Quantitative and Qualitative Changes in the Genetic Diversity of Bacterial Communities in Anaerobic Bioreactors with the Diatomaceous Earth/Peat Cell Carrier
Abstract
:1. Introduction
2. Materials and Methods
2.1. Feedstock and Carrier Material
2.2. Batch Anaerobic Digestion
2.3. Physicochemical Analysis
2.4. Analysis of the Total Bacterial Count
2.5. DNA Extraction and Next-Generation Sequencing (NGS)
2.6. Bioinformatics and Statistical Analysis
2.7. BioFlux Microfluidic Flow System
2.8. Visualisation of Microbiome Using Fluorescence Microscopy
3. Results and Discussion
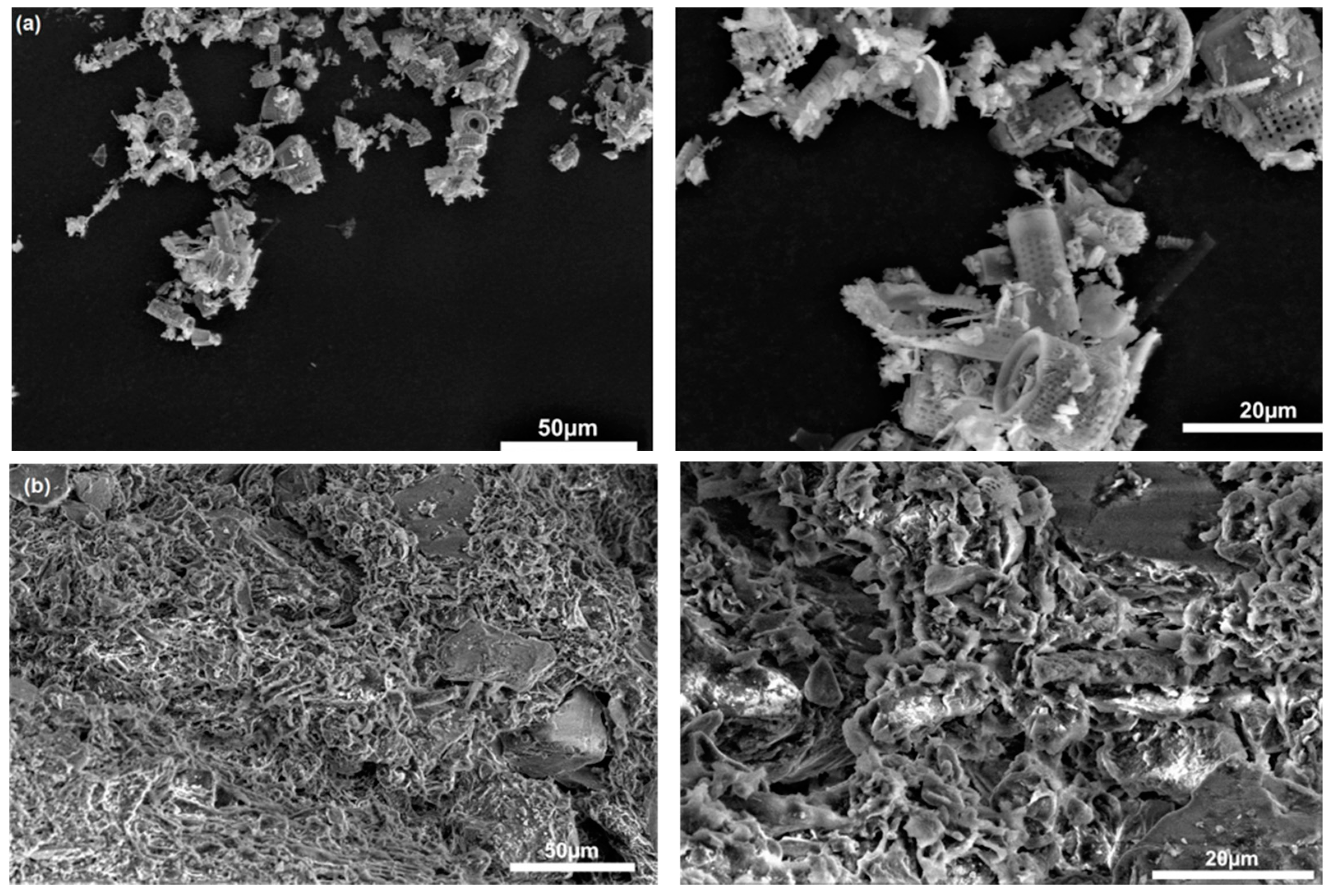
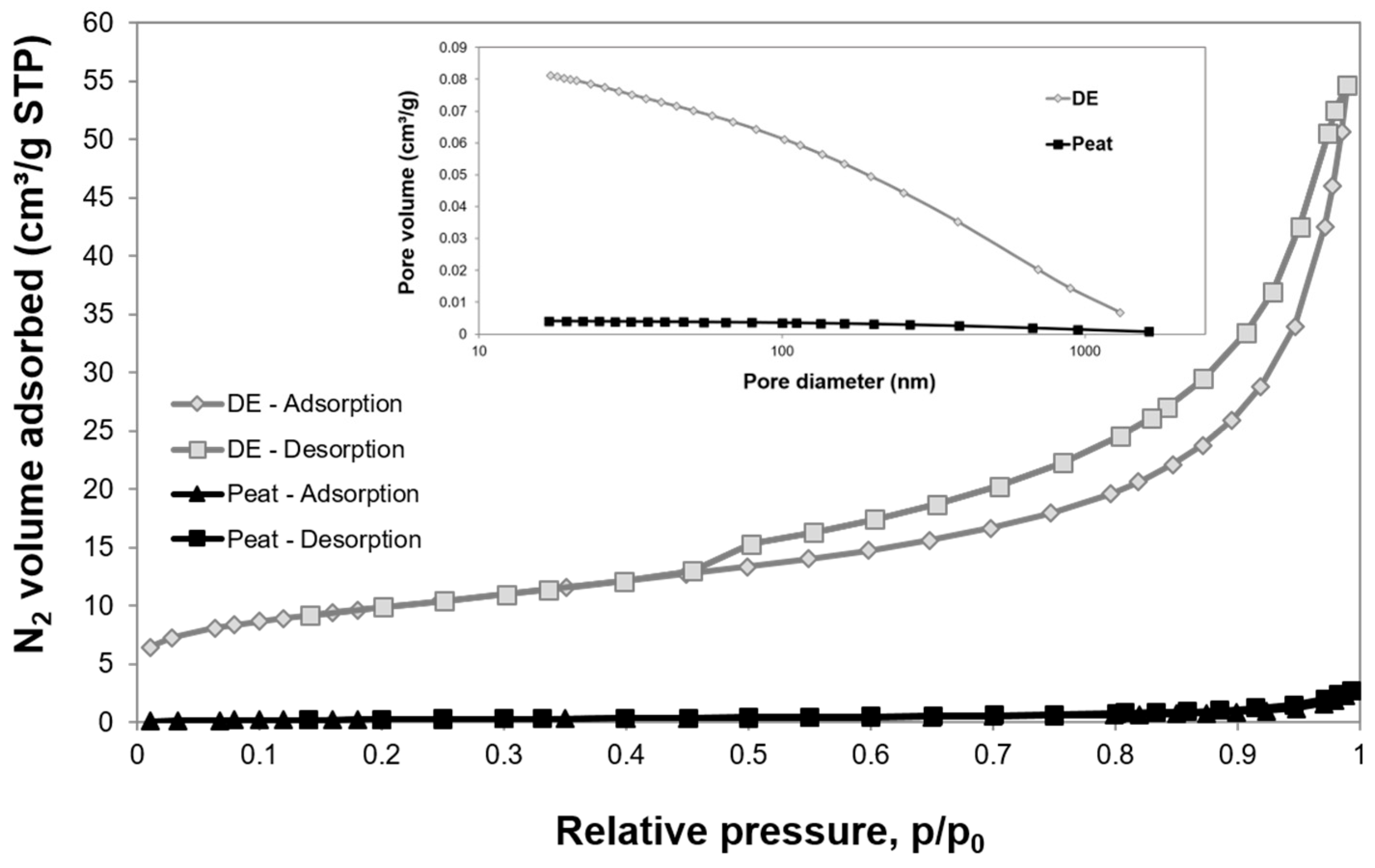
3.1. Characterisation of the Cell Carrier
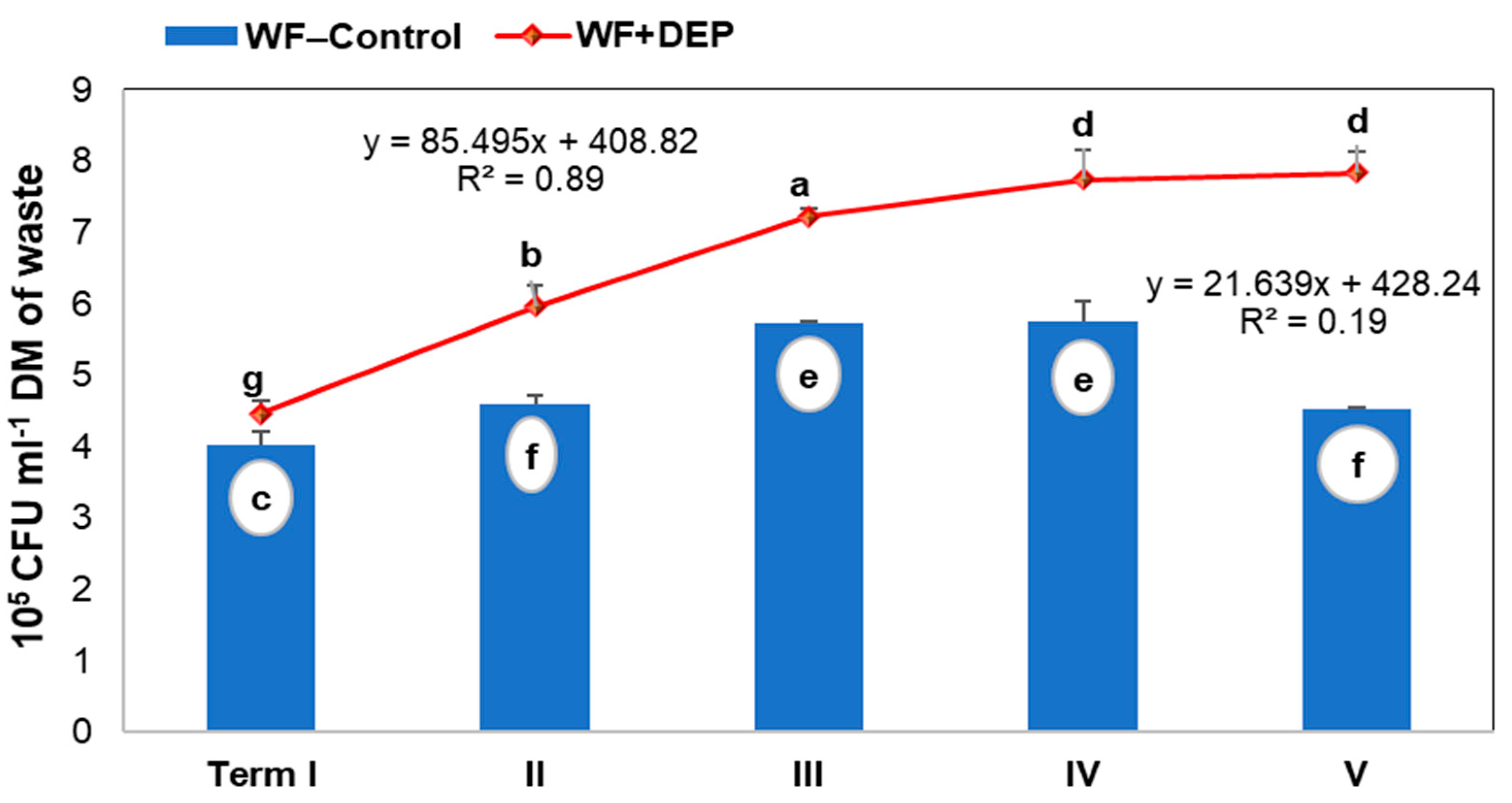
3.2. Total Bacterial Count in Digested Samples
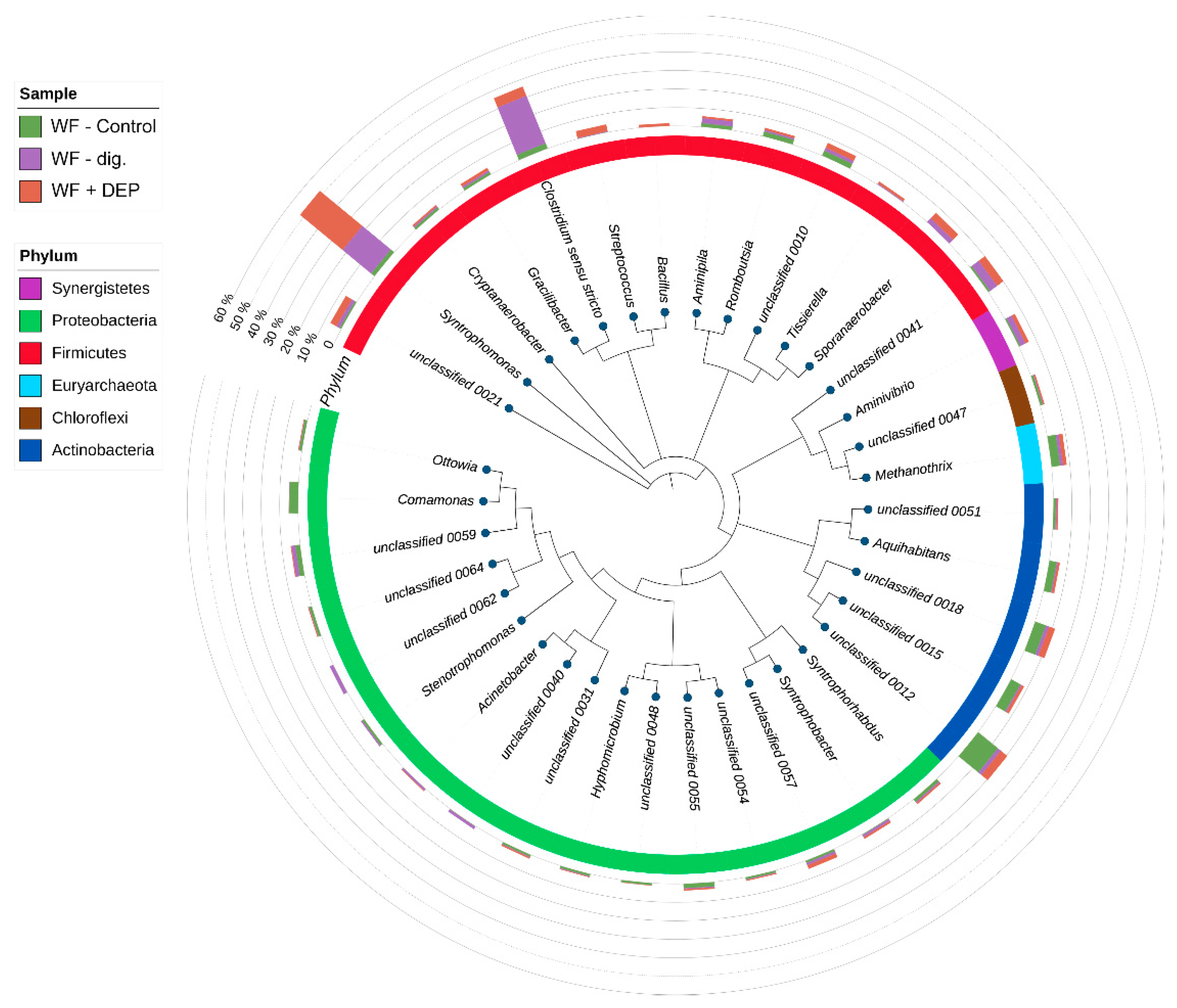
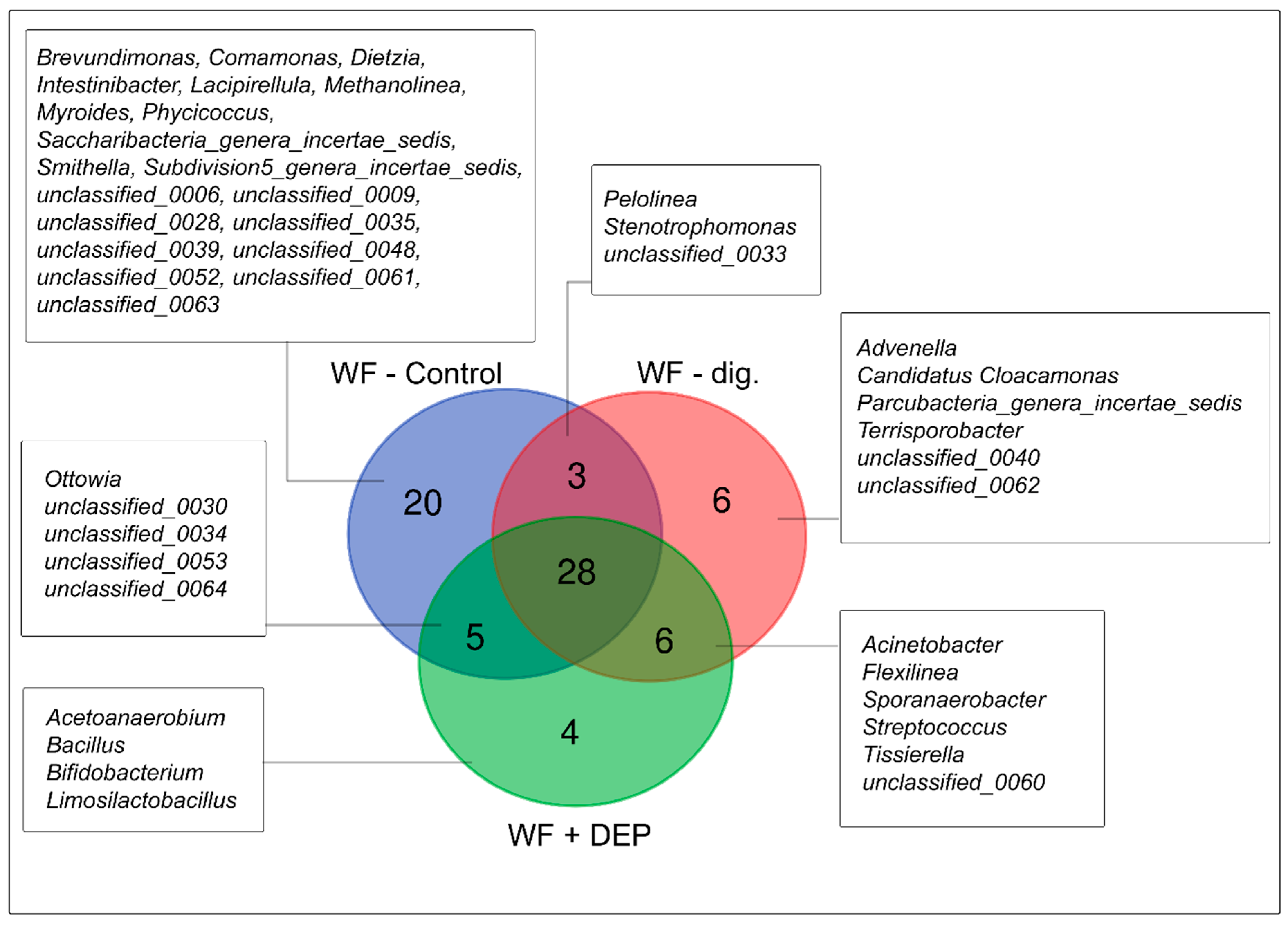
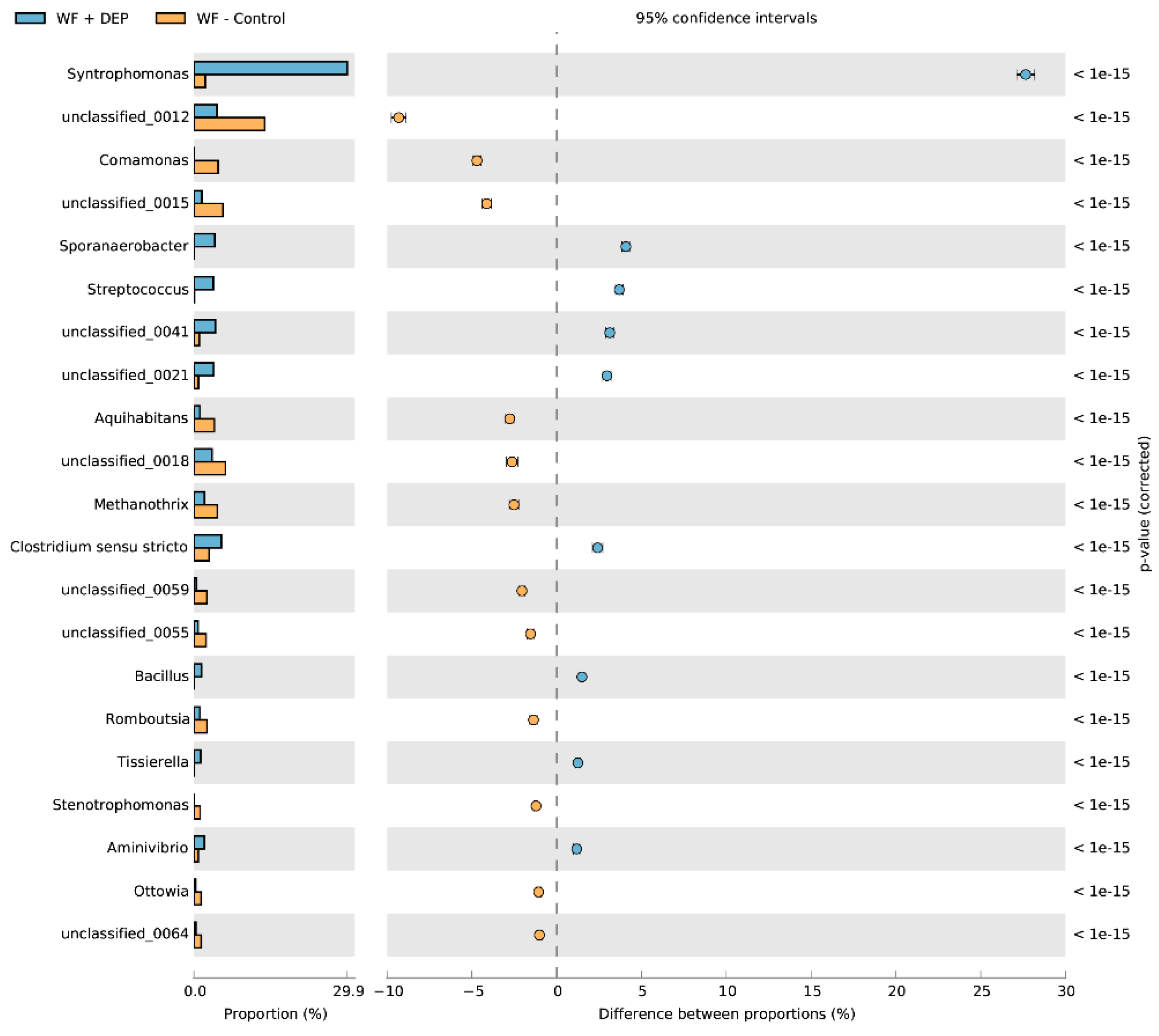
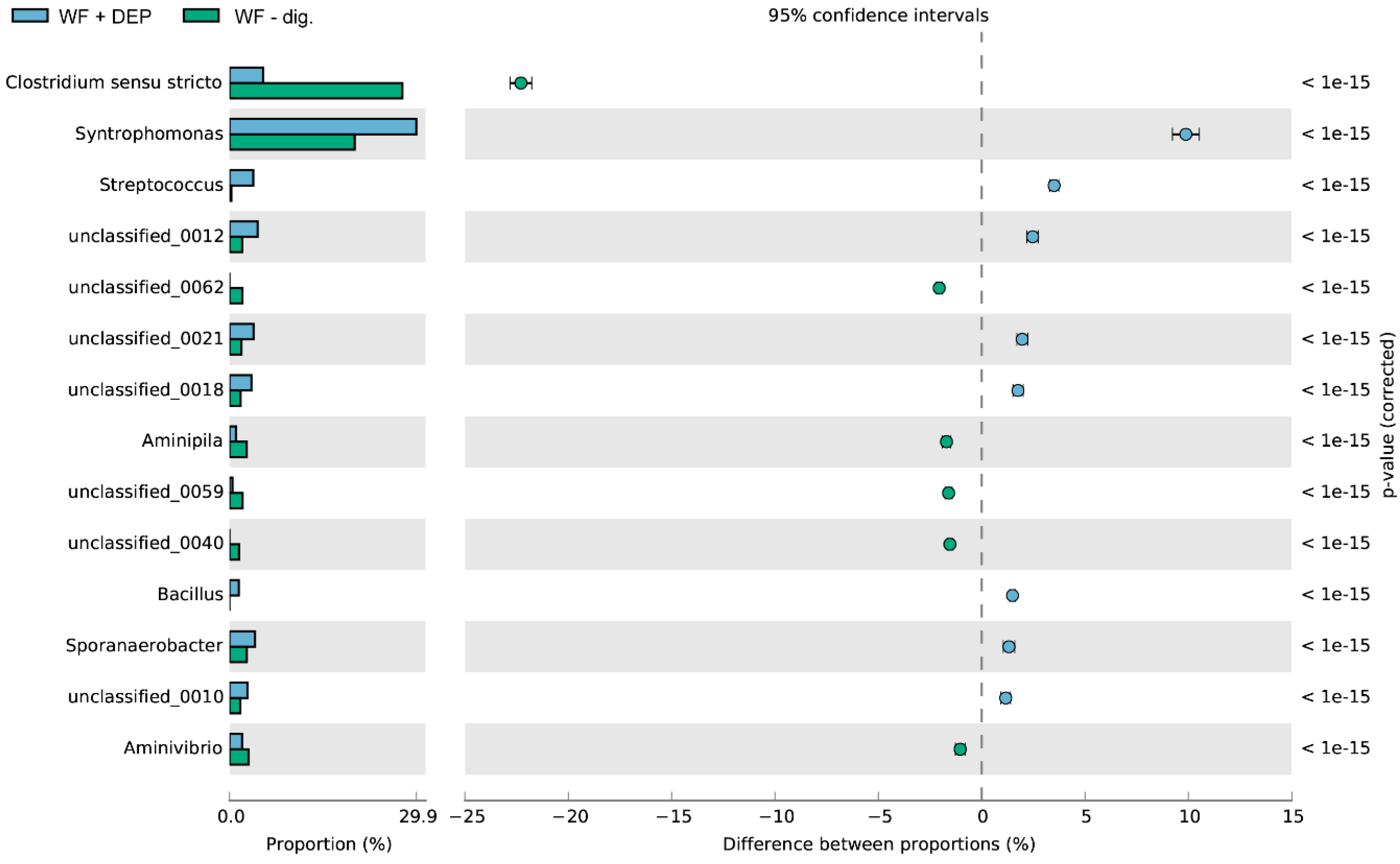
3.3. Bacterial Community Abundance and Composition
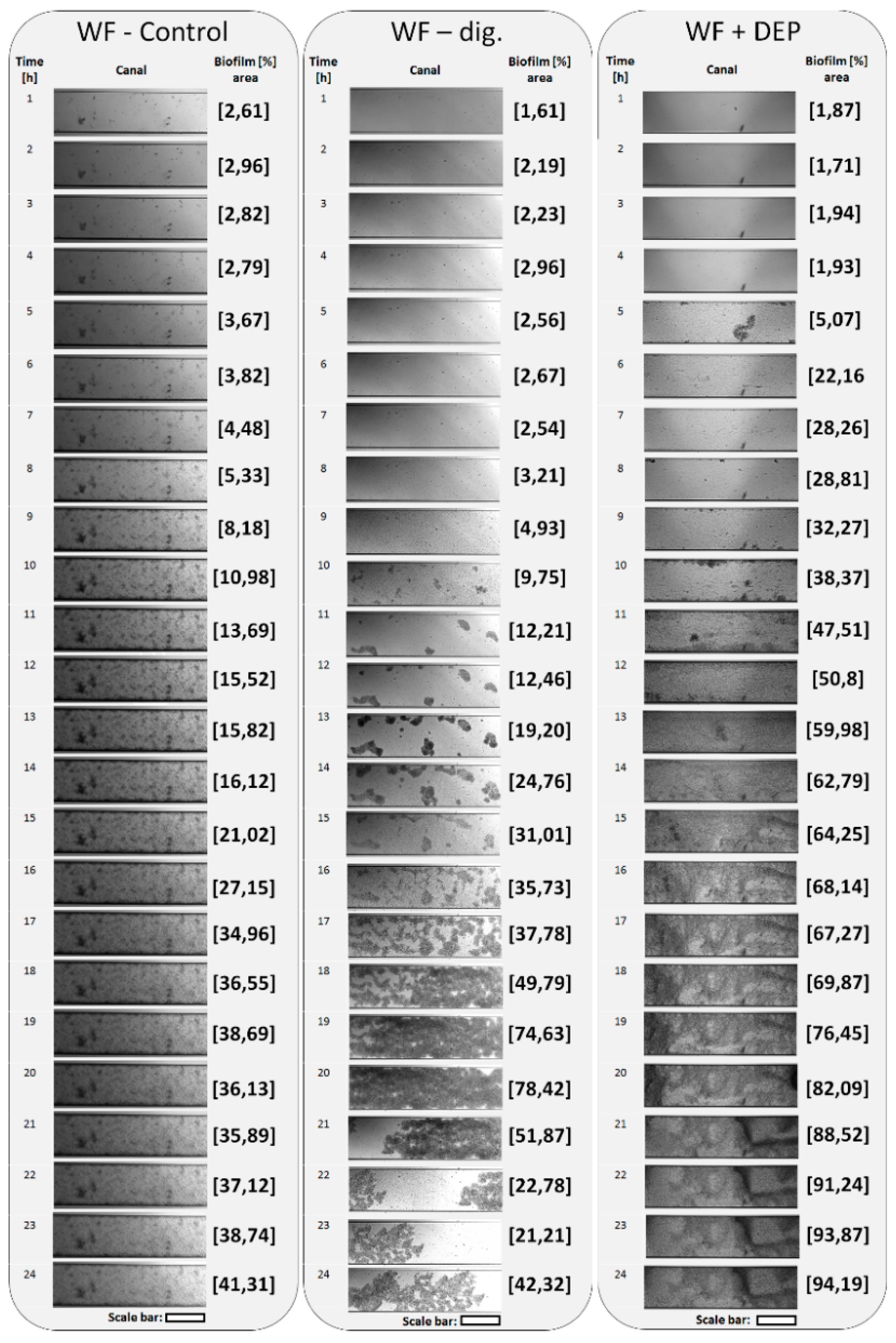
3.4. Adherence and Biofilm Formation Ability of Microorganisms
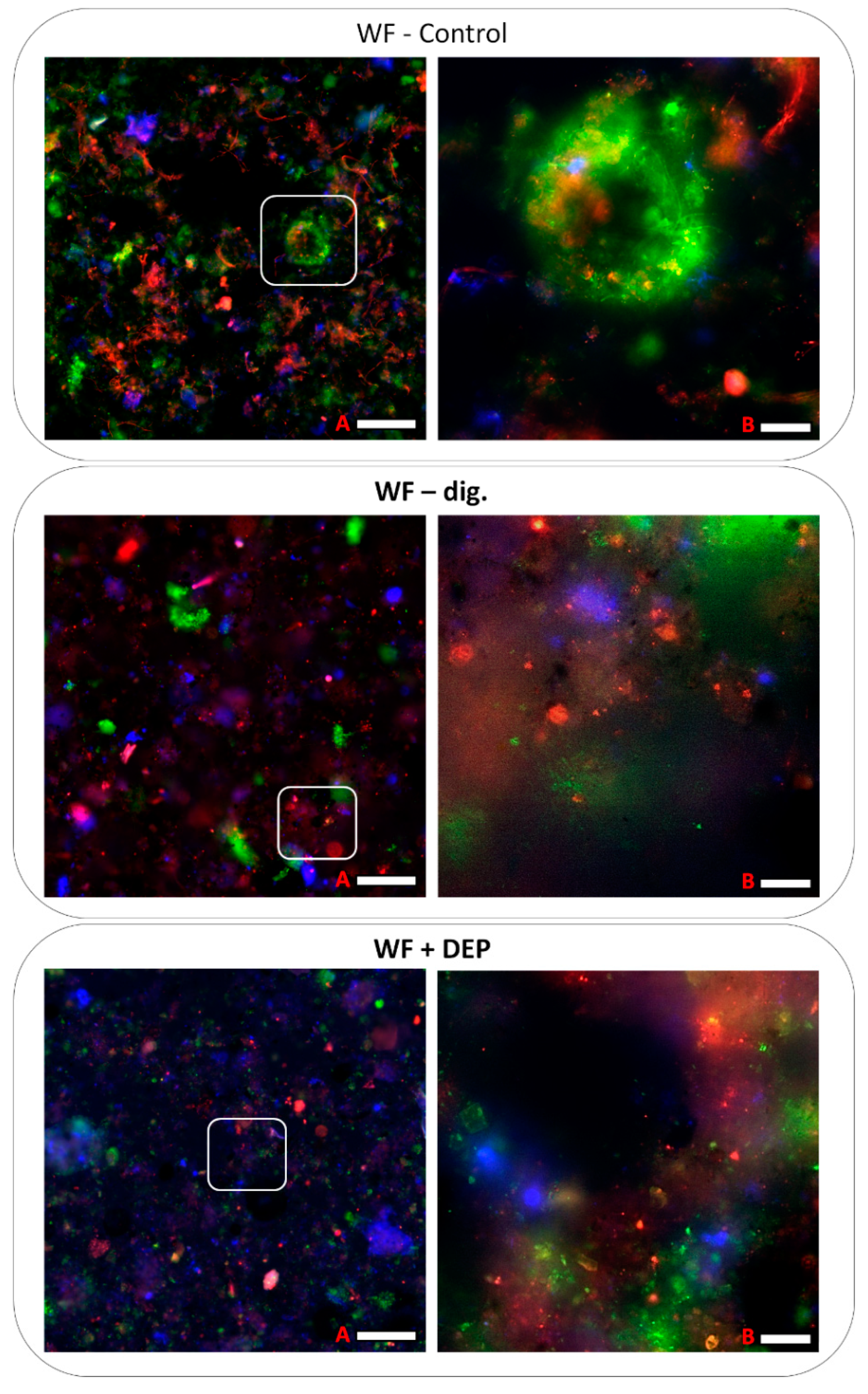
3.5. Visualisation of Microbiomes from Bioreactor Samples
3.6. Summary and a Look to the Future
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, F.; Li, Y.; Ge, X.; Yang, L.; Li, Y. Anaerobic digestion of food waste—Challenges and opportunities. Bioresour. Technol. 2018, 247, 1047–1058. [Google Scholar] [CrossRef]
- dos Santos, A.L.M.; Castro, A.L.S.; Salomon, K.R.; de Souza, T.S.O.; Vich, D.V. Global research trends on anaerobic digestion and biogas production from cassava wastewater: A bibliometric analysis. J. Chem. Technol. Biotechnol. 2021, 97, 1379–1389. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Wolna-Maruwka, A.; Pilarski, K.; Janczak, D.; Przybył, K.; Gawrysiak-Witulska, M. The use of lignin as a microbial carrier in the co-digestion of cheese and wafer waste. Polymers 2019, 11, 2073. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Wolna-Maruwka, A.; Boniecki, P.; Zaborowicz, M. Use of confectionery waste in biogas production by the anaerobic digestion process. Molecules 2019, 24, 37. [Google Scholar] [CrossRef]
- Cayetano, R.D.A.; Kim, G.B.; Park, J.; Yang, Y.H.; Jeon, B.H.; Jang, M.; Kim, S.H. Biofilm formation as a method of improved treatment during anaerobic digestion of organic matter for biogas recovery. Bioresour. Technol. 2022, 344, 126309. [Google Scholar] [CrossRef]
- Weiß, S.; Zankel, A.; Lebuhn, M.; Petrak, S.; Somitsch, W.; Guebitz, G.M. Investigation of microorganisms colonising activated zeolites during anaerobic biogas production from grass silage. Bioresour. Technol. 2011, 102, 4353–4359. [Google Scholar] [CrossRef] [PubMed]
- Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Pilarski, K.; Adamski, M.; Grzyb, A.; Grządziel, J.; Gałązka, A. Silica/lignin carrier as a factor increasing the process performance and genetic diversity of microbial communities in laboratory-scale anaerobic digesters. Energies 2021, 14, 4429. [Google Scholar] [CrossRef]
- Yang, H.J.; Yang, Z.M.; Xu, X.H.; Guo, R.B. Increasing the methane production rate of hydrogenotrophic methanogens using biochar as a biocarrier. Bioresour. Technol. 2020, 302, 122829. [Google Scholar] [CrossRef]
- Guo, X.; Li, B.; Zhao, R.; Zhang, J.; Lin, L.; Zhang, G.; Li, R.H.; Liu, J.; Li, P.; Li, Y.; et al. Performance and bacterial community of moving bed biofilm reactors with various biocarriers treating primary wastewater effluent with a low organic strength and low C/N ratio. Bioresour. Technol. 2019, 287, 121424. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Fang, Y.; Lai, W.; Xu, S.; Lichtfouse, E. Enhancing thermophilic anaerobic co-digestion of sewage sludge and food waste with biogas residue biochar. Renew. Energy 2022, 188, 465–475. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, L.; Huang, J.; Qu, Y.; Pan, Y.; Liu, L.; Zhu, H. Recovering short-chain fatty acids from waste sludge via biocarriers and microfiltration enhanced anaerobic fermentation. Resour. Conserv. Recycl. 2022, 182, 106342. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Adamski, M.; Zaborowicz, M.; Dorota Cais-Sokolińska, D.; Wolna-Maruwka, A.; Niewiadomska, A. Eco-friendly and effective diatomaceous earth/peat (DEP) microbial carriers in the anaerobic biodegradation of food waste products. Energies 2022, 15, 3442. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Waliszewska, B.; Zborowska, M.; Witaszek, K.; Waliszewska, H.; Kolasi´nski, M.; Szwarc-Rzepka, K. Evaluation of bio-methane yields for high-energy organic waste and sewage sludge: A pilot-scale study for a wastewater treatment plant. Environ. Eng. Manag. J. 2019, 18, 2019–2030. [Google Scholar] [CrossRef]
- Yadav, M.; Joshi, C.; Paritosh, K.; Thakur, J.; Pareek, N.; Masakapalli, S.K.; Vivekanand, V. Reprint of organic waste conversion through anaerobic digestion: A critical insight into the metabolic pathways and microbial interactions. Metab. Eng. 2022, 71, 62–76. [Google Scholar] [CrossRef]
- Wei, Y.; Gao, J.; Shi, Z.; Li, X.; Ma, W.; Yuan, H. Effect of hydrothermal pretreatment on two-stage anaerobic digestion of food waste and Enteromorpha: Digestion performance, bioenergy efficiency, and microbial community dynamics. Fuel 2022, 318, 123639. [Google Scholar] [CrossRef]
- De Vrieze, D.; Pinto, A.J.; Sloan, W.T.; Ijaz, U.Z. The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome 2018, 6, 63. [Google Scholar] [CrossRef]
