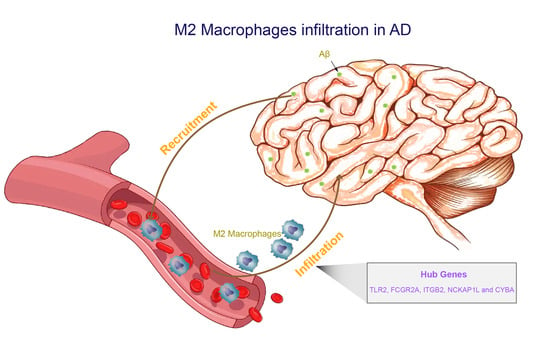
Identification of Biomarkers Related to M2 Macrophage Infiltration in Alzheimer’s Disease
Abstract
:1. Introduction
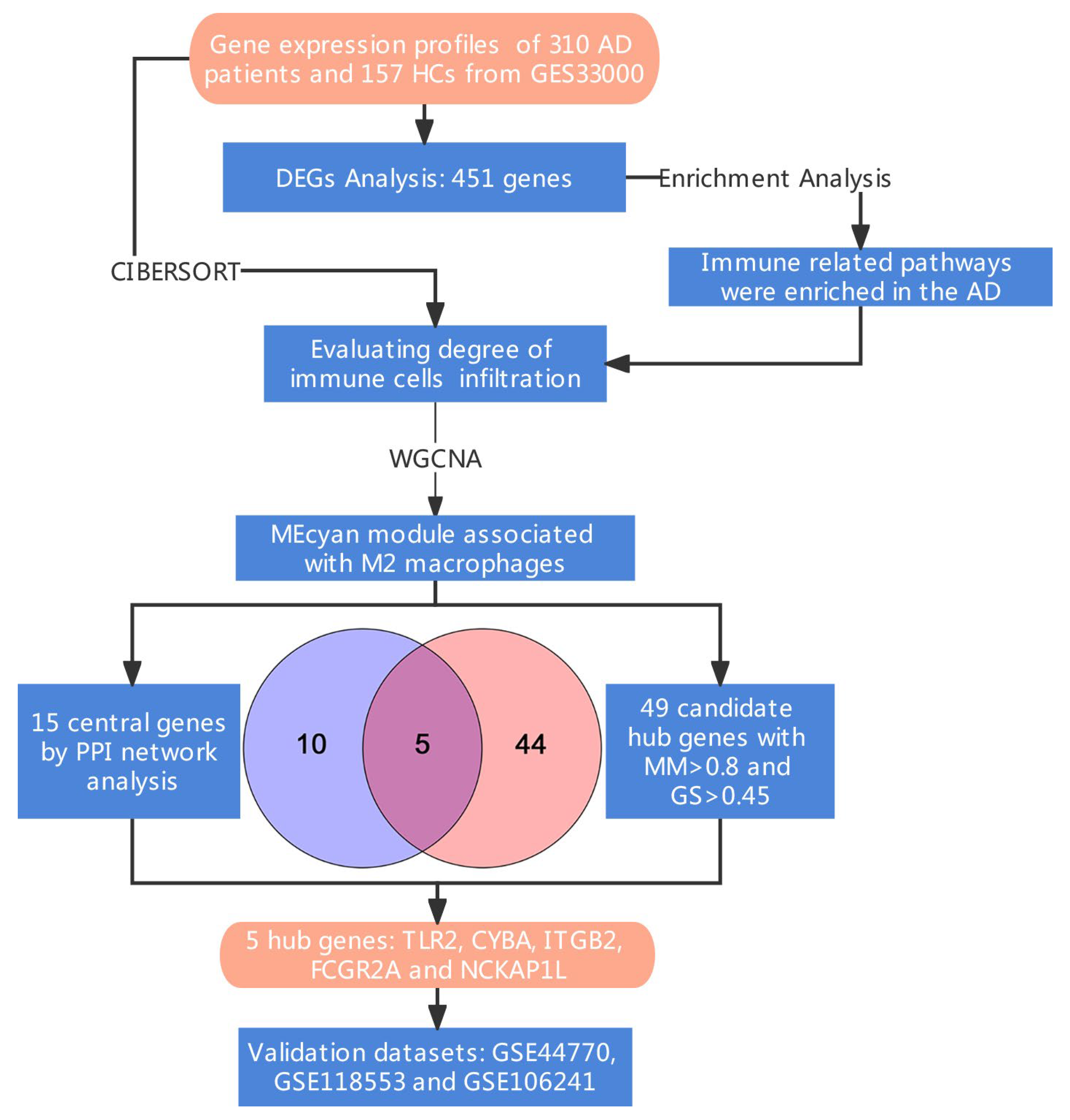
2. Materials and Methods
2.1. Data Source
2.2. Differentially Expressed Genes and Enrichment Analysis
2.3. Evaluation of Immune Infiltration
2.4. Weighted Gene Co-Expression Network Analysis
2.5. Identification of Hub Genes
2.6. Validation of Hub Genes
3. Results
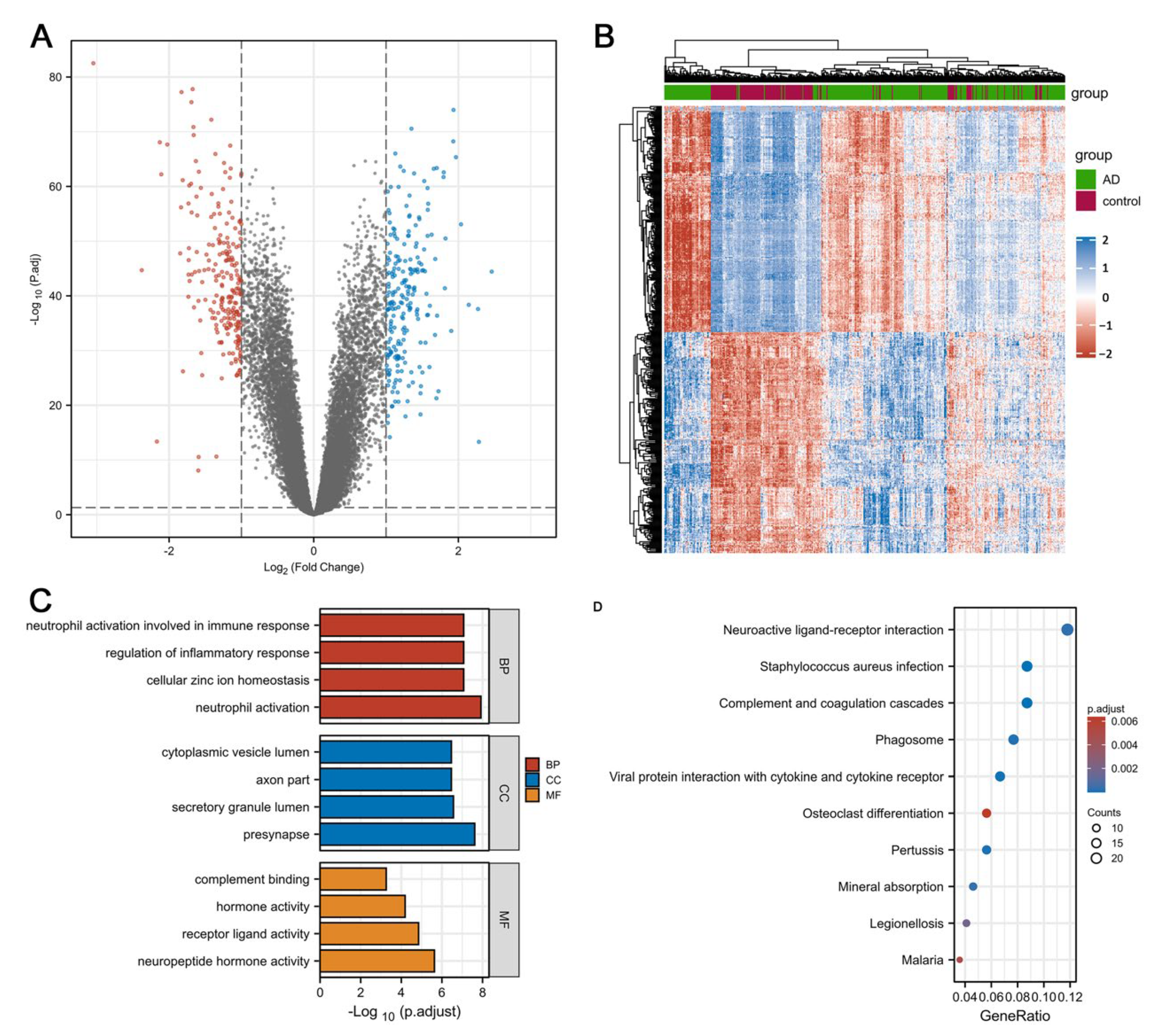
3.1. Identification of DEGs and Enrichment Analysis in Individuals with AD and HCs
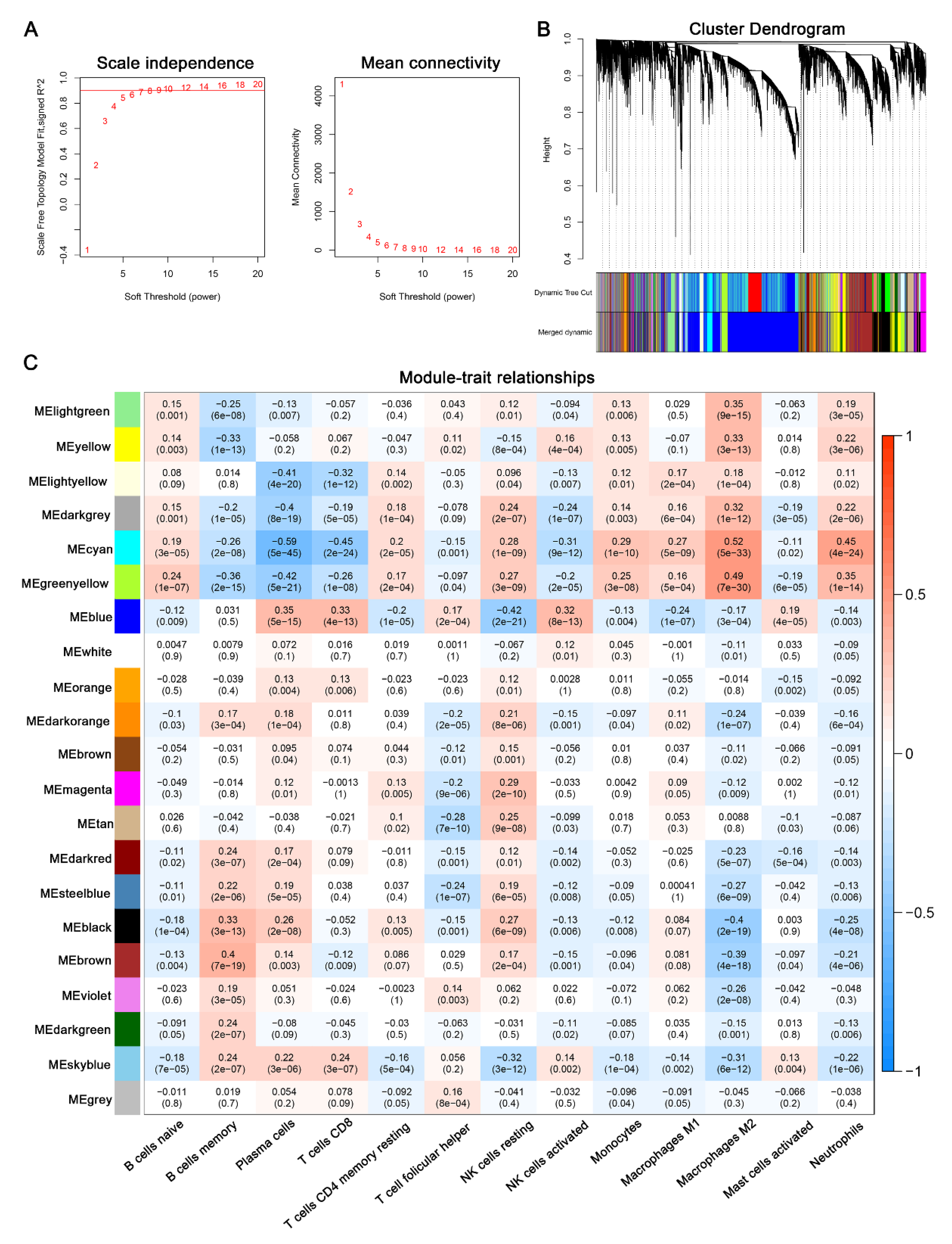
3.2. Identification of Immune Infiltration Pattern in AD
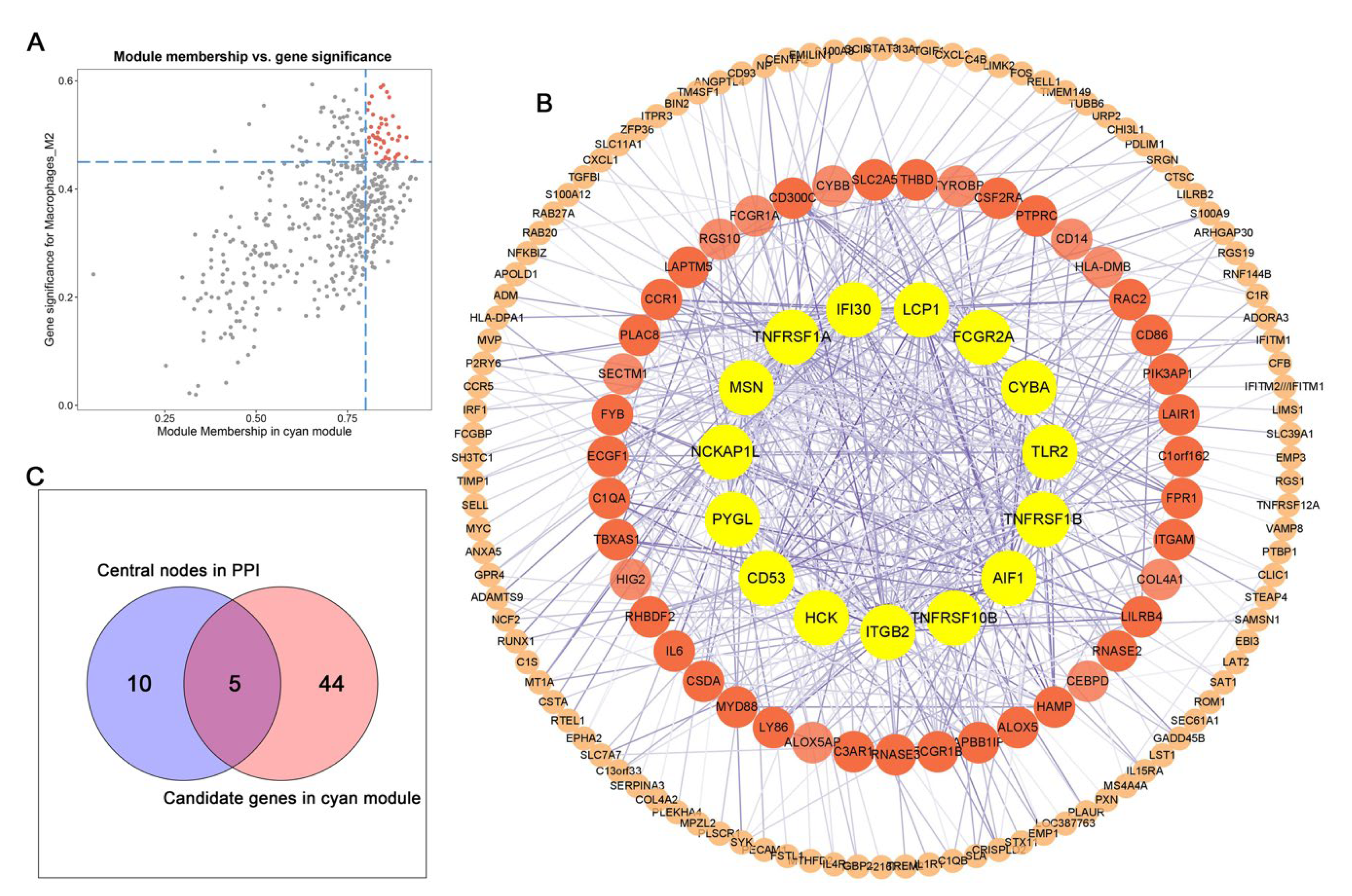
3.3. Identification of Key Modules via WGCNA
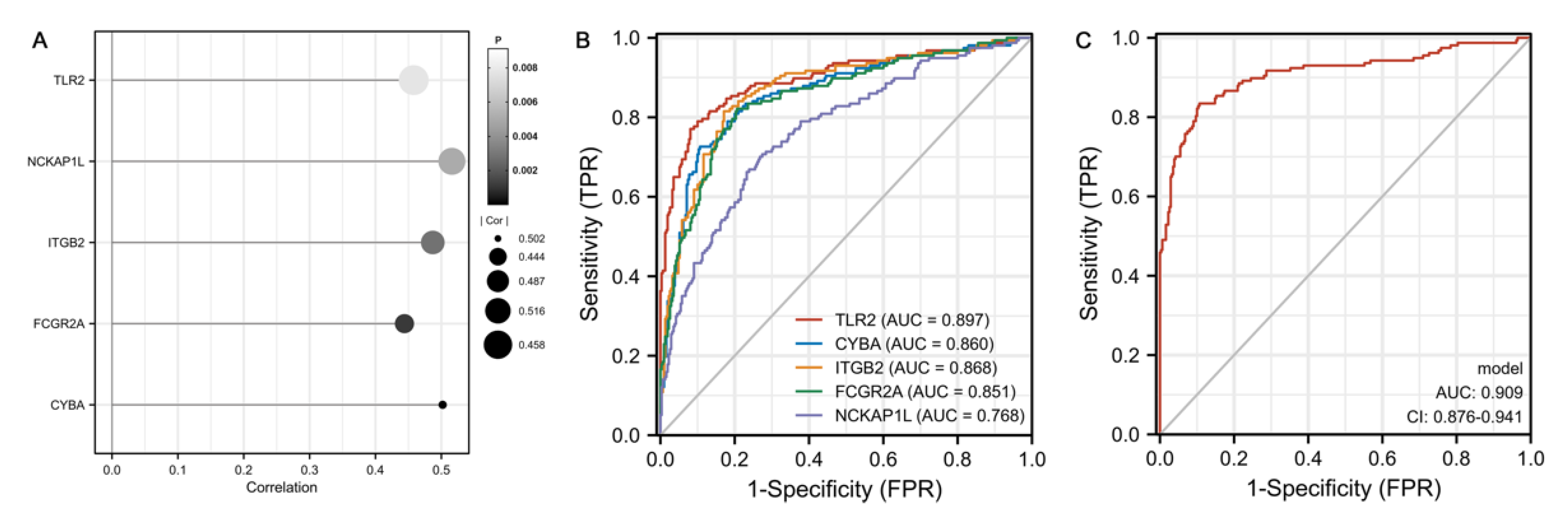
3.4. Identification of Hub Genes
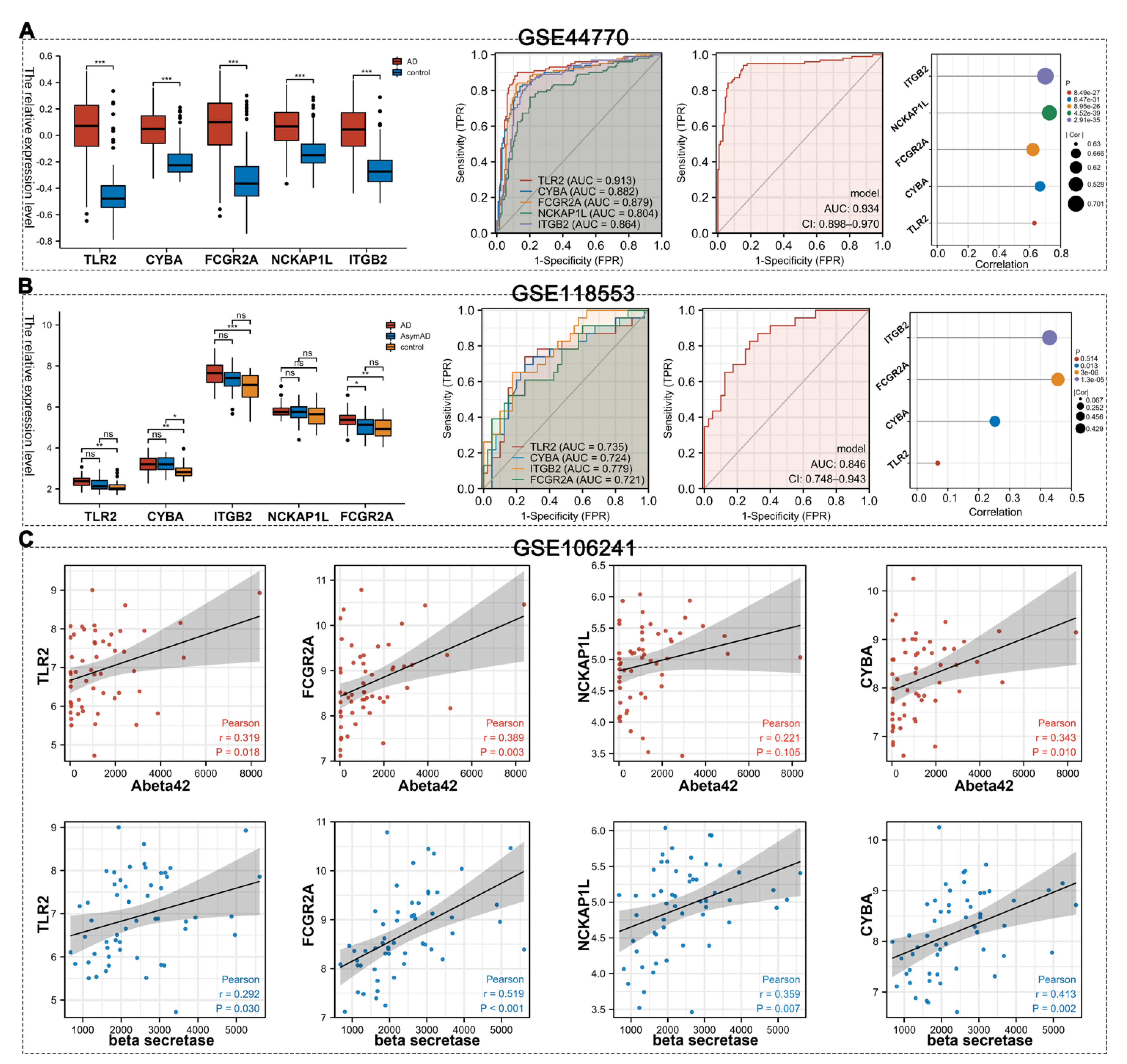
3.5. Validation of Hub Gens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimer’s Dement 2013, 9, 63–75. [Google Scholar] [CrossRef] [PubMed]
- 2022 Alzheimer’s disease facts and figures. Alzheimer’s Dement 2022, 18, 700–789. [CrossRef] [PubMed]
- Aisen, P.S.; Cummings, J.; Jack, C.R., Jr.; Morris, J.C.; Sperling, R.; Frölich, L.; Jones, R.W.; Dowsett, S.A.; Matthews, B.R.; Raskin, J.; et al. On the path to 2025: Understanding the Alzheimer’s disease continuum. Alzheimer’s Res. Ther. 2017, 9, 60. [Google Scholar] [CrossRef]
