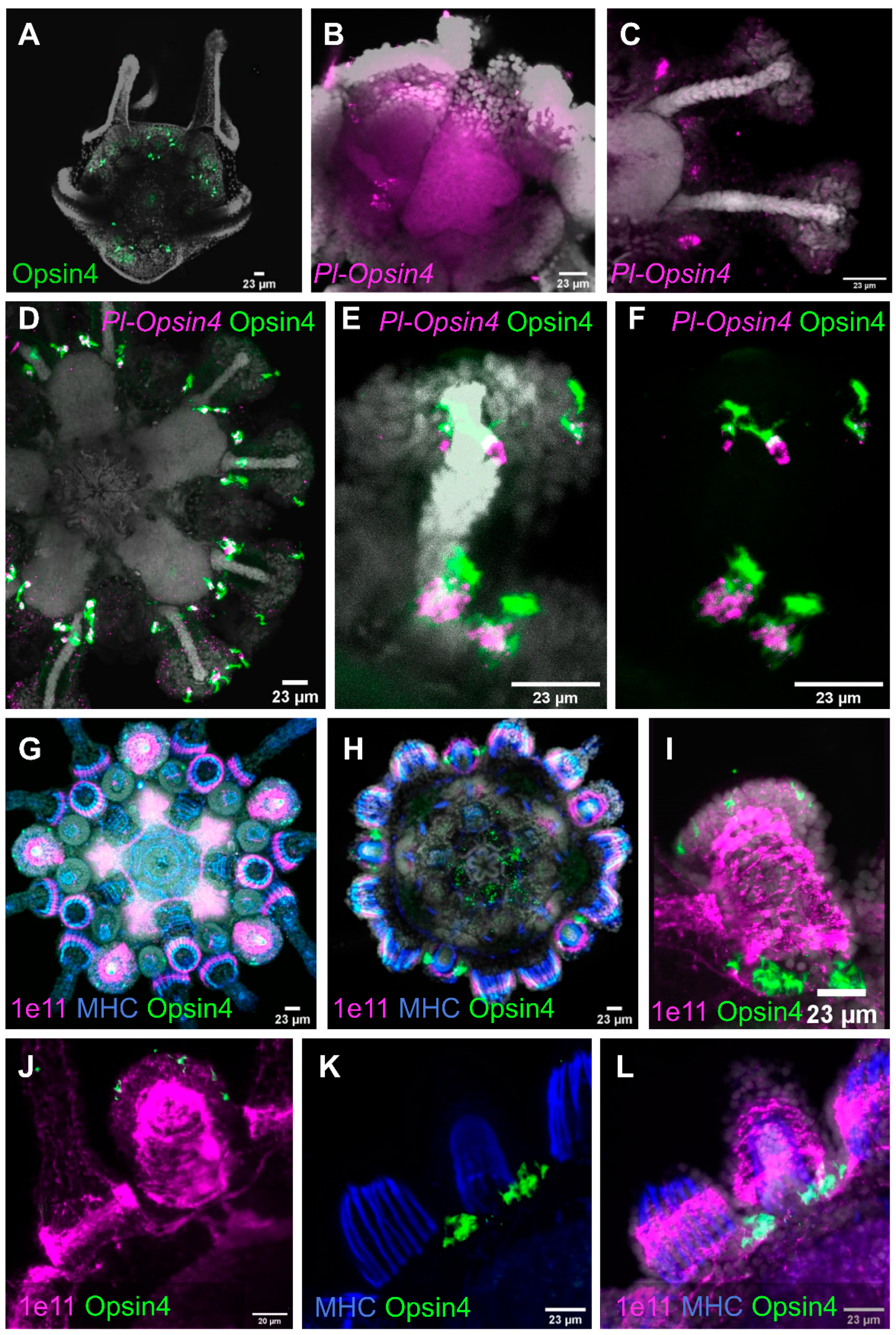
3.3.1. Opsin4
3.3.2. Pax6, NeuroD and Six3
3.4. Expression Profiles of Opsin Genes in Post-Metamorphic Juveniles
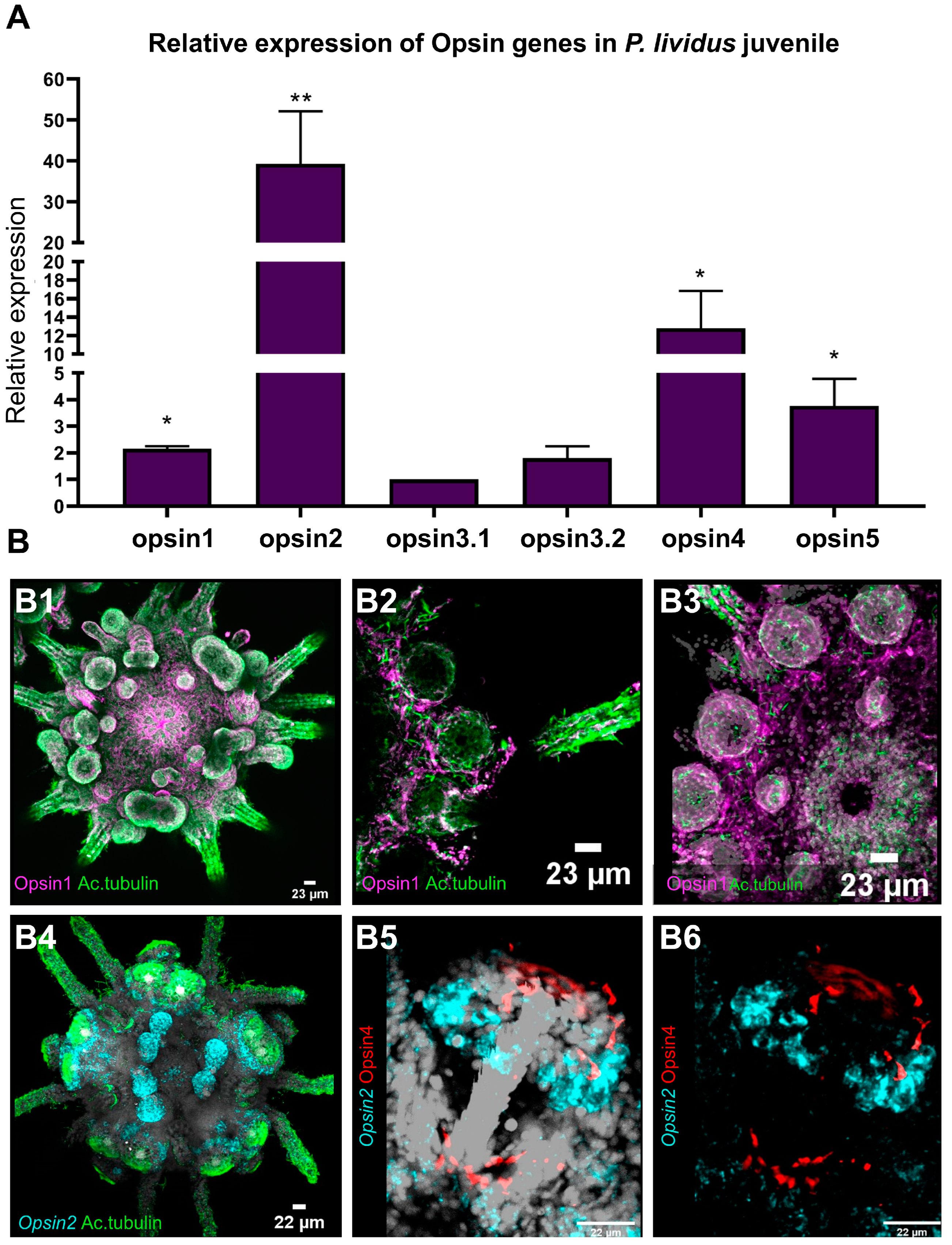
3.4.1. Relative Gene Expression of Opsin1, Opsin2, Opsin3.1, Opsin3.2, Opsin4 and Opsin5
3.4.2. Ciliary Opsin (opsin1)
3.4.3. Echinopsin (opsin2)
4. Discussion
4.1. Cell Type Identity
4.2. Retinal Gene Expression and Its Putative Function
4.3. Spatial and Temporal Analysis of Retinal Gene Products
4.3.1. Pax6, Six3, Otx
4.3.2. Opsins
4.4. Sea Urchin Photoreceptors
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arendt, D. Evolution of Eyes and Photoreceptor Cell Types. Int. J. Dev. Biol. 2003, 47, 563–571. [Google Scholar] [PubMed]
- Plachetzki, D.C.; Serb, J.M.; Oakley, T.H. New Insights into the Evolutionary History of Photoreceptor Cells. Trends Ecol. Evol. 2005, 20, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Gehring, W. New Perspectives on Eye Development and the Evolution of Eyes and Photoreceptors. J. Hered. 2005, 96, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Arendt, D.; Wittbrodt, J. Reconstructing the Eyes of Urbilateria. Philos. Trans. R. Soc. B Biol. Sci. 2001, 356, 1545–1563. [Google Scholar] [CrossRef]
- Arendt, D.; Tessmar-Raible, K.; Snyman, H.; Dorresteijn, A.W.; Wittbrodf, J. Ciliary Photoreceptors with a Vertebrate-Type Opsin in an Invertebrate Brain. Science 2004, 306, 869–871. [Google Scholar] [CrossRef]
- Panda, S.; Nayak, S.K.; Campo, B.; Walker, J.R.; Hogenesch, J.B.; Jegla, T. Illumination of the Melanopsin Signaling Pathway. Science 2005, 307, 600–604. [Google Scholar] [CrossRef]
- Mathers, P.H.; Grinberg, A.; Mahon, K.A.; Jamrich, M. The Rx Homeobox Gene Is Essential for Vertebrate Eye Development. Nature 1997, 387, 603–607. [Google Scholar] [CrossRef]
- Pan, Y.; Nekkalapudi, S.; Kelly, L.E.; El-Hodiri, H.M. The Rx-like Homeobox Gene (Rx-L) Is Necessary for Normal Photoreceptor Development. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4245–4253. [Google Scholar] [CrossRef]
- Burke, R.D.; Angerer, L.M.; Elphick, M.R.; Humphrey, G.W.; Yaguchi, S.; Kiyama, T.; Liang, S.; Mu, X.; Agca, C.; Klein, W.H.; et al. A Genomic View of the Sea Urchin Nervous System. Dev. Biol. 2006, 300, 434–460. [Google Scholar] [CrossRef]
- Raible, F.; Tessmar-Raible, K.; Arboleda, E.; Kaller, T.; Bork, P.; Arendt, D.; Arnone, M.I. Opsins and Clusters of Sensory G-Protein-Coupled Receptors in the Sea Urchin Genome. Dev. Biol. 2006, 300, 461–475. [Google Scholar] [CrossRef] [Green Version]