- Tsigkou, K.; Terpou, A.; Treu, L.; Panagiotis, G.; Kougias, P.G.; Kornaros, M. Thermophilic anaerobic digestion of olive mill wastewater in an upflow packed bed reactor: Evaluation of 16S rRNA amplicon sequencing for microbial analysis. J. Environ. Manag. 2022, 301, 113853. [Google Scholar] [CrossRef]
- Grosser, A.; Grobelak, A.; Rorat, A.; Courtois, P.; Vandenbulcke, F.; Lemière, S.; Guyoneaud, R.; Attard, E.; Celary, P. Effects of silver nanoparticles on performance of anaerobic digestion of sewage sludge and associated microbial communities. Renew. Energy 2021, 171, 1014–1025. [Google Scholar] [CrossRef]
- Jiang, X.; Xie, Y.; Liu, M.; Bin, S.; Liu, Y.; Huan, C.; Ji, G.; Wang, X.; Yan, Z.; Lyu, Q. Study on anaerobic co-digestion of municipal sewage sludge and fruit and vegetable wastes: Methane production, microbial community and three-dimension fluorescence excitation-emission matrix analysis. Bioresour. Technol. 2022, 347, 126748. [Google Scholar] [CrossRef]
- Banach, A.; Ciesielski, S.; Bacza, T.; Pieczykolan, M.; Ziembińska-Buczyńska, A. Microbial community composition and methanogens’ biodiversity during a temperature shift in a methane fermentation chamber. Environ. Technol. 2018, 40, 1–35. [Google Scholar] [CrossRef]
- Dyksma, S.; Jansen, L.; Gallert, C. Syntrophic acetate oxidation replaces acetoclastic methanogenesis during thermophilic digestion of biowaste. Microbiome 2020, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Norm VDI 4630; Fermentation of Organic Materials Characterization of the Substrate, Sampling, Collection of Material Data, Fermentation Tests. German Engineers Club: Düsseldorf, Germany, 2006.
- DIN Guideline 38 414-S8; Characterisation of the Substrate, Sampling, Collection of Material Data, Fermentation Tests. German Institute for Standardization: Berlin, Germany, 1985.
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Wright, E.S. RDP v16 Modified Training Set for 16S rRNA Classification. 2019. Available online: http://www2.decipher.codes/Classification/TrainingSets/RDP_v16-mod_March2018.RData (accessed on 2 January 2019).
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. cPhyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Paluch, E.; Okińczyc, P.; Zwyrzykowska-Wodzińska, A.; Szperlik, J.; Zarowska, B.; Duda-Madej, A.; Bąbelewski, P.; Włodarczyk, M.; Wojtasik, W.; Robert Kupczyński, R.; et al. Composition and antimicrobial activity of Ilex leaves water Extracts. Molecules 2021, 26, 7442. [Google Scholar] [CrossRef]
- Paluch, E.; Sobierajska, P.; Okińczyc, P.; Widelski, J.; Duda-Madej, A.; Krzyżanowska, B.; Krzyżek, P.; Ogórek, R.; Szperlik, J.; Chmielowiec, J.; et al. Nanoapatites doped and co-doped with noble metal ions as modern antibiofilm materials for biomedical applications against drug-resistant clinical strains of Enterococcus faecalis VRE and Staphylococcus aureus MRSA. Int. J. Mol. Sci. 2022, 23, 1533. [Google Scholar] [CrossRef]
