Regulation of the Cancer Stem Phenotype by Long Non-Coding RNAs
Abstract
:1. Introduction
2. Cancer Stem Cells
Characteristics of the Stem Phenotype
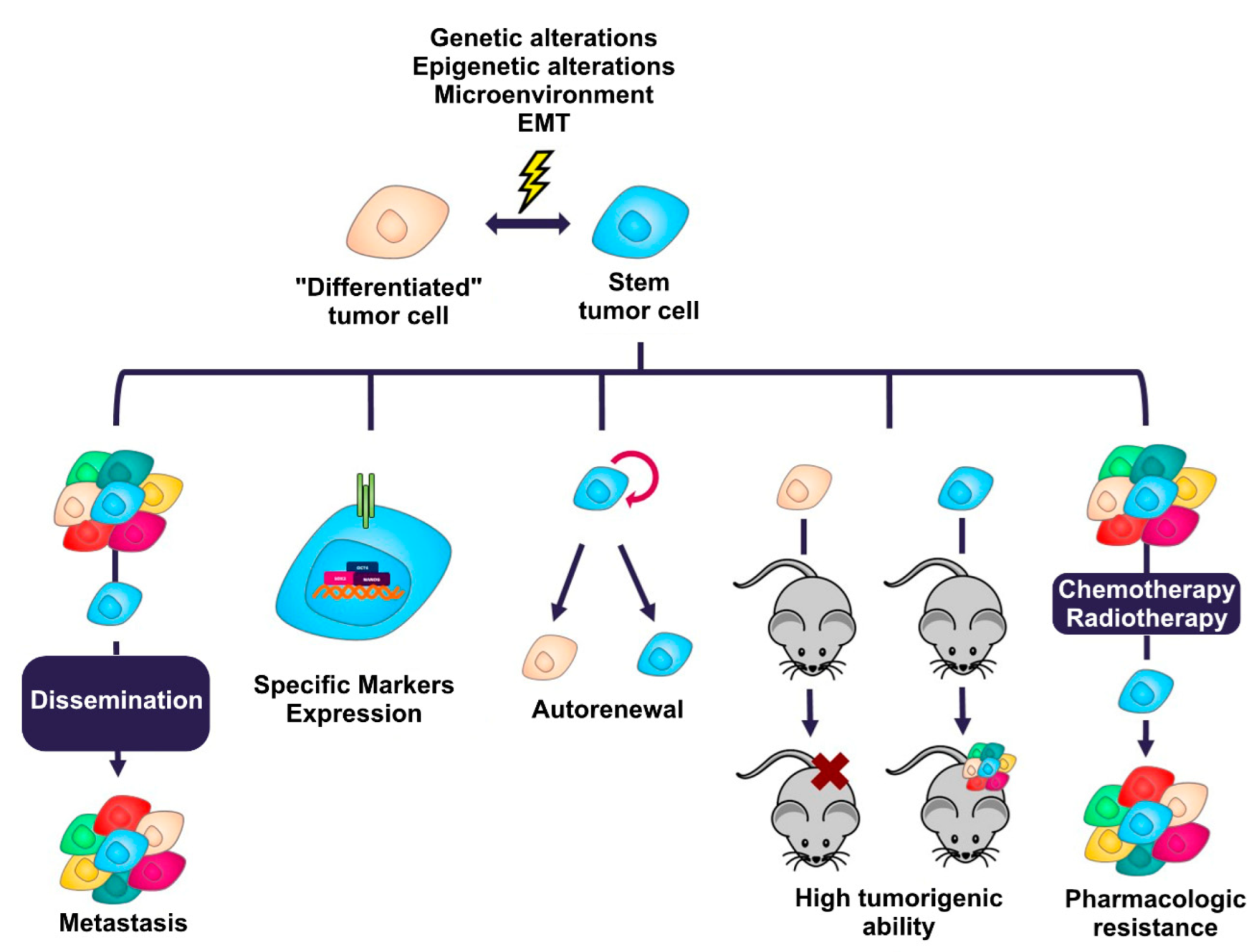
- Self-renewal. This is the ability of cancer stem cells to divide asymmetrically [40], causing one of the daughter cells resulting from cell division to maintain its stem characteristics [41] and the other resulting daughter cell to begin the process of a restricted differentiation program without the stem cell capabilities [42]. Cancer stem cells maintain their population at a constant level through this feature, allowing the malignant tumor to reform when, for example, non-stem cancer cells are pharmacologically eliminated [43].
- Tumorigenic ability. Cancer cells with a stem phenotype possess the ability to initiate and maintain a malignant tumor when transplanted into immunodeficient mice [4,5]. A small number of them is sufficient to recapitulate the malignant tumor from which they originate when transferred in this type of mouse [44,45], in contrast to non-stem cancer cells, which present a reduced tumor-forming ability [46].
- Drug resistance. Cancer stem cells are able to evade many conventional chemotherapeutic drugs [47,48]. These cells can remain quiescent for long periods of time, so drugs directed toward proliferating cells do not affect them, as they are not dividing [49]. Another mechanism involved in their drug resistance is the expression of ATP-dependent membrane transporters of the ABC family [50] or isoforms of aldehyde dehydrogenase enzymes, which allow the expulsion of drugs more efficiently than in cancer cells that do not express these proteins [51].
- Marker expression: The expression of specific molecules is one of the features that has allowed the identification, isolation and enrichment of cancer stem cells. The markers range from surface proteins, such as CD44 [32], CD133 [33], CD24 [5], EpCAM (epithelial cell adhesion molecule) [52], etc., to molecules with enzymatic activity, such as isoforms 1A1 and 1A3 of the enzyme aldehyde dehydrogenase (ALDH) [25,26]. In general, cancer stem cells usually express some of the transcription factors associated with pluripotency, such as SOX2, NANOG and OCT4) [53,54]. Table 1 lists the main markers that define the stem phenotype in the most common solid tumors.
| Markers | Tumor | References |
|---|---|---|
| CD44+ CD24−/low, CD44+ CD24−, CD44+ CD24− CD24 EpCAM+, ALDH enzyme activity (isoforms 1A1, 1A3) | Breast cancer | [5,25,26] |
| CD133, CD166, CD44, LGR5, ALDH1A1 enzyme activity | Colorectal cancer | [27,28,29,30,31,32,33] |
| CD44+ | Stomach cancer | [32] |
| CD133+ | Lung cancer | [33] |
| CD133+, CD44+ | Prostate cancer | [33,34] |
| CD44+ CD24+ EpCAM+, CD133+ CD44+, ALDH enzyme activity. | Pancreatic cancer | [52,55,56] |
| CD133+ | Brain cancer | [57] |
- Metastasis. It has been proposed that cancer stem cells are able to migrate to other regions of the body and form new malignant tumors (metastasis) by activating the EMT (epithelial-mesenchymal transition) program that allows the conversion of an epithelial cell to a mesenchymal cell [11,58]. To perform this process, cancer stem cells present activated signaling cascades that regulate transcription factors such as SNAIL (snail family transcriptional repressor 1), SLUG (snail family transcriptional repressor 2), ZEB1 (zinc finger E-Box binding homeobox 1), and ZEB2 (zinc finger E-Box binding homeobox 2) [59,60,61,62].