- Millott, N.; Coleman, R. The Podial Pit—A New Structure in the Echinoid Diadema antillarum Philippi. Zeitschrift für Zellforsch. und Mikroskopische Anat. 1969, 95, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Millott, N. The Photosensitivity of Echinoids. Adv. Mar. Biol. 1976, 13, 1–52. [Google Scholar] [CrossRef]
- Burke, R.D. Podial Sensory Receptors and the Induction of Metamorphosis in Echinoids. J. Exp. Mar. Bio. Ecol. 1980, 47, 223–234. [Google Scholar] [CrossRef]
- Agca, C.; Elhajj, M.C.; Klein, W.H.; Venuti, J.M. Neurosensory and Neuromuscular Organization in Tube Feet of the Sea Urchin Strongylocentrotus purpuratus. J. Comp. Neurol. 2011, 519, 3566–3579. [Google Scholar] [CrossRef]
- Lesser, M.P.; Carleton, K.L.; Böttger, S.A.; Barry, T.M.; Walker, C.W. Sea Urchin Tube Feet Are Photosensory Organs That Express a Rhabdomeric-like Opsin and PAX6. Proc. R. Soc. B Biol. Sci. 2011, 278, 3371–3379. [Google Scholar] [CrossRef]
- Ullrich-Lüter, E.M.; Dupont, S.; Arboleda, E.; Hausen, H.; Arnone, M.I. Unique System of Photoreceptors in Sea Urchin Tube Feet. Proc. Natl. Acad. Sci. USA 2011, 108, 8367–8372. [Google Scholar] [CrossRef]
- Ullrich-Lüter, E.M.; D’Aniello, S.; Arnone, M.I. C-Opsin Expressing Photoreceptors in Echinoderms. Integr. Comp. Biol. 2013, 53, 27–38. [Google Scholar] [CrossRef]
- D’Aniello, S.; Delroisse, J.; Valero-Gracia, A.; Lowe, E.K.; Byrne, M.; Cannon, J.T.; Halanych, K.M.; Elphick, M.R.; Mallefet, J.; Kaul-Strehlow, S.; et al. Opsin Evolution in the Ambulacraria. Mar. Genom. 2015, 24, 177–183. [Google Scholar] [CrossRef]
- Feuda, R.; Hamilton, S.C.; McInerney, J.O.; Pisani, D. Metazoan Opsin Evolution Reveals a Simple Route to Animal Vision. Proc. Natl. Acad. Sci. USA 2012, 109, 18868–18872. [Google Scholar] [CrossRef]
- Kojima, D.; Terakita, A.; Ishikawa, T.; Tsukahara, Y.; Maeda, A.; Shichida, Y. A Novel G(o)-Mediated Phototransduction Cascade in Scallop Visual Cells. J. Biol. Chem. 1997, 272, 22979–22982. [Google Scholar] [CrossRef] [Green Version]
- Ayers, T.; Tsukamoto, H.; Gühmann, M.; Veedin Rajan, V.B.; Tessmar-Raible, K. A Go-Type Opsin Mediates the Shadow Reflex in the Annelid Platynereis dumerilii. BMC Biol. 2018, 16, 41. [Google Scholar] [CrossRef] [PubMed]
- Czerny, T.; Busslinger, M. DNA-Binding and Transactivation Properties of Pax-6: Three Amino Acids in the Paired Domain Are Responsible for the Different Sequence Recognition of Pax-6 and BSAP (Pax-5). Mol. Cell. Biol. 1995, 15, 2858–2871. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.A.; Agca, C.; Mocko-Strand, J.A.; Wang, J.; Ullrich-Lüter, E.; Pan, P.; Wang, S.W.; Arnone, M.I.; Frishman, L.J.; Klein, W.H. Substituting Mouse Transcription Factor Pou4f2 with a Sea Urchin Orthologue Restores Retinal Ganglion Cell Development. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152978. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.; Koop, D.; Morris, V.B.; Chui, J.; Wray, G.A.; Cisternas, P. Expression of Genes and Proteins of the Pax-Six-Eya-Dach Network in the Metamorphic Sea Urchin: Insights into Development of the Enigmatic Echinoderm Body Plan and Sensory Structures. Dev. Dyn. 2018, 247, 239–249. [Google Scholar] [CrossRef]