- Tramontano, C.; Chianese, G.; Terracciano, M.; de Stefano, L.; Rea, I. Nanostructured biosilica of diatoms: From water world to biomedical applications. Appl. Sci. 2020, 10, 6811. [Google Scholar] [CrossRef]
- Albareda, M.; Rodríguez-Navarro, D.N.; Camacho, M.; Temprano, F.J. Alternatives to peat as a carrier for rhizobia inoculants: Solid and liquid formulations. Soil Biol. Biochem. 2008, 40, 2771–2779. [Google Scholar] [CrossRef]
- Benkacem, T.; Hamdi, B.; Chamayou, A.; Balard, H.; Calvet, R. Physicochemical characterization of a diatomaceous upon an acid treatment: A focus on surface properties by inverse gas chromatography. Powder Technol. 2016, 294, 498–507. [Google Scholar] [CrossRef]
- Paleckiene, R.; Navikaite, R.; Slinksiene, R. Peat as a raw material for plant nutrients and humic substances. Sustainability 2021, 13, 6354. [Google Scholar] [CrossRef]
- Janićijević, J.; Krajišnik, D.; Čalija, B.; Vasiljević, B.N.; Dobričić, V.; Daković, A.; Antonijević, M.D.; Milić, J. Modified local diatomite as potential functional drug carrier—A model study for diclofenac sodium. Int. J. Pharm. 2015, 496, 466–474. [Google Scholar] [CrossRef]
- Bartczak, B.; Norman, M.; Klapiszewski, Ł.; Karwańska, N.; Kawalec, M.; Baczyńska, M.; Wysokowski, M.; Zdarta, J.; Ciesielczyk, F.; Jesionowski, T. Removal of nickel(II) and lead(II) ions from aqueous solution using peat as a low-cost adsorbent: A kinetic and equilibrium study. Arab. J. Chem. 2018, 11, 1209–1222. [Google Scholar] [CrossRef]
- Stalin, N.; Prabhu, H.J. Effect of microbial growth on biogas generation using carrier material in the self circulating biogas plant. J. Eng. Appl. Sci. 2007, 2, 8–11. [Google Scholar]
- Tsai, W.T.; Hsien, K.J.; Lai, C.W. Chemical activation of spent diatomaceous earth by alkaline etching in the preparation of mesoporous adsorbents. Ind. Eng. Chem. Res. 2004, 43, 7513–7520. [Google Scholar] [CrossRef]
- Fu, X.; Liu, Z.; Xi, Y.; Wang, J.; Lei, J. Preparation and properties of lauric acid/diatomite composites as novel form-stable phase change materials for thermal energy storage. Energy Build. 2015, 104, 244–249. [Google Scholar] [CrossRef]
- Mishra, A.; Kumar, J.; Melo, J.S. Silica based bio-hybrid materials and their relevance to bionanotechnology. Austin J. Plant Biol. 2020, 6, 1024. [Google Scholar]
- Xu, R.; Zhang, K.; Liu, P.; Khan, A.; Xiong, J.; Tian, F.; Li, X. A critical review on the interaction of substrate nutrient balance and microbial community structure and function in anaerobic co-digestion. Bioresur. Biotechnol. 2018, 247, 1119–1127. [Google Scholar] [CrossRef]
- Chatellard, L.; Trably, E.; Carrère, H. The type of carbohydrates specifically selects microbial community structures and fermentation patterns. Bioresour Technol. 2016, 221, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Bedard, D.L.; Ritalahti, K.M.; Löffler, F.E. The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl. Environ. Microbiol. 2007, 73, 2513–2521. [Google Scholar] [CrossRef]