3. LncRNAs Regulate Gene Expression
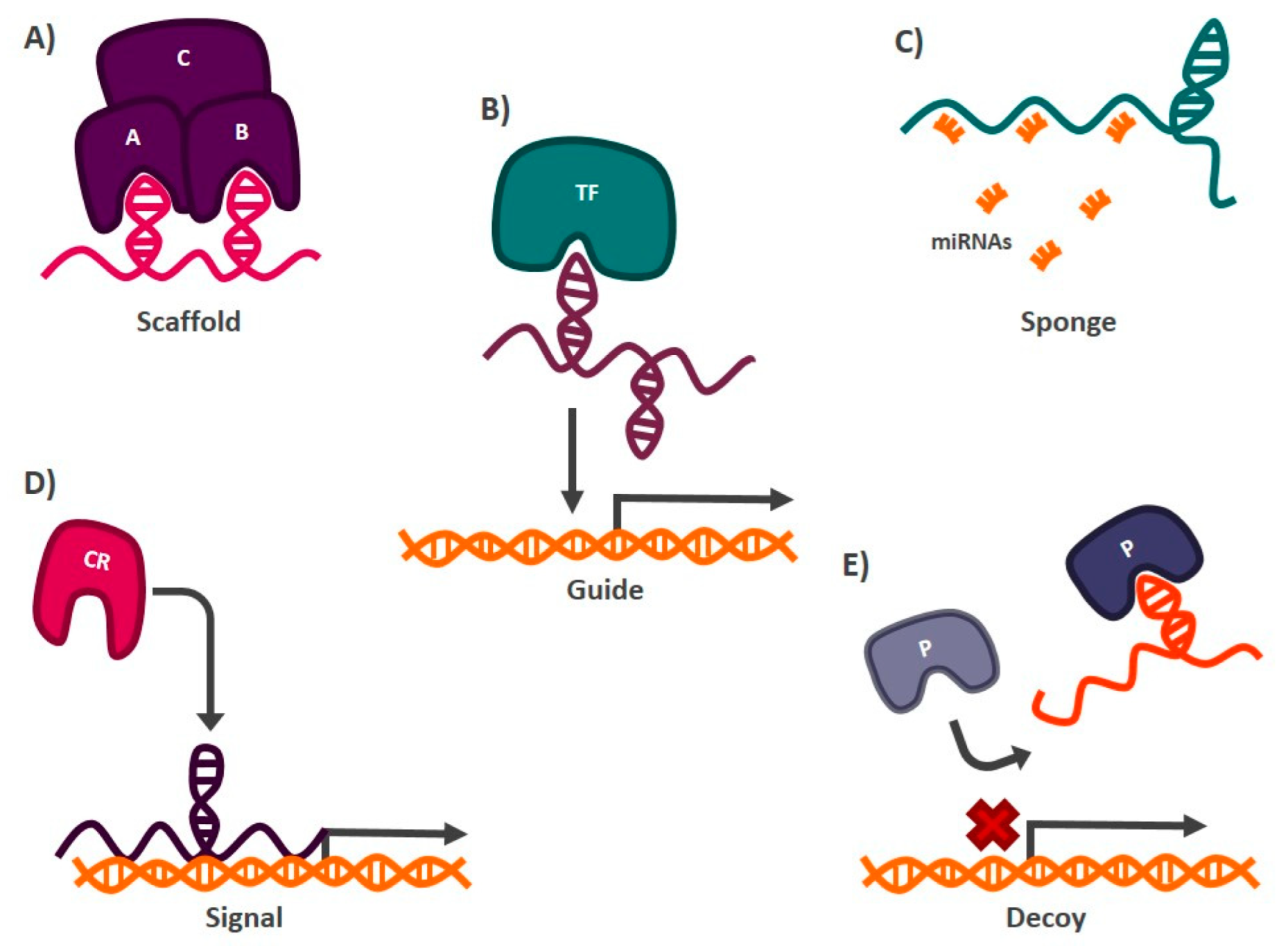
- Scaffolds: lncRNAs can form secondary structures that allow them to bind to proteins to form complexes that carry out particular functions in the cell [88]. An example of a transcript with this mechanism of action is lncRNA-NEAT1 (nuclear paraspeckle assembly transcript 1), which interacts with the protein subunit EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) of the chromatin remodeling complex PCR2 (polycomb repressive complex 2), which functions as a transcriptional repressor of different genes [89] (Figure 2).
- Sponges: lncRNAs with this mechanism of action present target miRNAs sequences, so they can prevent these miRNAs from carrying out their function by directly interacting with them. The miRNAs are complementary to the lncRNA sequence [90]. Among the transcripts with this mechanism of action is lncRNA-SNHG7 (small nucleolar RNA hostgene 7), which is found abundantly in the cell and hijacks miR-216b, blocking its activity by competing as a binding site against its target messenger RNA (mRNA) [91] (Figure 2).
- Guides: lncRNAs can interact with chromatin remodeling proteins and transcription factors to target specific DNA sequences where gene activation or silencing is required [92]. One of the best studied lncRNAs with this function is HOTAIR (HOX transcript antisense RNA), which mediates the silencing of tumor suppressor genes such as P21 in cancer. In this case, HOTAIR recruits the chromatin remodeling complex PRC2, through its interaction with the EZH2 subunit, to the promoter region of P21, establishing marks that allow the formation of heterochromatin and, therefore, the transcriptional repression of the tumor suppressor gene [93] (Figure 2).
- Signals: Acting as markers in different regions of the genome to signal chromatin remodeling complexes whether transcriptional activation or silencing of genes is required [92]. One of the lncRNAs reported to have this function is XIST (X inactive specific transcript). When this transcript is expressed, it functions as a mark indicating which X chromosome should be silenced, in females. XIST recruits chromatin remodelers, such as PRC2, to the X chromosome that will be repressed to generate heterochromatin [94] (Figure 2).
- Decoys: Preventing proteins or protein complexes such as the transcription initiator complex from carrying out their function, thus impacting gene expression. LncRNAs with decoy activity bind to proteins and DNA, preventing other molecules from interacting with them. In this way, the hijacked protein or DNA sequence cannot interact with its target. The lncRNA TERRA (telomeric-repeat-containing RNA) is an example of a transcript with this mechanism of action. This lncRNA binds to the active site of the enzyme telomerase, blocking its activity and preventing it from carrying out its function of protecting the telomeric regions of chromosomes [95] (Figure 2).