- Musser, J.M.; Schippers, K.J.; Nickel, M.; Mizzon, G.; Kohn, A.B.; Pape, C.; Ronchi, P.; Papadopoulos, N.; Tarashansky, A.J.; Hammel, J.U.; et al. Profiling Cellular Diversity in Sponges Informs Animal Cell Type and Nervous System Evolution. Science 2021, 374, 717–723. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Saudemont, B.; Chomsky, E.; Plessier, F.; Mailhé, M.P.; Renno, J.; Loe-Mie, Y.; Lifshitz, A.; Mukamel, Z.; Schmutz, S.; et al. Cnidarian Cell Type Diversity and Regulation Revealed by Whole-Organism Single-Cell RNA-Seq. Cell 2018, 173, 1520–1534.e20. [Google Scholar] [CrossRef]
- Rentzsch, F.; Juliano, C.; Galliot, B. Modern Genomic Tools Reveal the Structural and Cellular Diversity of Cnidarian Nervous Systems. Curr. Opin. Neurobiol. 2019, 56, 87–96. [Google Scholar] [CrossRef]
- Meyer, A.; Ku, C.; Hatleberg, W.; Telmer, C.A.; Hinman, V. New Hypotheses of Cell Type Diversity and Novelty from Comparative Single Cell and Nuclei Transcriptomics in Echinoderms. bioRxiv 2022. [Google Scholar] [CrossRef]
- Massri, A.J.; Greenstreet, L.; Afanassiev, A.; Berrio, A.; Wray, G.A.; Schiebinger, G.; McClay, D.R. Developmental Single-Cell Transcriptomics in the Lytechinus variegatus Sea Urchin Embryo. Development 2021, 148, dev198614. [Google Scholar] [CrossRef]
- Paganos, P.; Voronov, D.; Musser, J.; Arendt, D.; Arnone, M.I. Single Cell Rna Sequencing of the Strongylocentrotus purpuratus Larva Reveals the Blueprint of Major Cell Types and Nervous System of a Nonchordate Deuterostome. eLife 2021, 10, e70416. [Google Scholar] [CrossRef]
- Zeisel, A.; Muñoz-Manchado, A.B.; Codeluppi, S.; Lönnerberg, P.; Manno, G.L.; Juréus, A.; Marques, S.; Munguba, H.; He, L.; Betsholtz, C.; et al. Cell Types in the Mouse Cortex and Hippocampus Revealed by Single-Cell RNA-Seq. Science 2015, 347, 1138–1142. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shiau, F.; Yi, W.; Lu, S.; Wu, Q.; Pearson, J.D.; Kallman, A.; Zhong, S.; Hoang, T.; Zuo, Z.; et al. Single-Cell Analysis of Human Retina Identifies Evolutionarily Conserved and Species-Specific Mechanisms Controlling Development. Dev. Cell 2020, 53, 473–491.e9. [Google Scholar] [CrossRef] [PubMed]
- Marletaz, F. The Paracentrotus lividus Genome. Cell Genom. 2022; manuscript under revision. [Google Scholar]