- van Doesburg, W.; van Eekert, M.H.A.; Middeldorp, P.J.M.; Balk, M.; Schraa, G.; Stams, A.J.M. Reductive dechlorination of beta-hexachlorocyclohexane (beta-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 2005, 54, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.S.; Clark, C.M.; Ómarsdóttir, S.; Sanchez, L.M.; Murphy, B.T. Minimizing taxonomic and natural product redundancy in microbial libraries using MALDI-TOF MS and the bioinformatics pipeline IDBac. J. Nat. Prod. 2019, 82, 2167–2173. [Google Scholar] [CrossRef] [PubMed]
- Gärtner, A.; Wiese, J.; Imhoff, J.F. Diversity of Micromonospora strains from the deep Mediterranean Sea and their potential to produce bioactive compounds. AIMS Microbiol. 2016, 2, 205–221. [Google Scholar] [CrossRef]
- Yamada, T.; Imachi, H.; Ohashi, A.; Harada, H.; Hanada, S.; Kamagata, Y.; Sekiguchi, Y. Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int. J. Syst. Evol. Microbiol. 2007, 57, 2299–2306. [Google Scholar] [CrossRef]
- Watanabe, K.; Teramoto, M.; Harayama, S. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 1999, 65, 2813–2819. [Google Scholar] [CrossRef]
- Tanaka, Y.H.; Matsuzawa, H.; Nigaya, M.; Mori, K.; Kamagata, Y. Microbial community analysis in the roots of aquatic plants and isolation of novel microbes including an organism of the candidate phylum OP10. Microbes Environ. 2012, 27, 149–157. [Google Scholar] [CrossRef]
- Arulmani, S.R.B.; Jayaraj, V.; Jebakumar, S.R.D. Long-term electricity production from soil electrogenic bacteria and high-content screening of biofilm formation on the electrodes. J. Soils Sediments 2016, 16, 831–841. [Google Scholar] [CrossRef]
- Rivière, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef]
- Jin, B.; Niu, J.; Wang, L.; Zhao, J.; Li, Y.; Pang, L.; Zhang, M. Effect of sodium dichloroisocyanurate treatment on enhancing the biodegradability of waste-activated sludge anaerobic fermentation. J. Environ. Manag. 2021, 287, 112353. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, J.; Song, T.; Lu, Y. Stimulation of Smithella-dominating propionate oxidation in a sediment enrichment by magnetite and carbon nanotubes. Environ. Microbial. Rep. 2019, 11, 236–248. [Google Scholar] [CrossRef]
- Hauben, L.; Vauterin, L.; Moore, E.R.B.; Hoste, B.; Swings, J. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Evol. Microbiol. 1999, 49, 1749–1760. [Google Scholar] [CrossRef]