- Heneka, M.T.; Carson, M.J.; Khoury, J.E.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Calsolaro, V.; Edison, P. Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimer’s Dement 2016, 12, 719–732. [Google Scholar] [CrossRef]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef]
- Simard, A.R.; Soulet, D.; Gowing, G.; Julien, J.-P.; Rivest, S. Bone Marrow-Derived Microglia Play a Critical Role in Restricting Senile Plaque Formation in Alzheimer’s Disease. Neuron 2006, 49, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Zenaro, E.; Pietronigro, E.; Bianca, V.D.; Piacentino, G.; Marongiu, L.; Budui, S.; Turano, E.; Rossi, B.; Angiari, S.; Dusi, S.; et al. Neutrophils promote Alzheimer’s disease–like pathology and cognitive decline via LFA-1 integrin. Nat. Med. 2015, 21, 880–886. [Google Scholar] [CrossRef]
- Unger, M.S.; Li, E.; Scharnagl, L.; Poupardin, R.; Altendorfer, B.; Mrowetz, H.; Hutter-Paier, B.; Weiger, T.M.; Heneka, M.T.; Attems, J.; et al. CD8(+) T-cells infiltrate Alzheimer’s disease brains and regulate neuronal- and synapse-related gene expression in APP-PS1 transgenic mice. Brain Behav. Immun. 2020, 89, 67–86. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newcombe, E.A.; Camats-Perna, J.; Silva, M.L.; Valmas, N.; Huat, T.J.; Medeiros, R. Inflammation: The link between comorbidities, genetics, and Alzheimer’s disease. J. Neuroinflammation 2018, 15, 276. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Jung, S.; Priller, J. Microglia Biology: One Century of Evolving Concepts. Cell 2019, 179, 292–311. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, J.B.; Moore, K.J.; Means, T.K.; Leung, J.; Terada, K.; Toft, M.; Freeman, M.W.; Luster, A.D. CD36 mediates the innate host response to beta-amyloid. J. Exp. Med. 2003, 197, 1657–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, C.R.; Stuart, L.M.; Wilkinson, K.; van Gils, J.M.; Deng, J.; Halle, A.; Rayner, K.J.; Boyer, L.; Zhong, R.; Frazier, W.A.; et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 2010, 11, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Tahara, K.; Kim, H.-D.; Jin, J.-J.; Maxwell, J.A.; Li, L.; Fukuchi, K.-I. Role of toll-like receptor signalling in Aβ uptake and clearance. Brain 2006, 129, 3006–3019. [Google Scholar] [CrossRef] [Green Version]
- Webster, S.D.; Yang, A.J.; Margol, L.; Garzon-Rodriguez, W.; Glabe, C.G.; Tenner, A.J. Complement component C1q modulates the phagocytosis of Abeta by microglia. Exp. Neurol. 2000, 161, 127–138. [Google Scholar] [CrossRef]