4. The Role of LncRNAs in the Regulation of the Stem Phenotype in Malignant Tumors
4.1. Breast Cancer
4.2. Colorectal Cancer
4.3. Gastric Cancer
4.4. LncRNAs in the Regulation of the Stem Phenotype of Other Types of Cancer
| LncRNA | Genes/Signaling Pathway Involved | Tumor | References |
|---|---|---|---|
| LncRNA-AGAP2-AS1 | SOX2, OCT4 | Breast | [108] |
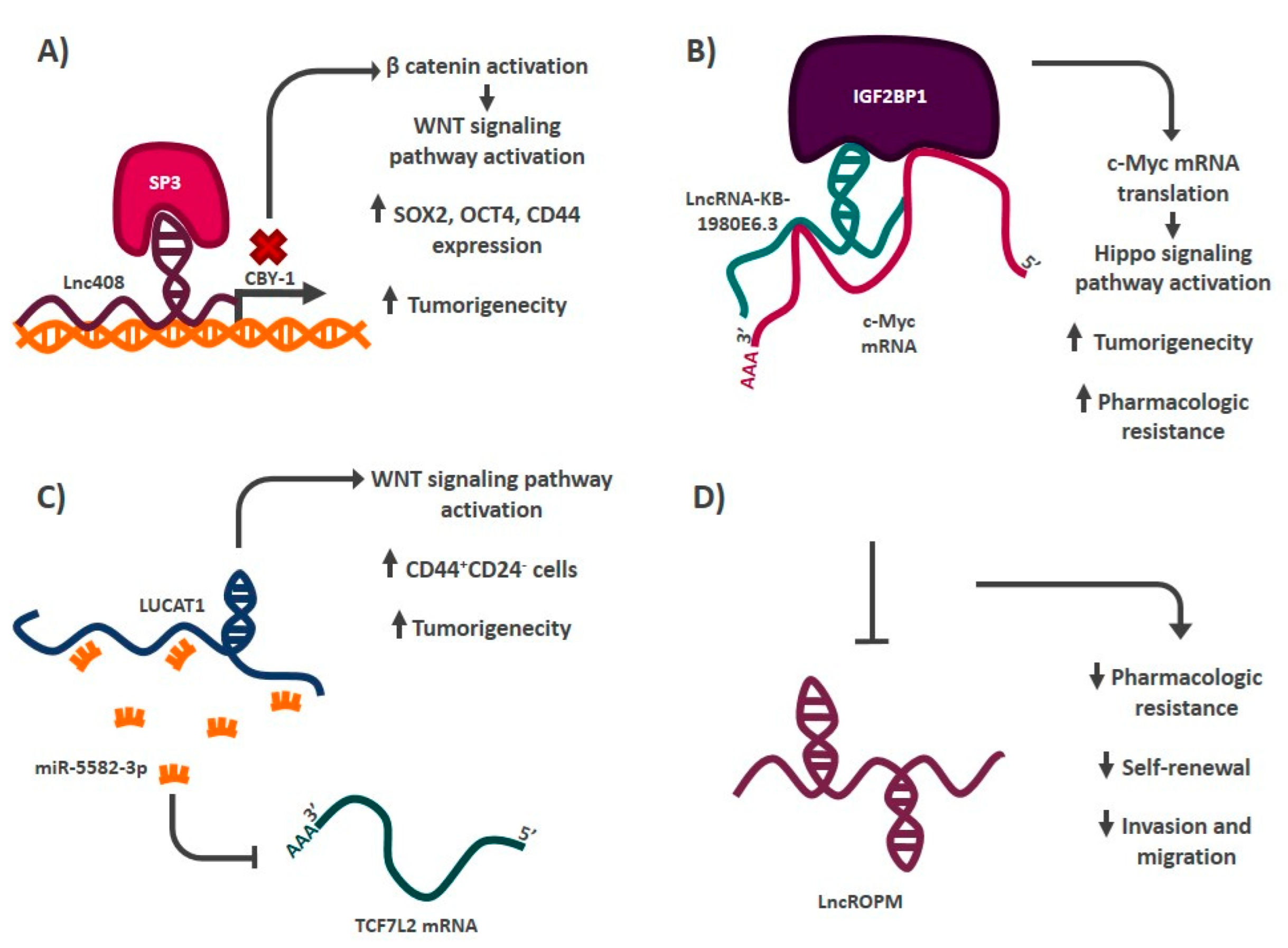
| Lnc408 | CBY-1/WNT signaling pathway | Breast | [147] |
| LncRNA-HOTTIP | miR-148a-3p/Wnt pathway | Breast | [112] |
| LncRNA-ROPM | Not addressed | Breast | [106] |
| LINC00617 | CD44+ CD24− | Breast | [107] |
| lncRNA-HAL | CD44+ CD24− via NANOG and ALDH1A3. | Breast | [109] |
| LncRNA-KB-1980E6.3 | IGF2BP1-c-Myc | Breast | [104] |
| HOTAIR | miR-34a-SOX2 | Breast | [102] |
| miRNA-590-sp-JAK/STAT3 signaling pathway | Prostate | [142] | |
| miR-34a-JAK2/STAT3 | Pancreas | [143] | |
| LncRNA-FEZF1-AS1 | miR-30a-NANOG | Breast | [113] |
| miR-363-3p-SOX2, NANOG and OCT4 | Stomach | [132] | |
| LINC00261 | miRNA-550a-3p | Breast | [114] |
| LUCAT1 | miR-5582-3p-TCF7L2/WNT signaling pathway | Breast | [105] |
| LncRNA-Hh | Not addressed | Breast | [115] |
| LncRNA-SOX21-AS1 | Hippo YAP/TAZ signaling pathway | Breast | [116] |
| LncRNA-NEAT1 | SOX2 | Breast | [117] |
| LincRNA-ROR | EMT program (ZEB1, ZEB2, TWIST, SLUG and SNAIL) | Breast | [118] |
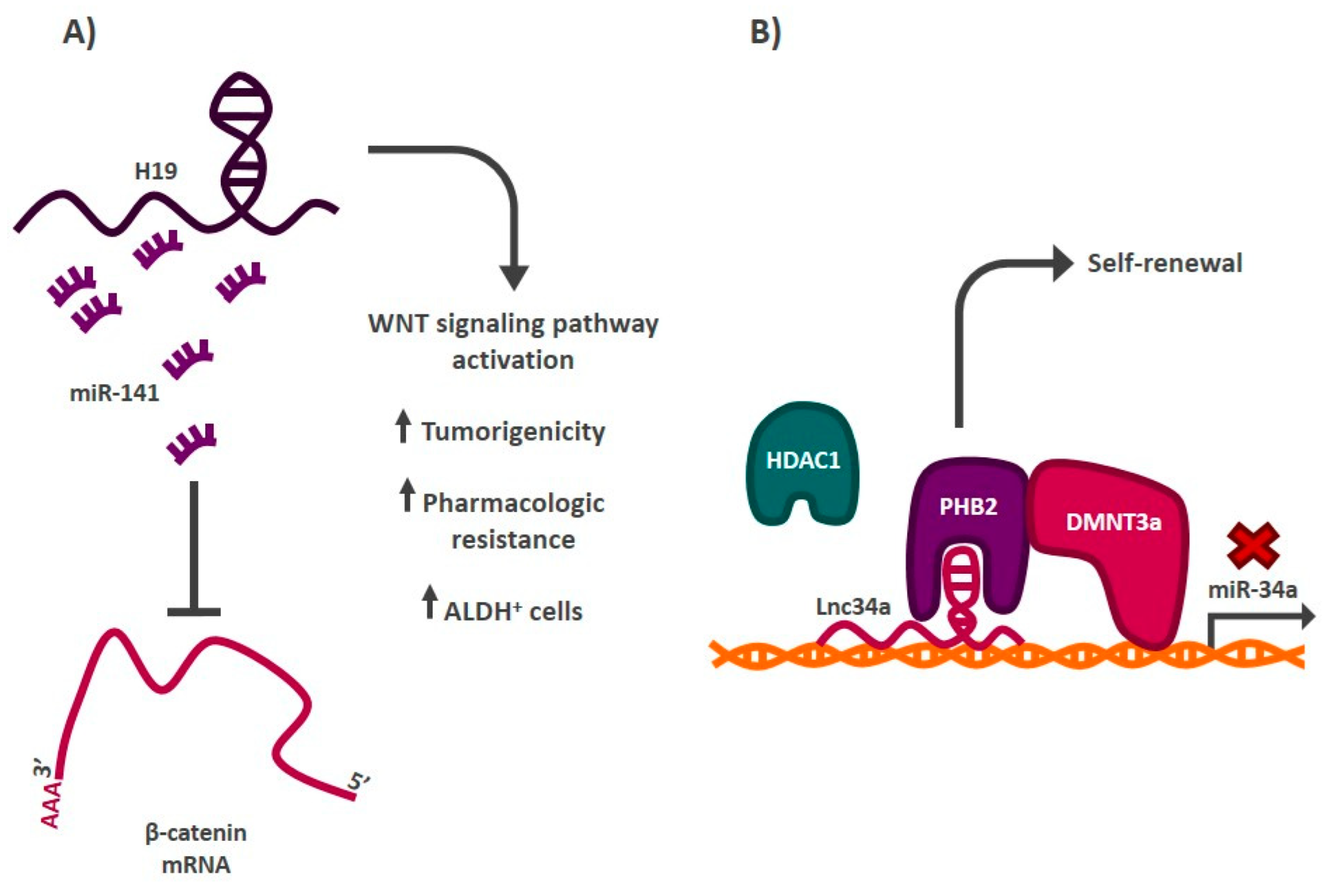
| LncRNA-H19 | let-7 | Breast | [111] |
| miR-141-β-catenin | Colorectal | [121] | |
| LINC00511 | miR-185-3p-E2F | Breast | [110] |
| MALAT-1 | SOX2 | Breast | [119] |
| Bmi1, β-catenin, C-MYC, NANOG, SOX2 and OCT4. | Pancreas | [148] | |
| LNCRNA-TUG 1 | Not addressed | Colorectal | [124] |
| LncRNA-BCAR4 | miR-665 | Colorectal | [125] |
| LncRNA SLCO4A1-AS1 | miR-150-3p-SLCO4A1 | Colorectal | [126] |
| LncRNA-cCSC1 | SMO/GLI1 (Hedgehog signaling pathway) | Colorectal | [122] |
| LncRNA KLK8 | SOX2, OCT4 and NANOG | Colorectal | [127] |
| Lnc34a | miR-34a-DNMT3a/HDAC1 | Colorectal | [123] |
| LncRNA-RP11-567G11.1 | JAGGED1, HES1 and HES5 (Notch signaling pathway) | Pancreas | [149] |
| GAS5 | miR-221-SOCS3 | Pancreas | [144] |
| LncRNA STXBP5-AS1 | EZH2-ADGB | Pancreas | [150] |
| LncRNA-uc.345 | SOX2, OCT4 and NANOG | Pancreas | [151] |
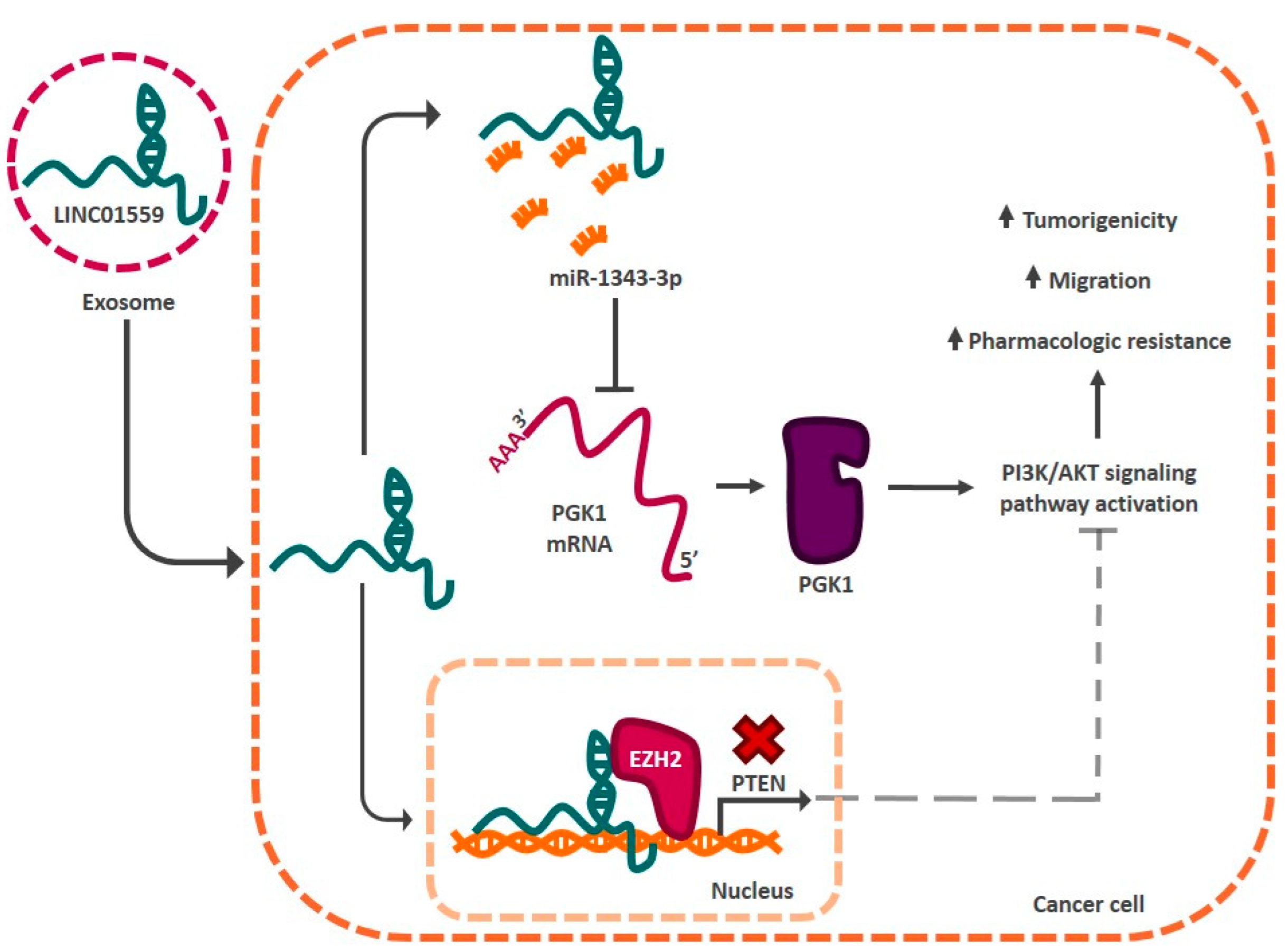
| LINC01559 | miR-1343-3p-PTEN-PI3K/AKT signaling pathway | Stomach | [131] |
| LncRNA-MACC1-AS1 | TGF-β signaling pathway-miR-145-5P | Stomach | [130] |
| LncRNA-MEG3 | miR-708-SOCS3-JAK/STAT3 pathway | Stomach | [139] |
| miR-421-WNT signaling pathway | Oral | [138] | |
| FMR1-AS1 | NF-κB/c-Myc signaling pathway | Esophagus | [146] |
| LncRNA HAND2-AS1 | INO80 chromatin remodeling complex-BMPR1A-BMP signaling pathway | Liver | [145] |
| LncTCF7 | SWI/SNF chromatin remodeling complex-TCF7-WNT signaling pathway | Liver | [140] |
| miR-200c-EpCAM | Brain (Glioma) | [141] | |
| LncRNA-UCA1 | miRNA-122-5p-SOX2 | Cervical | [152] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Marzagalli, M.; Fontana, F.; Raimondi, M.; Limonta, P. Cancer Stem Cells-Key Players in Tumor Relapse. Cancers 2021, 13, 376. [Google Scholar] [CrossRef] [PubMed]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Shenghui, H.; Nakada, D.; Morrison, S.J. Mechanisms of Stem Cell Self-Renewal. Annu. Rev. Cell Dev. Biol. 2009, 25, 377–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumon, K.; Afify, S.M.; Hassan, G.; Ueno, S.; Monzur, S.; Nawara, H.M.; Quora, H.A.A.; Sheta, M.; Xu, Y.; Fu, X.; et al. Differentiation of cancer stem cells into erythroblasts in the presence of CoCl(2). Sci. Rep. 2021, 11, 23977. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Z.; Ajani, J.A.; Song, S. Drug resistance and Cancer stem cells. Cell Commun. Signal. 2021, 19, 19. [Google Scholar] [CrossRef]
- Leng, Z.; Xia, Q.; Chen, J.; Li, Y.; Xu, J.; Zhao, E.; Zheng, H.; Ai, W.; Dong, J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell. Physiol. Biochem. 2018, 46, 860–872. [Google Scholar] [CrossRef]
- Zhang, H.; Brown, R.L.; Wei, Y.; Zhao, P.; Liu, S.; Liu, X.; Deng, Y.; Hu, X.; Zhang, J.; Gao, X.D.; et al. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019, 33, 166–179. [Google Scholar] [CrossRef]
- Liao, W.T.; Ye, Y.P.; Deng, Y.J.; Bian, X.W.; Ding, Y.Q. Metastatic cancer stem cells: From the concept to therapeutics. Am. J. Stem Cells 2014, 3, 46–62. [Google Scholar] [PubMed]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef]
- Xiao, X.; Li, L.; Cui, J.C.; Wang, Y. LncRNA FALEC promotes proliferation, migration, and invasion of PTC cells through regulating Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4361–4367. [Google Scholar]
- Chen, L.; Yang, W.; Guo, Y.; Chen, W.; Zheng, P.; Zeng, J.; Tong, W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE 2017, 12, e0185406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.-T. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Kataoka, M.; Wang, Y.; Pu, L.; Dong, X.; Fu, X.; Zhang, F.; Gao, F.; Liang, T.; Pei, J.; et al. LncRNA LncHrt preserves cardiac metabolic homeostasis and heart function by modulating the LKB1-AMPK signaling pathway. Basic Res. Cardiol. 2021, 116, 48. [Google Scholar] [CrossRef] [PubMed]