- Li, F.; Cheng, C.C.; Zheng, J.; Liu, J.; Quevedo, R.M.; Li, J.; Roos, S.; Gänzle, M.G.; Walter, J. Limosilactobacillus balticus sp. nov., Limosilactobacillus agrestis sp. nov., Limosilactobacillus albertensis sp. nov., Limosilactobacillus rudii sp. nov. and Limosilactobacillus fastidiosus sp. nov., five novel Limosilactobacillus species isolated from the vertebrate gastrointestinal tract, and proposal of six subspecies of Limosilactobacillus reuteri adapted to the gastrointestinal tract of specific vertebrate hosts. Int. J. Syst. Evol. Microbiol. 2021, 71, 004644. [Google Scholar]
- Campanaro, S.; Treu, L.; Rodriguez, R.-L.M.; Kovalovszki, A.; Ziels, R.M.; Maus, I.; Zhu, X.; Kougias, P.G.; Basile, A.; Luo, G.; et al. The anaerobic digestion microbiome: A collection of 1600 metagenome-assembled genomes shows high species diversity related to methane production. BioRxiv 2019, 680553. [Google Scholar] [CrossRef]
- Theuerl, S.; Klang, J.; Hülsemann, B.; Mächtig, T.; Hassa, J. Microbiome diversity and community-level change points within manure-based small biogas plants. Microorganisms 2020, 8, 1169. [Google Scholar] [CrossRef]
- Cong, S.; Xu, Y.; Lu, Y. Growth coordination between butyrate-oxidizing syntrophs and hydrogenotrophic methanogens. Front. Microbiol. 2021, 27, 2061–2075. [Google Scholar] [CrossRef]
- Granja-Travez, R.S.; Persinoti, G.F.; Squina, F.M.; Bugg, T.D. Functional genomic analysis of bacterial lignin degraders: Diversity in mechanisms of lignin oxidation and metabolism. Appl. Microbiol. Biotechnol. 2020, 104, 3305–3320. [Google Scholar] [CrossRef]
- Zhang, L.; Ban, Q.; Li, J.; Wang, T. Simultaneous production of hydrogen-methane and spatial community succession in an anaerobic baffled reactor treating corn starch processing wastewater. Chemosphere 2022, 300, 134503. [Google Scholar] [CrossRef]
- Yang, G.; Wang, J. Changes in microbial community structure during dark fermentative hydrogen production. Int. J. Hydrogen Energy 2019, 44, 25542–25550. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, A.; Sihvonen, M.; Muñoz-Palazon, B.; Rodriguez-Sanchez, A.; Mikola, A.; Vaha, R. Microbial ecology of full-scale wastewater treatment systems in the Polar Arctic Circle: Archaea, Bacteria and Fungi. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Dong, F.; Zhang, D.; Zhang, J.; Wang, X. Effect of microfluidic channel geometry on Bacillus subtilis biofilm formation. Biomed. Microdevices 2022, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Besemer, K. Biodiversity, community structure and function of biofilms in stream ecosystems. Res. Microbiol. 2015, 166, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Lukumbuzya, M.; Schmid, M.; Pjevac, P.; Daims, H. A multicolor fluorescence in situ hybridization approach using an extended set of fluorophores to visualize. Microorganisms 2019, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Habouzit, F.; Hamelin, J.; Santa-Catalina, G.; Steyer, J.P.; Bernet, N. Biofilm development during the start-up period of anaerobic biofilm reactors: The biofilm Archaea community is highly dependent on the support material. Microbial Biotechnol. 2014, 7, 257–264. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y. Application of ethanol-type fermentation in establishment of direct interspecies electron transfer: A practical engineering case study. Renew. Energy 2019, 136, 846–855. [Google Scholar] [CrossRef]