- Edler, M.K.; Mhatre-Winters, I.; Richardson, J.R. Microglia in Aging and Alzheimer’s Disease: A Comparative Species Review. Cells 2021, 10, 1138. [Google Scholar] [CrossRef]
- Gate, D.; Rezai-Zadeh, K.; Jodry, D.; Rentsendorj, A.; Town, T. Macrophages in Alzheimer’s disease: The blood-borne identity. J. Neural Transm. 2010, 117, 961–970. [Google Scholar] [CrossRef] [Green Version]
- Dionisio-Santos, D.A.; Olschowka, J.A.; O’Banion, M.K. Exploiting microglial and peripheral immune cell crosstalk to treat Alzheimer’s disease. J. Neuroinflammation 2019, 16, 74. [Google Scholar] [CrossRef] [Green Version]
- Stalder, A.K.; Ermini, F.; Bondolfi, L.; Krenger, W.; Burbach, G.J.; Deller, T.; Coomaraswamy, J.; Staufenbiel, M.; Landmann, R.; Jucker, M. Invasion of hematopoietic cells into the brain of amyloid precursor protein transgenic mice. J. Neurosci. 2005, 25, 11125–11132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Khoury, J.; Toft, M.; Hickman, S.E.; Means, T.K.; Terada, K.; Geula, C.; Luster, A.D. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 2007, 13, 432–438. [Google Scholar] [CrossRef]
- Rentsendorj, A.; Sheyn, J.; Fuchs, D.-T.; Daley, D.; Salumbides, B.C.; Schubloom, H.E.; Hart, N.J.; Li, S.; Hayden, E.Y.; Teplow, D.B.; et al. A novel role for osteopontin in macrophage-mediated amyloid-β clearance in Alzheimer’s models. Brain Behav. Immun. 2018, 67, 163–180. [Google Scholar] [CrossRef] [Green Version]