- Cros, J.; Raffenne, J.; Couvelard, A.; Poté, N. Tumor Heterogeneity in Pancreatic Adenocarcinoma. Pathobiology 2017, 85, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, D.; Meng, Q.; Liu, Z.; Xie, H.; Liu, L.; Xu, F.; Chen, X. Precision treatment exploration of breast cancer based on heterogeneity analysis of lncRNAs at the single-cell level. BMC Cancer 2021, 21, 918. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Kim, H.J.; Kang, E.; Kim, E.-K.; Kim, S.H.; Kim, J.H.; Kim, I.A.; Park, S.Y. Genomic profiling of multiple breast cancer reveals inter-lesional heterogeneity. Br. J. Cancer 2020, 122, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Grosselin, K.; Durand, A.; Marsolier, J.; Poitou, A.; Marangoni, E.; Nemati, F.; Dahmani, A.; Lameiras, S.; Reyal, F.; Frenoy, O.; et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 2019, 51, 1060–1066. [Google Scholar] [CrossRef]
- Lei, X.; Lei, Y.; Li, J.-K.; Du, W.-X.; Li, R.-G.; Yang, J.; Li, J.; Li, F.; Tan, H.-B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef]
- Khaniki, S.H.; Fakoor, V.; Sales, S.S.; Esmaily, H.; Miri, H.H. Risk of relapse and death from colorectal cancer and its related factors using non-Markovian Multi-State model. Gastroenterol. Hepatol. Bed Bench 2020, 13, 200–208. [Google Scholar]
- Walcher, L.; Kistenmacher, A.-K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.-R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcato, P.; Dean, C.A.; Pan, D.; Araslanova, R.; Gillis, M.; Joshi, M.; Helyer, L.; Pan, L.; Leidal, A.; Gujar, S.; et al. Aldehyde Dehydrogenase Activity of Breast Cancer Stem Cells Is Primarily Due To Isoform ALDH1A3 and Its Expression Is Predictive of Metastasis. Stem Cells 2011, 29, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2006, 445, 106–110. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, T.G.; Powell, A.E.; Davies, P.S.; Silk, A.D.; Dismuke, A.D.; Anderson, E.C.; Swain, J.R.; Wong, M.H. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 2010, 139, 2072–2082.e2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, E.H.; Hynes, M.J.; Zhang, T.; Ginestier, C.; Dontu, G.; Appelman, H.; Fields, J.Z.; Wicha, M.S.; Boman, B.M. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009, 69, 3382–3389. [Google Scholar] [CrossRef] [Green Version]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Takaishi, S.; Okumura, T.; Tu, S.; Wang, S.S.W.; Shibata, W.; Vigneshwaran, R.; Gordon, S.A.K.; Shimada, Y.; Wang, T.C. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells 2009, 27, 1006–1020. [Google Scholar] [CrossRef] [Green Version]
- Leung, E.L.-H.; Fiscus, R.R.; Tung, J.W.; Tin, V.P.-C.; Cheng, L.C.; Sihoe, A.D.-L.; Fink, L.M.; Ma, Y.; Wong, M.P. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS ONE 2010, 5, e14062. [Google Scholar] [CrossRef] [Green Version]
- Patrawala, L.; Calhoun, T.; Schneider-Broussard, R.; Li, H.; Bhatia, B.; Tang, S.; Reilly, J.G.; Chandra, D.; Zhou, J.; Claypool, K.; et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 2006, 25, 1696–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreso, A.; Dick, J.E. Evolution of the Cancer Stem Cell Model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, M.C.; Hollingsworth, R.E.; Hurt, E.M. Cancer stem cell plasticity and tumor hierarchy. World J. Stem Cells 2015, 7, 27–36. [Google Scholar] [CrossRef]
- Butti, R.; Gunasekaran, V.P.; Kumar, T.V.S.; Banerjee, P.; Kundu, G.C. Breast cancer stem cells: Biology and therapeutic implications. Int. J. Biochem. Cell Biol. 2019, 107, 38–52. [Google Scholar] [CrossRef]
- Papaccio, F.; Paino, F.; Regad, T.; Papaccio, G.; Desiderio, V.; Tirino, V. Concise Review: Cancer Cells, Cancer Stem Cells, and Mesenchymal Stem Cells: Influence in Cancer Development. Stem Cells Transl. Med. 2017, 6, 2115–2125. [Google Scholar] [CrossRef]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Koguchi, M.; Nakahara, Y.; Ito, H.; Wakamiya, T.; Yoshioka, F.; Ogata, A.; Inoue, K.; Masuoka, J.; Izumi, H.; Abe, T. BMP4 induces asymmetric cell division in human glioma stem-like cells. Oncol. Lett. 2020, 19, 1247–1254. [Google Scholar] [CrossRef] [Green Version]
- Winquist, R.J.; Hall, A.B.; Eustace, B.K.; Furey, B.F. Evaluating the immortal strand hypothesis in cancer stem cells: Symmetric/self-renewal as the relevant surrogate marker of tumorigenicity. Biochem. Pharmacol. 2014, 91, 129–134. [Google Scholar] [CrossRef]
- Lee, S.I.; Roney, M.S.I.; Park, J.H.; Baek, J.Y.; Park, J.; Kim, S.K.; Park, S.K. Dopamine receptor antagonists induce differentiation of PC-3 human prostate cancer cell-derived cancer stem cell-like cells. Prostate 2019, 79, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Mirshahidi, S.; Simental, A.; Lee, S.C.; De Andrade Filho, P.A.; Peterson, N.R.; Duerksen-Hughes, P.; Yuan, X. Cancer stem cell self-renewal as a therapeutic target in human oral cancer. Oncogene 2019, 38, 5440–5456. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Stepputtis, S.S.; Strietz, J.; Preca, B.-T.; Urban, S.; Willmann, D.; Allen, A.; Zenk, F.; Iovino, N.; Bronsert, P.; et al. KDM4 Inhibition Targets Breast Cancer Stem–like Cells. Cancer Res. 2017, 77, 5900–5912. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Santillan, K.; Melendez-Zajgla, J.; Jimenez-Hernandez, L.E.; Gaytan-Cervantes, J.; Muñoz-Galindo, L.; Piña-Sanchez, P.; Martinez-Ruiz, G.; Torres, J.; Garcia-Lopez, P.; Gonzalez-Torres, C.; et al. NF-kappaΒ-inducing kinase regulates stem cell phenotype in breast cancer. Sci. Rep. 2016, 6, 37340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, T.; Li, B.; Feng, X.; Fan, S.; Liu, L.; Liu, D.; Mao, J.; Lu, Y.; Yang, J.; Yu, X.; et al. Abnormally elevated USP37 expression in breast cancer stem cells regulates stemness, epithelial-mesenchymal transition and cisplatin sensitivity. J. Exp. Clin. Cancer Res. 2018, 37, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Zhang, H.; Ghia, E.M.; Huang, J.; Wu, L.; Zhang, J.; Lam, S.; Lei, Y.; He, J.; Cui, B.; et al. Inhibition of chemotherapy resistant breast cancer stem cells by a ROR1 specific antibody. Proc. Natl. Acad. Sci. USA 2019, 116, 1370–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Reed-Newman, T.; Anant, S.; Ramasamy, T.S. Regulatory Role of Quiescence in the Biological Function of Cancer Stem Cells. Stem Cell Rev. Rep. 2020, 16, 1185–1207. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G.; Macleod, K.F. Autophagy, cancer stem cells and drug resistance. J. Pathol. 2019, 247, 708–718. [Google Scholar] [CrossRef] [Green Version]
- Begicevic, R.-R.; Falasca, M. ABC Transporters in Cancer Stem Cells: Beyond Chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef] [Green Version]
- Januchowski, R.; Wojtowicz, K.; Zabel, M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed. Pharmacother. 2013, 67, 669–680. [Google Scholar] [CrossRef]
- Nomura, A.; Banerjee, S.; Chugh, R.; Dudeja, V.; Yamamoto, M.; Vickers, S.M.; Saluja, A.K. CD133 initiates tumors, induces epithelial-mesenchymal transition and increases metastasis in pancreatic cancer. Oncotarget 2015, 6, 8313–8322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Xie, Y.; Tran, L.; Lan, J.; Yang, Y.; Murugan, N.L.; Wang, R.; Wang, Y.J.; Semenza, G.L. Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. J. Clin. Investig. 2020, 130, 4607–4623. [Google Scholar] [CrossRef] [PubMed]
- Kaufhold, S.; Garbán, H.; Bonavida, B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J. Exp. Clin. Cancer Res. 2016, 35, 84. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of Pancreatic Cancer Stem Cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.P.; Fleming, J.B.; Wang, H.; Abbruzzese, J.L.; Choi, W.; Kopetz, S.; McConkey, D.J.; Evans, D.B.; Gallick, G.E. ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma. PLoS ONE 2011, 6, e20636. [Google Scholar] [CrossRef]
- Brescia, P.; Ortensi, B.; Fornasari, L.; Levi, D.; Broggi, G.; Pelicci, G. CD133 Is Essential for Glioblastoma Stem Cell Maintenance. Stem Cells 2013, 31, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S.; Quader, S.; Cabral, H.; Ono, R. Interplay of EMT and CSC in Cancer and the Potential Therapeutic Strategies. Front. Pharmacol. 2020, 11, 904. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lv, R.; Qi, W.; Wu, D.; Xu, Y.; Liu, W.; Mou, Y.; Wang, L. Snail contributes to the maintenance of stem cell-like phenotype cells in human pancreatic cancer. PLoS ONE 2014, 9, e87409. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, D.; Sheng, D.; Xu, J.; Chen, W.; Qin, Y.; Du, R.; Yang, X.; He, X.; Xie, N.; et al. NOTCH4 maintains quiescent mesenchymal-like breast cancer stem cells via transcriptionally activating SLUG and GAS1 in triple-negative breast cancer. Theranostics 2020, 10, 2405–2421. [Google Scholar] [CrossRef] [PubMed]
- Pérez, G.; López-Moncada, F.; Indo, S.; Torres, M.; Castellón, E.; Contreras, H. Knockdown of ZEB1 reverses cancer stem cell properties in prostate cancer cells. Oncol. Rep. 2021, 45, 58. [Google Scholar] [CrossRef]
- Li, N.; Babaei-Jadidi, R.; Lorenzi, F.; Spencer-Dene, B.; Clarke, P.; Domingo, E.; Tulchinsky, E.; Vries, R.G.J.; Kerr, D.; Pan, Y.; et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Yu, C.; Xu, M. Linking Tumor Microenvironment to Plasticity of Cancer Stem Cells: Mechanisms and Application in Cancer Therapy. Front. Oncol. 2021, 11, 678333. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Semenza, G.L. Hypoxia-inducible factors promote breast cancer stem cell specification and maintenance in response to hypoxia or cytotoxic chemotherapy. Adv. Cancer Res. 2019, 141, 175–212. [Google Scholar] [CrossRef]
- El-Sahli, S.; Wang, L. Cancer Stem Cell-Associated Pathways in the Metabolic Reprogramming of Breast Cancer. Int. J. Mol. Sci. 2020, 21, 9125. [Google Scholar] [CrossRef]
- Sultan, M.; Coyle, K.M.; Vidovic, D.; Thomas, M.L.; Gujar, S.; Marcato, P. Hide-and-seek: The interplay between cancer stem cells and the immune system. Carcinogenesis 2016, 38, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Ren, Y.; Zhang, J.; Chen, J.; Zhou, W.; Guo, W.; Wang, X.; Chen, H.; Li, M.; et al. Cisplatin-enriching cancer stem cells confer multidrug resistance in non-small cell lung cancer via enhancing TRIB1/HDAC activity. Cell Death Dis. 2017, 8, e2746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Zhang, J.; Yang, H. OCT4, SOX2, and NANOG positive expression correlates with poor differentiation, advanced disease stages, and worse overall survival in HER2(+) breast cancer patients. Onco. Targets Ther. 2018, 11, 7873–7881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeulen, L.; De Sousa E Melo, F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhou, C.; Zhang, Z.; Wang, Q.; Wei, H.; Shi, W.; Li, J.; Wang, Z.; Ou, Y.; Wang, W.; et al. Jagged1-Notch1-deployed tumor perivascular niche promotes breast cancer stem cell phenotype through Zeb1. Nat. Commun. 2020, 11, 5129. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; De, R.; Javed, S.; Srinivasan, R.; Pal, A.; Bhattacharyya, S. Sonic hedgehog pathway activation regulates cervical cancer stem cell characteristics during epithelial to mesenchymal transition. J. Cell. Physiol. 2019, 234, 15726–15741. [Google Scholar] [CrossRef]