- Sieber, J.R.; Crable, B.R.; Sheik, C.S.; Hurst, G.B.; Rohlin, L.; Gunsalus, R.P.; McInerney, M.J. Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. Front. Microbiol. 2015, 6, 115. [Google Scholar] [CrossRef]
- Li, Y.; Liu, M.; Che, X.; Li, C.; Liang, D.; Zhou, H.; Liu, L.; Zhao, Z.; Zhang, Y. Biochar stimulates growth of novel species capable of direct interspecies electron transfer in anaerobic digestion via ethanol-type fermentation. Environ. Res. 2020, 189, 109983. [Google Scholar] [CrossRef]
- Ma, K.; Wang, W.; Liu, Y.; Bao, L.; Cui, Y.; Kang, W.; Wu, Q.; Xin, X. Insight into the performance and microbial community profiles of magnetite-amended anaerobic digestion: Varying promotion effects at increased loads. Bioresour. Technol. 2021, 329, 124928. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Mukherjee, M.; Zhou, Y. Direct interspecies electron transfer (DIET) can be suppressed under ammonia-stressed condition—Reevaluate the role of conductive materials. Water Res. 2020, 183, 116094. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Yan, Y.; Nie, Y.; Lu, A.; Wu, X.; Li, Y.; Wang, C.; Ding, H. Natural extracellular electron transfer between semiconducting minerals and electroactive bacterial communities occurred on the rock varnish. Front. Microbiol. 2019, 10, 293. [Google Scholar] [CrossRef] [PubMed]

| Batches | WF (g) | Carrier (g) | Inoculum (g) | pH | TS (%) | VS (%) |
|---|---|---|---|---|---|---|
| WF-control | 9.8 | - | 830.2 | 7.15 | 4.00 | 72.50 |
| WF + DEP | 9.8 | 20.0 | 830.3 | 7.03 | 3.96 | 70.62 |
| Unclassified Symbol (in This Research) | NCBI Accession Numbers (% of Sequence Identity) | Source/Environment | References (If Available) | Closest Relative |
|---|---|---|---|---|
| unclassified_0010 | EF059533 (97.2%) | PCB-dechlorinating enrichment culture | Bedard et al. (2007) [46] | Sedimentibacter sp. |
| AY766467 (96.5%) | Anaerobic coculture enriched from a hexachlorocyclohexane (HCH) polluted soil. | Wim van Doesburg et al. (2005) [47] | Sedimentibacter sp. | |
| unclassified_0015 | MK143173 (98.8%) | Algae (Iceland) | Costa et al. (2019) [48] | Knoellia sp. |
| KX256211 (98.8%) | Eastern Mediterranean sea sediment | Gärtner et al. (2016) [49] | Intrasporangium sp. | |
| unclassified_0047 | NR_041354 (97%) | Thermophilic digester sludge, methanogenic propionate-degrading consortia | Yamada et al. (2007) [50] | Bellilinea caldifistulae |