- Colton, C.A. Heterogeneity of microglial activation in the innate immune response in the brain. J. Neuroimmune Pharm. 2009, 4, 399–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geissmann, F.; Jung, S.; Littman, D.R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003, 19, 71–82. [Google Scholar] [CrossRef] [Green Version]
- Davies, L.C.; Taylor, P.R. Tissue-resident macrophages: Then and now. Immunology 2015, 144, 541–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Zhu, D.; Yang, N.; Liu, Y.Y.; Zheng, J.; Ji, C.; Zuo, P.P. M2 Macrophage Transplantation Ameliorates Cognitive Dysfunction in Amyloid-β-Treated Rats Through Regulation of Microglial Polarization. J. Alzheimers Dis. 2016, 52, 483–495. [Google Scholar] [CrossRef]
- Aluri, J.; Cooper, M.A.; Schuettpelz, L.G. Toll-Like Receptor Signaling in the Establishment and Function of the Immune System. Cells 2021, 10, 1374. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Rangasamy, S.B.; Jana, M.; Roy, A.; Corbett, G.T.; Kundu, M.; Chandra, S.; Mondal, S.; Dasarathi, S.; Mufson, E.J.; Mishra, R.K.; et al. Selective disruption of TLR2-MyD88 interaction inhibits inflammation and attenuates Alzheimer’s pathology. J. Clin. Invest 2018, 128, 