- Hu, J.K.-H.; Du, W.; Shelton, S.J.; Oldham, M.C.; DiPersio, C.M.; Klein, O.D. An FAK-YAP-mTOR Signaling Axis Regulates Stem Cell-Based Tissue Renewal in Mice. Cell Stem Cell 2017, 21, 91–106.e106. [Google Scholar] [CrossRef] [Green Version]
- Ghuwalewala, S.; Ghatak, D.; Das, S.; Roy, S.; Das, P.; Butti, R.; Gorain, M.; Nath, S.; Kundu, G.C.; Roychoudhury, S. MiRNA-146a/AKT/β-Catenin Activation Regulates Cancer Stem Cell Phenotype in Oral Squamous Cell Carcinoma by Targeting CD24. Front. Oncol. 2021, 11, 651692. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Zajgla, J.; Maldonado, V. The Role of lncRNAs in the Stem Phenotype of Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2021, 22, 6374. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; pp. 1–46. [Google Scholar] [CrossRef]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Zhuang, C.; Ma, Q.; Zhuang, C.; Ye, J.; Zhang, F.; Gui, Y. LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. FASEB J. 2019, 33, 11045–11059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Zheng, X.; Cheng, J.; Zhang, K.; Ma, C. LncRNA TUG1 regulates proliferation and apoptosis by regulating miR-148b/IGF2 axis in ox-LDL-stimulated VSMC and HUVEC. Life Sci. 2020, 243, 117287. [Google Scholar] [CrossRef]
- Hon, C.-C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [Green Version]
- Perez, C.A.G.; Adachi, S.; Nong, Q.D.; Adhitama, N.; Matsuura, T.; Natsume, T.; Wada, T.; Kato, Y.; Watanabe, H. Sense-overlapping lncRNA as a decoy of translational repressor protein for dimorphic gene expression. PLoS Genet. 2021, 17, e1009683. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Chen, J.; Zhang, X.; Wang, Z.; Chen, J.; Lin, X.; Huang, H.; Fu, W.; Liang, J.; Wu, W.; et al. The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis. Nat Commun 2021, 12, 1341. [Google Scholar] [CrossRef]
- Li, G.; Zhang, T.; Huang, K.; Zhu, Y.; Xu, K.; Gu, J.; Huang, S.; Gu, C.; Zhan, R.; Shen, J. Long noncoding RNA GAS8-AS1: A novel biomarker in human diseases. Biomed. Pharmacother. 2021, 139, 111572. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, A.-S.; Ørom, U.A. Bidirectional expression of long ncRNA/protein-coding gene pairs in cancer. Brief. Funct. Genom. 2015, 15, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.R.; Messenger, Z.J.; Tam, H.W.; Phillips, S.L.; Recio, L.; Smart, R.C. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015, 6, e1700. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cel. 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.B.; Kobayashi, N.; Todo, Y.; Watari, H. Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front. Genet. 2018, 9, 471. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Liu, C.; Li, H.; Zhang, L.; Luo, G.; Liang, S.; Lü, M. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol. Lett. 2020, 19, 595–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Y.; Ma, J.; Pan, Y.; Hu, J.; Liu, B.; Jia, L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018, 9, 722. [Google Scholar] [CrossRef] [Green Version]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, B.; Liu, X.; Lu, L.; Luo, F.; Lu, X.; Shi, L.; Xu, W.; Liu, Q. Epigenetic silencing of p21 by long non-coding RNA HOTAIR is involved in the cell cycle disorder induced by cigarette smoke extract. Toxicol. Lett. 2016, 240, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Mlynarczyk-Evans, S.; Nusinow, D.A.; Panning, B. Xist RNA and the Mechanism of X Chromosome Inactivation. Annu. Rev. Genet. 2002, 36, 233–278. [Google Scholar] [CrossRef] [PubMed]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Ma, S.; Deng, X.; Yang, Y.; Zhang, Q.; Zhou, T.; Liu, Z. The lncRNA LINC00675 regulates cell proliferation, migration, and invasion by affecting Wnt/β-catenin signaling in cervical cancer. Biomed. Pharmacother. 2018, 108, 1686–1693. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Xiang, W.; Liu, J.; Tang, J.; Wang, J.; Liu, B.; Long, Z.; Wang, L.; Yin, G.; Liu, J. The regulatory role of antisense lncRNAs in cancer. Cancer Cell Int. 2021, 21, 459. [Google Scholar] [CrossRef]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.; Wang, X.; Wang, Y.; Ma, D. Wnt/β-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem. Biophys. Res. Commun. 2010, 392, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Kahn, M. The role of the Wnt signaling pathway in cancer stem cells: Prospects for drug development. Res. Rep. Biochem. 2014, 4, 1–12. [Google Scholar] [PubMed] [Green Version]
- Deng, J.; Yang, M.; Jiang, R.; An, N.; Wang, X.; Liu, B. Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem-Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS ONE 2017, 12, e0170860. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. Role of hypoxia inducible factor-1 in cancer stem cells (Review). Mol. Med. Rep. 2021, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; He, F.; Hou, Y.; Tu, G.; Li, Q.; Jin, T.; Zeng, H.; Qin, Y.; Wan, X.; Qiao, Y.; et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 2021, 40, 1609–1627. [Google Scholar] [CrossRef]