- Cellario, C.; Fenaux, L. Paracentrotus lividus (Lamarck) in Culture (Larval and Benthic Phases): Parameters of Growth Observed during Two Years Following Metamorphosis. Aquaculture 1990, 84, 173–188. [Google Scholar] [CrossRef]
- Gosselin, P.; Jangoux, M. Induction of Metamorphosis in Paracentrotus lividus Larvae (Echinodermata, Echinoidea). Oceanol. Acta 1996, 19, 293–296. [Google Scholar]
- Kirwan, J.D.; Bok, M.J.; Smolka, J.; Foster, J.J.; Hernández, J.C.; Nilsson, D.E. The Sea Urchin Diadema africanum Uses Low Resolution Vision to Find Shelter and Deter Enemies. J. Exp. Biol. 2018, 221, jeb176271. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Paganos, P.; Caccavale, F.; La Vecchia, C.; D’Aniello, E.; D’Aniello, S.; Arnone, M.I. FISH for All: A Fast and Efficient Fluorescent In Situ Hybridization (FISH) Protocol for Marine Embryos and Larvae. Front. Physiol. 2022, 13, 742. [Google Scholar] [CrossRef]
- Perillo, M.; Paganos, P.; Spurrell, M.; Arnone, M.I.; Wessel, G.M. Methodology for Whole Mount and Fluorescent RNA In Situ Hybridization in Echinoderms: Single, Double, and Beyond. Methods Mol. Biol. 2021, 2219, 195–216. [Google Scholar]
- Tsironis, I.; Paganos, P.; Gouvi, G.; Tsimpos, P.; Stamopoulou, A.; Arnone, M.I.; Flytzanis, C.N. Coup-TF: A Maternal Factor Essential for Differentiation along the Embryonic Axes in the Sea Urchin Paracentrotus lividus. Dev. Biol. 2021, 475, 131–144. [Google Scholar] [CrossRef]
- Burke, R.D.; Osborne, L.; Wang, D.; Murabe, N.; Yaguchi, S.; Nakajima, Y. Neuron-Specific Expression of a Synaptotagmin Gene in the Sea Urchin Strongylocentrotus purpuratus. J. Comp. Neurol. 2006, 496, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Caccavale, F.; Annona, G.; Subirana, L.; Escriva, H.; Bertrand, S.; D’aniello, S. Crosstalk between Nitric Oxide and Retinoic Acid Pathways Is Essential for Amphioxus Pharynx Development. eLife 2021, 10, e58295. [Google Scholar] [CrossRef] [PubMed]
- Milito, A.; Murano, C.; Castellano, I.; Romano, G.; Palumbo, A. Antioxidant and Immune Response of the Sea Urchin Paracentrotus lividus to Different Re-Suspension Patterns of Highly Polluted Marine Sediments. Mar. Environ. Res. 2020, 160, 104978. [Google Scholar] [CrossRef] [PubMed]
- Perillo, M.; Oulhen, N.; Foster, S.; Spurrell, M.; Calestani, C.; Wessel, G. Regulation of Dynamic Pigment Cell States at Single-Cell Resolution. eLife 2020, 9, e60388. [Google Scholar] [CrossRef]
- Hyman, L.H. The Invertebrates: Echinodermata; McGraw-HillBook Co., Inc.: New York, NY, USA, 1955; Volume 4, pp. 1–763. [Google Scholar]