4297–4312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Iribarren, P.; Hu, J.; Chen, J.; Gong, W.; Cho, E.H.; Lockett, S.; Dunlop, N.M.; Wang, J.M. Activation of Toll-like Receptor 2 on Microglia Promotes Cell Uptake of Alzheimer Disease-associated Amyloid β Peptide. J. Biol. Chem. 2006, 281, 3651–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Sun, X.; Hu, Y.; Song, J.; Dong, S.; Kong, D.; Wang, Y.; Hua, X.; Han, J.; Zhou, Y.; et al. Genomic deletion of TLR2 induces aggravated white matter damage and deteriorated neurobehavioral functions in mouse models of Alzheimer’s disease. Aging 2019, 11, 7257–7273. [Google Scholar] [CrossRef]
- Richard, K.L.; Filali, M.; Préfontaine, P.; Rivest, S. Toll-like receptor 2 acts as a natural innate immune receptor to clear amyloid beta 1-42 and delay the cognitive decline in a mouse model of Alzheimer’s disease. J. Neurosci. 2008, 28, 5784–5793. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.; Zhang, Y.; Yang, X.; Li, M.; Hu, H.; Xiong, J.; Wang, N.; Jin, J.; Zhang, Y.; Song, Y.; et al. Hepatitis B Core Antigen Impairs the Polarization While Promoting the Production of Inflammatory Cytokines of M2 Macrophages via the TLR2 Pathway. Front. Immunol. 2020, 11, 535. [Google Scholar] [CrossRef]