- Zheng, A.; Song, X.; Zhang, L.; Zhao, L.; Mao, X.; Wei, M.; Jin, F. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 305. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Sun, Y.; Hou, Y.; Yang, L.; Wan, X.; Qin, Y.; Liu, Y.; Wang, R.; Zhu, P.; Teng, Y.; et al. A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J. Hematol. Oncol. 2021, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, L.; Xu, L.; Qin, K.; Liu, C.; Yu, Y.; Su, D.; Wu, K.; Sheng, Y. Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol. Carcinog. 2015, 56, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Qu, H.; Han, M.; Ding, Y.; Xie, M.; Hu, J.; Chen, Y.; Dong, H. MSC-induced lncRNA AGAP2-AS1 promotes stemness and trastuzumab resistance through regulating CPT1 expression and fatty acid oxidation in breast cancer. Oncogene 2020, 40, 833–847. [Google Scholar] [CrossRef]
- García-Venzor, A.; Mandujano-Tinoco, E.A.; Lizarraga, F.; Zampedri, C.; Krötzsch, E.; Salgado, R.M.; Dávila-Borja, V.M.; Encarnación-Guevara, S.; Melendez-Zajgla, J.; Maldonado, V. Microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2019, 1866, 118523. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, Y.; Ma, Y.; Lu, J.; Chen, Y.; Jiang, Q.; Qin, Q.; Zhao, L.; Huang, Q.; Luo, Z.; et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 2018, 37, 289. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Li, T.-T.; Wang, K.-L.; Xiao, G.-Q.; Wang, J.-H.; Zhao, H.-D.; Kang, Z.-J.; Fan, W.-J.; Zhu, L.-L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Yan, Y.; Zhao, L.; Liu, Y.; Lv, X.; Zhang, L.; Zhao, Y.; Zhao, H.; He, M.; Wei, M. LncRNA HOTTIP facilitates the stemness of breast cancer via regulation of miR-148a-3p/WNT1 pathway. J. Cell. Mol. Med. 2020, 24, 6242–6252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Sun, L.; Zhang, Y.; Lu, G.; Li, Y.; Wei, Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J. Cell. Physiol. 2018, 233, 8630–8638. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, C. LINC00261/microRNA-550a-3p/SDPR axis> axis affects the biological characteristics of breast cancer stem cells. IUBMB Life 2020, 73, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.-E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Meng, D.; Wang, R. Long non-coding RNA SOX21-AS1 enhances the stemness of breast cancer cells via the Hippo pathway. FEBS Open Bio 2021, 11, 251–264. [Google Scholar] [CrossRef]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.-Y.; Siu, M.-T.; Ho, C.-W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef] [Green Version]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Cen, Y.; Chen, J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 2018, 15, 2117–2122. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.Y.; Yi, G.Q.; Tang, Z.L.; Chen, B. Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements. Yi Chuan 2020, 42, 435–443. [Google Scholar]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef]
- Zhou, H.; Xiong, Y.; Peng, L.; Wang, R.; Zhang, H.; Fu, Z. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. J. Cell. Biochem. 2019, 121, 2510–2524. [Google Scholar] [CrossRef]
- Wang, L.; Bu, P.; Ai, Y.; Srinivasan, T.; Chen, H.J.; Xiang, K.; Lipkin, S.M.; Shen, X. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. eLife 2016, 5, e14620. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Zhou, H.; Bao, X.; Wu, Y.; Jia, H.; Zhao, H.; Liu, G. lncRNA TUG1 Facilitates Colorectal Cancer Stem Cell Characteristics and Chemoresistance by Enhancing GATA6 Protein Stability. Stem Cells Int. 2021, 2021, 1075481. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhou, X.; Chen, Z.; Wang, M.; Zheng, X.; Xie, M. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019, 19, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Xu, T.; Song, X.; Shen, J.; Zheng, S.; Zhang, L.; Tao, G.; Jiang, B. LncRNA SLCO4A1-AS1 modulates colon cancer stem cell properties by binding to miR-150-3p and positively regulating SLCO4A1. Lab. Investig. 2021, 101, 908–920. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Song, W.; Shen, Y.; Wang, H.; Fan, Z. LncRNA KLK8 modulates stem cell characteristics in colon cancer. Pathol. Res. Pract. 2021, 224, 153437. [Google Scholar] [CrossRef]
- Das, M.; Law, S. Role of tumor microenvironment in cancer stem cell chemoresistance and recurrence. Int. J. Biochem. Cell Biol. 2018, 103, 115–124. [Google Scholar] [CrossRef]
- Korkaya, H.; Liu, S.; Wicha, M.S. Regulation of cancer stem cells by cytokine networks: Attacking cancer’s inflammatory roots. Clin. Cancer Res. 2011, 17, 6125–6129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, W.; Liang, B.; Wang, C.; Li, S.; Zhao, Y.; Huang, Q.; Liu, Z.; Yao, Z.; Wu, Q.; Liao, W.; et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 2019, 38, 4637–4654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Bo, X.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L.; Li, B. Exosome-transferred LINC01559 promotes the progression of gastric cancer via PI3K/AKT signaling pathway. Cell Death Dis. 2020, 11, 723. [Google Scholar] [CrossRef]
- Hui, Y.; Yang, Y.; Li, D.; Wang, J.; Di, M.; Zhang, S.; Wang, S. LncRNA FEZF1-AS1 Modulates Cancer Stem Cell Properties of Human Gastric Cancer through miR-363-3p/HMGA2. Cell Transpl. 2020, 29, 963689720925059. [Google Scholar] [CrossRef]
- Rothe, K.; Porter, V.; Jiang, X. Current Outlook on Autophagy in Human Leukemia: Foe in Cancer Stem Cells and Drug Resistance, Friend in New Therapeutic Interventions. Int. J. Mol. Sci. 2019, 20, 461. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Zhu, G.; Xu, J.; Lai, Q.; Yan, B.; Guo, Y.; Fung, T.K.; Zeisig, B.B.; Cui, Y.; Zha, J.; et al. HOTTIP lncRNA Promotes Hematopoietic Stem Cell Self-Renewal Leading to AML-like Disease in Mice. Cancer Cell 2019, 36, 645–659.e648. [Google Scholar] [CrossRef]