- Cary, G.A.; McCauley, B.S.; Zueva, O.; Pattinato, J.; Longabaugh, W.; Hinman, V.F. Systematic Comparison of Sea Urchin and Sea Star Developmental Gene Regulatory Networks Explains How Novelty Is Incorporated in Early Development. Nat. Commun. 2020, 11, 6235. [Google Scholar] [CrossRef]
- Smith, M.K.; Bose, U.; Mita, M.; Hall, M.R.; Elizur, A.; Motti, C.A.; Cummins, S.F. Differences in Small Molecule Neurotransmitter Profiles from the Crown-of-Thorns Seastar Radial Nerve Revealed between Sexes and Following Food-Deprivation. Front. Endocrinol. 2018, 9, 551. [Google Scholar] [CrossRef]
- Díaz-Balzac, C.A.; Mejías, W.; Jiménez, L.B.; García-Arrarás, J.E. The Catecholaminergic Nerve Plexus of Holothuroidea. Zoomorphology 2010, 129, 99–109. [Google Scholar] [CrossRef]
- Kingston, A.C.N.; Cronin, T.W. Diverse Distributions of Extraocular Opsins in Crustaceans, Cephalopods, and Fish. Integr. Comp. Biol. 2016, 56, 820–833. [Google Scholar] [CrossRef]
- Halford, S.; Freedman, M.S.; Bellingham, J.; Inglis, S.L.; Poopalasundaram, S.; Soni, B.G.; Foster, R.G.; Hunt, D.M. Characterization of a Novel Human Opsin Gene with Wide Tissue Expression and Identification of Embedded and Flanking Genes on Chromosome 1q43. Genomics 2001, 72, 203–208. [Google Scholar] [CrossRef]
- Yim, P.D.; Gallos, G.; Perez-Zoghbi, J.F.; Zhang, Y.; Xu, D.; Wu, A.; Berkowitz, D.E.; Emala, C.W. Airway Smooth Muscle Photorelaxation via Opsin Receptor Activation. Am. J. Physiol.—Lung Cell. Mol. Physiol. 2019, 316, L82–L93. [Google Scholar] [CrossRef]
- Moutsaki, P.; Whitmore, D.; Bellingham, J.; Sakamoto, K.; David-Gray, Z.K.; Foster, R.G. Teleost Multiple Tissue (Tmt) Opsin: A Candidate Photopigment Regulating the Peripheral Clocks of Zebrafish? Mol. Brain Res. 2003, 112, 135–145. [Google Scholar] [CrossRef]
- Wu, A.D.; Dan, W.; Zhang, Y.; Vemaraju, S.; Upton, B.A.; Lang, R.A.; Buhr, E.D.; Berkowitz, D.E.; Gallos, G.; Emala, C.W.; et al. Opsin 3–Gas Promotes Airway Smooth Muscle Relaxation Modulated by G Protein Receptor Kinase 2. Am. J. Respir. Cell Mol. Biol. 2021, 64, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Yim, P.D.; Hyuga, S.; Wu, A.D.; Dan, W.; Vink, J.Y.; Gallos, G. Activation of an Endogenous Opsin 3 Light Receptor Mediates Photo-Relaxation of Pre-Contracting Late Gestation Human Uterine Smooth Muscle Ex Vivo. Reprod. Sci. 2020, 27, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Buscone, S.; Mardaryev, A.N.; Raafs, B.; Bikker, J.W.; Sticht, C.; Gretz, N.; Farjo, N.; Uzunbajakava, N.E.; Botchkareva, N.V. A New Path in Defining Light Parameters for Hair Growth: Discovery and Modulation of Photoreceptors in Human Hair Follicle. Lasers Surg. Med. 2017, 49, 705–718. [Google Scholar] [CrossRef]