| KX261406 (93.8%) | Sludge and beet sugar industrial wastewater | - | Levilinea saccharolytica | |
| unclassified_0018 | AB021325 (98%) | Activated sludge with phenol as the sole carbon source | Watanebe et al. (1999) [51] | Uncultured/unclassified |
| JQ899231 (97.5%) | Marine soil sediment | - | Streptomyces aomiensis | |
| unclassified_054 | AB529706 (98%) | Rhizoplane | Tanaka et al. (2012) [52] | Uncultured/unclassified |
| HM124367 (96.8%) | Lake sediment | - | Hyphomicrobium sp. | |
| unclassified_0012 | MN826598 (98.52%) | Rhizosphere in fertilised and degraded soils | - | Janibacter limosus |
| unclassified_0041 | KX876303 (100%) | Dark fermentation reactor | Chatellard et al. (2016) [45] | Uncultured/unclassified |
| unclassified_0059 | KM675947 (100%) | Rhizosphere soil | Arulmani and Jebakumar (2015) [53] | Hydrogenophaga sp. |
| unclassified_0062 | MH553022 (99.76%) | Bioreactor | - | Alcaligenaceae bacterium |
| HQ670757 (99.53%) | Sugarcane molasses-based distillery | - | Pusillimonas sp. | |
| unclassified_0021 | MG910712 (100%) | Bioreactor | - | Uncultured/unclassified |
| AB997768 (100%) | Sludge from full scale anaerobic digester | - | Uncultured/unclassified | |
| unclassified_0057 | MG910621 (100%) | Bioreactor | - | Uncultured/unclassified |
| CU922732 (100%) | Wastewater sludge | Rivière et al. (2009) [54] | Uncultured Deltaproteobacteria |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Grządziel, J.; Gałązka, A.; Paluch, E.; Borowiak, K.; Pilarski, K. Quantitative and Qualitative Changes in the Genetic Diversity of Bacterial Communities in Anaerobic Bioreactors with the Diatomaceous Earth/Peat Cell Carrier. Cells 2022, 11, 2571. https://doi.org/10.3390/cells11162571
Pilarska AA, Wolna-Maruwka A, Niewiadomska A, Grządziel J, Gałązka A, Paluch E, Borowiak K, Pilarski K. Quantitative and Qualitative Changes in the Genetic Diversity of Bacterial Communities in Anaerobic Bioreactors with the Diatomaceous Earth/Peat Cell Carrier. Cells. 2022; 11(16):2571. https://doi.org/10.3390/cells11162571
Chicago/Turabian StylePilarska, Agnieszka A., Agnieszka Wolna-Maruwka, Alicja Niewiadomska, Jarosław Grządziel, Anna Gałązka, Emil Paluch, Klaudia Borowiak, and Krzysztof Pilarski. 2022. "Quantitative and Qualitative Changes in the Genetic Diversity of Bacterial Communities in Anaerobic Bioreactors with the Diatomaceous Earth/Peat Cell Carrier" Cells 11, no. 16: 2571. https://doi.org/10.3390/cells11162571
APA StylePilarska, A. A., Wolna-Maruwka, A., Niewiadomska, A., Grządziel, J., Gałązka, A., Paluch, E., Borowiak, K., & Pilarski, K. (2022). Quantitative and Qualitative Changes in the Genetic Diversity of Bacterial Communities in Anaerobic Bioreactors with the Diatomaceous Earth/Peat Cell Carrier. Cells, 11(16), 2571. https://doi.org/10.3390/cells11162571