- Al-Kershi, S.; Bhayadia, R.; Ng, M.; Verboon, L.; Emmrich, S.; Gack, L.; Schwarzer, A.; Strowig, T.; Heckl, D.; Klusmann, J.H. The stem cell-specific long noncoding RNA HOXA10-AS in the pathogenesis of KMT2A-rearranged leukemia. Blood Adv. 2019, 3, 4252–4263. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Fan, X.; Zhu, J.; Chen, X.; Liu, Y.; Zhou, H. LncRNA MAGI2-AS3 inhibits the self-renewal of leukaemic stem cells by promoting TET2-dependent DNA demethylation of the LRIG1 promoter in acute myeloid leukaemia. RNA Biol. 2020, 17, 784–793. [Google Scholar] [CrossRef]
- Bill, M.; Papaioannou, D.; Karunasiri, M.; Kohlschmidt, J.; Pepe, F.; Walker, C.J.; Walker, A.E.; Brannan, Z.; Pathmanathan, A.; Zhang, X.; et al. Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia 2019, 33, 2169–2182. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-Y.; Hsieh, P.-L.; Peng, C.-Y.; Liao, Y.-W.; Yu, C.-H.; Yu, C.-C. LncRNA MEG3 inhibits self-renewal and invasion abilities of oral cancer stem cells by sponging miR-421. J. Formos. Med. Assoc. 2021, 120, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ji, W.-W.; Wei, W.; Zhan, L.-X.; Huang, X. Long noncoding RNA Meg3 sponges miR-708 to inhibit intestinal tumorigenesis via SOCS3-repressed cancer stem cells growth. Cell Death Dis. 2021, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P.; et al. The Long Noncoding RNA lncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells through Activation of Wnt Signaling. Cell Stem Cell 2015, 16, 413–425. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhang, L.; Zheng, L.; Hong, Y.; Zhao, L. LncRNATCF7 promotes the growth and self-renewal of glioma cells via suppressing the miR-200c-EpCAM axis. Biomed. Pharmacother. 2018, 97, 203–208. [Google Scholar] [CrossRef]
- Wang, N.; Jiang, Y.; Lv, S.; Wen, H.; Wu, D.; Wei, Q.; Dang, Q. HOTAIR expands the population of prostatic cancer stem-like cells and causes Docetaxel resistance via activating STAT3 signaling. Aging 2020, 12, 12771–12782. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Wang, J.; Zhang, L.; Li, J.; Jin, Y. LncRNA HOTAIR Promotes Cancer Stem-Like Cells Properties by Sponging miR-34a to Activate the JAK2/STAT3 Pathway in Pancreatic Ductal Adenocarcinoma. Onco Targets Ther. 2021, 14, 1883–1893. [Google Scholar] [CrossRef]
- Liu, B.; Wu, S.; Ma, J.; Yan, S.; Xiao, Z.; Wan, L.; Zhang, F.; Shang, M.; Mao, A. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 2018, 13, 472–482. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhu, P.; Luo, J.; Wang, J.; Liu, Z.; Wu, W.; Du, Y.; Ye, B.; Wang, D.; He, L.; et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019, 38, e101110. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, L.; Guo, B.; Deng, J.; Wu, S.; Li, F.; Wang, Y.; Lu, J.; Zhou, Y. Exosomal FMR1-AS1 facilitates maintaining cancer stem-like cell dynamic equilibrium via TLR7/NFκB/c-Myc signaling in female esophageal carcinoma. Mol. Cancer 2019, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Qin, Y.; Wang, R.; Yang, L.; Zeng, H.; Zhu, P.; Li, Q.; Qiu, Y.; Chen, S.; Liu, Y.; et al. A novel Lnc408 maintains breast cancer stem cell stemness by recruiting SP3 to suppress CBY1 transcription and increasing nuclear β-catenin levels. Cell Death Dis. 2021, 12, 437. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Hu, H.; Han, T.; Yuan, C.; Wang, L.; Jin, Z.; Guo, Z.; Wang, L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015, 16, 6677–6693. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Nie, W.; Yao, K.; Chou, J. Depletion of the lncRNA RP11-567G11.1 inhibits pancreatic cancer progression. Biomed. Pharmacother. 2019, 112, 108685. [Google Scholar] [CrossRef]
- Chen, S.; Huang, L.; Li, G.; Qiu, F.; Wang, Y.; Yang, C.; Pan, J.; Wu, Z.; Chen, J.; Tian, Y. LncRNA STXBP5-AS1 suppresses stem cell-like properties of pancreatic cancer by epigenetically inhibiting neighboring androglobin gene expression. Clin. Epigenet. 2020, 12, 168. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, J.; Yuan, X.; Qian, W.; Zhang, B.; Shi, M.; Xie, J.; Shen, B.; Xu, H.; Hou, Z.; et al. Long noncoding RNA uc.345 promotes tumorigenesis of pancreatic cancer by upregulation of hnRNPL expression. Oncotarget 2016, 7, 71556–71566. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Wang, Q.; Ji, M.; Guo, X.; Li, L.; Su, X. Exosomal lncRNA UCA1 modulates cervical cancer stem cell self-renewal and differentiation through microRNA-122-5p/SOX2 axis. J. Transl. Med. 2021, 19, 229. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutierrez-Cruz, J.A.; Maldonado, V.; Melendez-Zajgla, J. Regulation of the Cancer Stem Phenotype by Long Non-Coding RNAs. Cells 2022, 11, 2352. https://doi.org/10.3390/cells11152352
Gutierrez-Cruz JA, Maldonado V, Melendez-Zajgla J. Regulation of the Cancer Stem Phenotype by Long Non-Coding RNAs. Cells. 2022; 11(15):2352. https://doi.org/10.3390/cells11152352
Chicago/Turabian StyleGutierrez-Cruz, Jose Adan, Vilma Maldonado, and Jorge Melendez-Zajgla. 2022. "Regulation of the Cancer Stem Phenotype by Long Non-Coding RNAs" Cells 11, no. 15: 2352. https://doi.org/10.3390/cells11152352
APA StyleGutierrez-Cruz, J. A., Maldonado, V., & Melendez-Zajgla, J. (2022). Regulation of the Cancer Stem Phenotype by Long Non-Coding RNAs. Cells, 11(15), 2352. https://doi.org/10.3390/cells11152352