- Kitamura, K.; Yanazawa, M.; Sugiyama, N.; Miura, H.; Iizuka-Kogo, A.; Kusaka, M.; Omichi, K.; Suzuki, R.; Kato-Fukui, Y.; Kamiirisa, K.; et al. Mutation of ARX Causes Abnormal Development of Forebrain and Testes in Mice and X-Linked Lissencephaly with Abnormal Genitalia in Humans. Nat. Genet. 2002, 32, 359–369. [Google Scholar] [CrossRef]
- Elphick, M.R.; Rowe, M.L. NGFFFamide and Echinotocin: Structurally Unrelated Myoactive Neuropeptides Derived from Neurophysin-Containing Precursors in Sea Urchins. J. Exp. Biol. 2009, 212, 1067–1077. [Google Scholar] [CrossRef]
- Wood, N.J.; Mattiello, T.; Rowe, M.L.; Ward, L.; Perillo, M.; Arnone, M.I.; Elphick, M.R.; Oliveri, P. Neuropeptidergic Systems in Pluteus Larvae of the Sea Urchin Strongylocentrotus purpuratus: Neurochemical Complexity in a “Simple” Nervous System. Front. Endocrinol. 2018, 9, 628. [Google Scholar] [CrossRef]
- Schmidt, T.M.; Chen, S.K.; Hattar, S. Intrinsically Photosensitive Retinal Ganglion Cells: Many Subtypes, Diverse Functions. Trends Neurosci. 2011, 34, 572–580. [Google Scholar] [CrossRef]
- Zhu, M.Y.; Gasperowicz, M.; Chow, R.L. The Expression of NOTCH2, HES1 and SOX9 during Mouse Retinal Development. Gene Expr. Patterns 2013, 13, 78–83. [Google Scholar] [CrossRef]
- Bernardos, R.L.; Lentz, S.I.; Wolfe, M.S.; Raymond, P.A. Notch–Delta Signaling Is Required for Spatial Patterning and Müller Glia Differentiation in the Zebrafish Retina. Dev. Biol. 2005, 278, 381–395. [Google Scholar] [CrossRef]
- Motahari, Z.; Martinez-De Luna, R.I.; Viczian, A.S.; Zuber, M.E. Tbx3 Represses Bmp4 Expression and, with Pax6, Is Required and Sufficient for Retina Formation. Dev. 2016, 143, 3560–3572. [Google Scholar] [CrossRef] [PubMed]
- Angueyra, J.; Kunze, V.P.; Patak, L.K.; Kim, H.; Kindt, K.S.; Li, W. Identification of Transcription Factors Involved in the Specification of Photoreceptor Subtypes. bioRxiv 2022. [Google Scholar] [CrossRef]
- Rorick, A.M.; Mei, W.; Liette, N.L.; Phiel, C.; El-Hodiri, H.M.; Yang, J. PP2A:B56ε Is Required for Eye Induction and Eye Field Separation. Dev. Biol. 2007, 302, 477–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Q.; Chen, H.; Xie, X.; Libby, R.T.; Tian, N.; Gan, L. BARHL2 Differentially Regulates the Development of Retinal Amacrine and Ganglion Neurons. J. Neurosci. 2009, 29, 3992–4003. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Morales, J.R.; Signore, M.; Acampora, D.; Simeone, A.; Bovolenta, P. Otx Genes Are Required for Tissue Specification in the Developing Eye. Development 2001, 128, 2019–2030. [Google Scholar] [CrossRef]
- Frankfort, B.J.; Pepple, K.L.; Mamlouk, M.; Rose, M.F.; Mardon, G. Senseless Is Required for Pupal Retinal Development in Drosophila. Genesis 2004, 38, 182–194. [Google Scholar] [CrossRef]