- Shiau, D.J.; Kuo, W.T.; Davuluri, G.V.N.; Shieh, C.C.; Tsai, P.J.; Chen, C.C.; Lin, Y.S.; Wu, Y.Z.; Hsiao, Y.P.; Chang, C.P. Hepatocellular carcinoma-derived high mobility group box 1 triggers M2 macrophage polarization via a TLR2/NOX2/autophagy axis. Sci. Rep. 2020, 10, 13582. [Google Scholar] [CrossRef]
- Wang, J.; Shirota, Y.; Bayik, D.; Shirota, H.; Tross, D.; Gulley, J.L.; Wood, L.V.; Berzofsky, J.A.; Klinman, D.M. Effect of TLR Agonists on the Differentiation and Function of Human Monocytic Myeloid-Derived Suppressor Cells. J. Immunol. 2015, 194, 4215. [Google Scholar] [CrossRef] [Green Version]
- Anania, J.C.; Chenoweth, A.M.; Wines, B.D.; Hogarth, P.M. The Human FcγRII (CD32) Family of Leukocyte FcR in Health and Disease. Front. Immunol. 2019, 10, 464. [Google Scholar] [CrossRef]
- Akiyama, H.; McGeer, P.L. Brain microglia constitutively express beta-2 integrins. J. Neuroimmunol. 1990, 30, 81–93. [Google Scholar] [CrossRef]
- Mukherjee, S.; Klaus, C.; Pricop-Jeckstadt, M.; Miller, J.A.; Struebing, F.L. A Microglial Signature Directing Human Aging and Neurodegeneration-Related Gene Networks. Front. Neurosci. 2019, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Stasia, M.J. CYBA encoding p22(phox), the cytochrome b558 alpha polypeptide: Gene structure, expression, role and physiopathology. Gene 2016, 586, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.S.; Lee, J.S.; Rodgers, M.; Min, C.K.; Lee, J.Y.; Kim, H.J.; Lee, K.H.; Kim, C.J.; Oh, B.; Zandi, E.; et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe 2012, 11, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, C.N.; Rosenzwajg, M.; Carapito, R.; Shahrooei, M.; Konantz, M.; Khan, A.; Miao, Z.; Groß, M.; Tranchant, T.; Radosavljevic, M.; et al. NCKAP1L defects lead to a novel syndrome combining immunodeficiency, lymphoproliferation, and hyperinflammation. J. Exp. Med. 2020, 217, e20192275. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Xu, C.; Zhou, Y.; Chen, A.; Jin, B. Identification of Biomarkers Related to M2 Macrophage Infiltration in Alzheimer’s Disease. Cells 2022, 11, 2365. https://doi.org/10.3390/cells11152365
Lin C, Xu C, Zhou Y, Chen A, Jin B. Identification of Biomarkers Related to M2 Macrophage Infiltration in Alzheimer’s Disease. Cells. 2022; 11(15):2365. https://doi.org/10.3390/cells11152365
Chicago/Turabian StyleLin, Caixiu, Congcong Xu, Yongji Zhou, Anqi Chen, and Baiye Jin. 2022. "Identification of Biomarkers Related to M2 Macrophage Infiltration in Alzheimer’s Disease" Cells 11, no. 15: 2365. https://doi.org/10.3390/cells11152365
APA StyleLin, C., Xu, C., Zhou, Y., Chen, A., & Jin, B. (2022). Identification of Biomarkers Related to M2 Macrophage Infiltration in Alzheimer’s Disease. Cells, 11(15), 2365. https://doi.org/10.3390/cells11152365