- Liu, W.; Khare, S.L.; Liang, X.; Peters, M.A.; Liu, X.; Cepko, C.L.; Xiang, M. All Brn3 Genes Can Promote Retinal Ganglion Cell Differentiation in the Chick. Development 2000, 127, 3237–3247. [Google Scholar] [CrossRef]
- Silver, S.J.; Rebay, I. Signaling Circuitries in Development: Insights from the Retinal Determination Gene Network. Development 2005, 132, 3–13. [Google Scholar] [CrossRef]
- Marquardt, T.; Ashery-Padan, R.; Andrejewski, N.; Scardigli, R.; Guillemot, F.; Gruss, P. Pax6 Is Required for the Multipotent State of Retinal Progenitor Cells. Cell 2001, 105, 43–55. [Google Scholar] [CrossRef]
- Conte, I.; Marco-Ferreres, R.; Beccari, L.; Cisneros, E.; Ruiz, J.M.; Tabanera, N.; Bovolenta, P. Proper Differentiation of Photoreceptors and Amacrine Cells Depends on a Regulatory Loop between NeuroD and Six6. Development 2010, 137, 2307–2317. [Google Scholar] [CrossRef]
- Zagozewski, J.L.; Zhang, Q.; Pinto, V.I.; Wigle, J.T.; Eisenstat, D.D. The Role of Homeobox Genes in Retinal Development and Disease. Dev. Biol. 2014, 393, 195–208. [Google Scholar] [CrossRef]
- Martín-Partido, G.; Francisco-Morcillo, J. The Role of Islet-1 in Cell Specification, Differentiation, and Maintenance of Phenotypes in the Vertebrate Neural Retina. Neural Regen. Res. 2015, 10, 1951. [Google Scholar] [PubMed]
- Hughes, S.; Edwards, J.K.; Wilcox, A.G.; Pothecary, C.A.; Barnard, A.R.; Joynson, R.; Joynson, G.; Hankins, M.W.; Peirson, S.N.; Banks, G.; et al. Zfhx3 Modulates Retinal Sensitivity and Circadian Responses to Light. FASEB J. 2021, 35, e21802. [Google Scholar] [CrossRef] [PubMed]
- Furimsky, M.; Wang, Y.P.; Wallace, V.A. Gli3 Controls Precursor Cell Proliferation and Differentiation in the Developing Mouse Retina. Investig. Ophthalmol. Vis. Sci. 2005, 46, 578. [Google Scholar]
- Brown, N.L.; Patel, S.; Brzezinski, J.; Glaser, T. Math5 Is Required for Retinal Ganglion Cell and Optic Nerve Formation. Development 2001, 128, 2497–2508. [Google Scholar] [CrossRef]
- Valencia, J.E.; Feuda, R.; Mellott, D.O.; Burke, R.D.; Peter, I.S. Ciliary photoreceptors in sea urchin larvae indicate pan-deuterostome cell type conservation. BMC Biol. 2021, 19, 257. [Google Scholar] [CrossRef]
- Frankfort, B.J.; Mardon, G. R8 Development in the Drosophila Eye: A Paradigm for Neural Selection and Differentiation. Development 2002, 129, 1295–1306. [Google Scholar] [CrossRef]
- Hsiung, F.; Moses, K. Retinal Development in Drosophila: Specifying the First Neuron. Hum. Mol. Genet. 2002, 11, 1207–1214. [Google Scholar] [CrossRef]
- Inoue, T.; Hojo, M.; Bessho, Y.; Tano, Y.; Lee, J.E.; Kageyama, R. Math3 and NeuroD Regulate Amacrine Cell Fate Specification in the Retina. Development 2002, 129, 831–842. [Google Scholar] [CrossRef]
- Pappu, K.S.; Mardon, G. Genetic Control of Retinal Specification and Determination in Drosophila. Int. J. Dev. Biol. 2004, 48, 913–924. [Google Scholar] [CrossRef]
- Belecky-Adams, T.; Tomarev, S.; Li, H.S.; Ploder, L.; McInnes, R.R.; Sundin, O.; Adler, R. Pax-6, Prox 1, and Chx10 Homeobox Gene Expression Correlates with Phenotypic Fate of Retinal Precursor Cells. Investig. Ophthalmol. Vis. Sci. 1997, 38, 1293–1303. [Google Scholar]
- Hsieh, Y.W.; Yang, X.J. Dynamic Pax6 Expression during the Neurogenic Cell Cycle Influences Proliferation and Cell Fate Choices of Retinal Progenitors. Neural Dev. 2009, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Eakin, R.M. Evolutionary Significance of Photoreceptors: In Retrospect. Integr. Comp. Biol. 1979, 19, 647–653. [Google Scholar] [CrossRef]
- Ullrich-Lüter, E.M.; (Museum für Naturkunde, Berlin, Germany). Personal communication, 2022.
- Land, M.F.; Nilsson, D.-E. Animal Eyes, 2nd ed.; OUP: Oxford, UK, 2012; pp. 1–271. [Google Scholar]
- Zelhof, A.C.; Hardy, R.W.; Becker, A.; Zuker, C.S. Transforming the Architecture of Compound Eyes. Nature 2006, 443, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paganos, P.; Ullrich-Lüter, E.; Caccavale, F.; Zakrzewski, A.; Voronov, D.; Fournon-Berodia, I.; Cocurullo, M.; Lüter, C.; Arnone, M.I. A New Model Organism to Investigate Extraocular Photoreception: Opsin and Retinal Gene Expression in the Sea Urchin Paracentrotus lividus. Cells 2022, 11, 2636. https://doi.org/10.3390/cells11172636
Paganos P, Ullrich-Lüter E, Caccavale F, Zakrzewski A, Voronov D, Fournon-Berodia I, Cocurullo M, Lüter C, Arnone MI. A New Model Organism to Investigate Extraocular Photoreception: Opsin and Retinal Gene Expression in the Sea Urchin Paracentrotus lividus. Cells. 2022; 11(17):2636. https://doi.org/10.3390/cells11172636
Chicago/Turabian StylePaganos, Periklis, Esther Ullrich-Lüter, Filomena Caccavale, Anne Zakrzewski, Danila Voronov, Inés Fournon-Berodia, Maria Cocurullo, Carsten Lüter, and Maria Ina Arnone. 2022. "A New Model Organism to Investigate Extraocular Photoreception: Opsin and Retinal Gene Expression in the Sea Urchin Paracentrotus lividus" Cells 11, no. 17: 2636. https://doi.org/10.3390/cells11172636
APA StylePaganos, P., Ullrich-Lüter, E., Caccavale, F., Zakrzewski, A., Voronov, D., Fournon-Berodia, I., Cocurullo, M., Lüter, C., & Arnone, M. I. (2022). A New Model Organism to Investigate Extraocular Photoreception: Opsin and Retinal Gene Expression in the Sea Urchin Paracentrotus lividus. Cells, 11(17), 2636. https://doi.org/10.3390/cells11172636






