Immunorthodontics: PD-L1, a Novel Immunomodulator in Cementoblasts, Is Regulated by HIF-1α under Hypoxia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Hypoxic Conditioning of Cementoblasts
2.3. Pharmacological Inhibitor
2.4. CRISPR/Cas9 Plasmid Transfection
2.5. RNA Isolation and Quality Measurement
2.6. Reverse Transcription and RT-qPCR Analysis
2.7. Immunofluorescence (IF)
2.8. Flow Cytometry Analysis
2.9. Protein Extraction and Western Blot Analysis
2.10. Statistical Analysis
3. Results
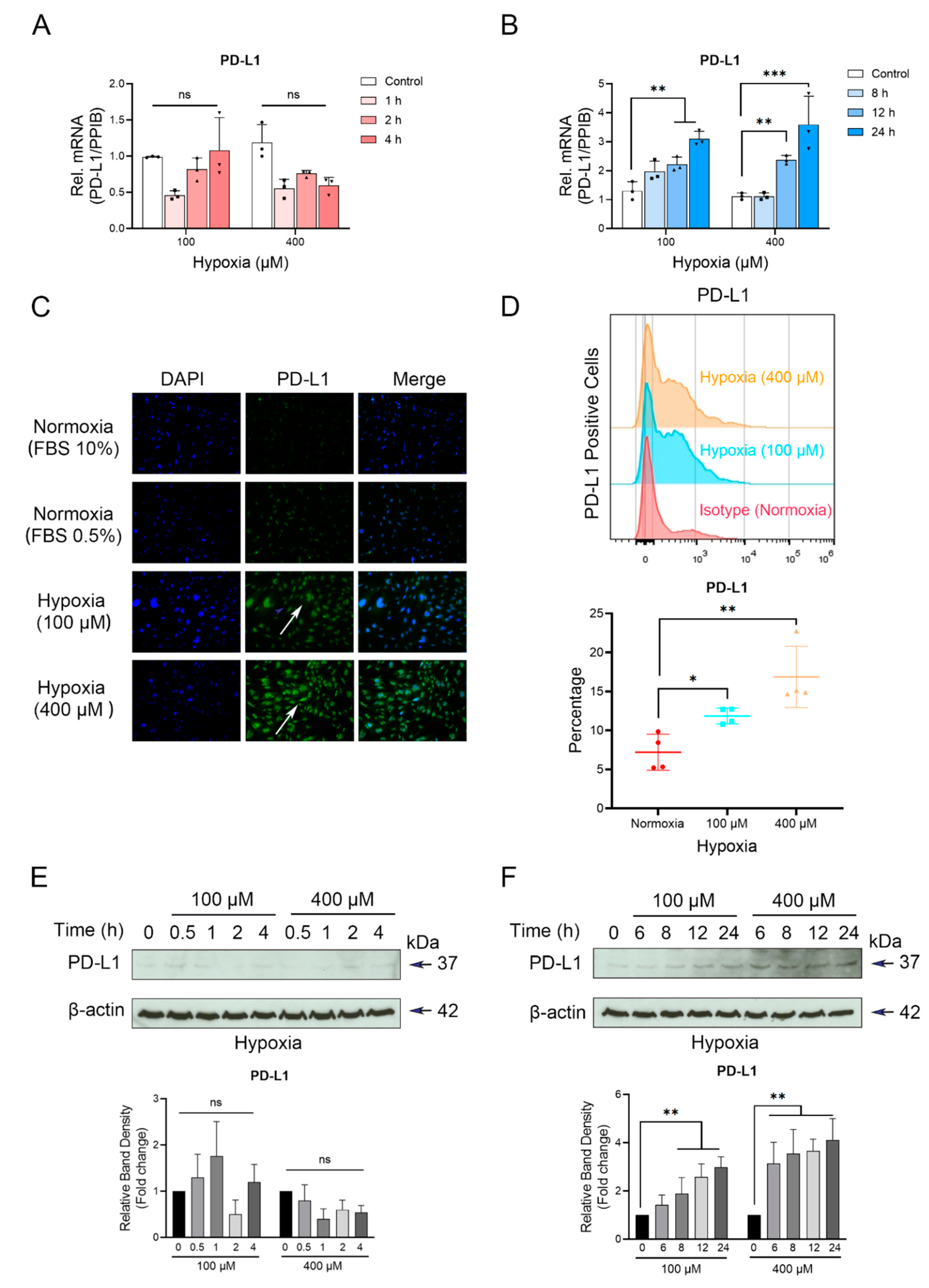
3.1. Up-Regulation of PD-L1 in Cementoblasts under Hypoxic Conditions
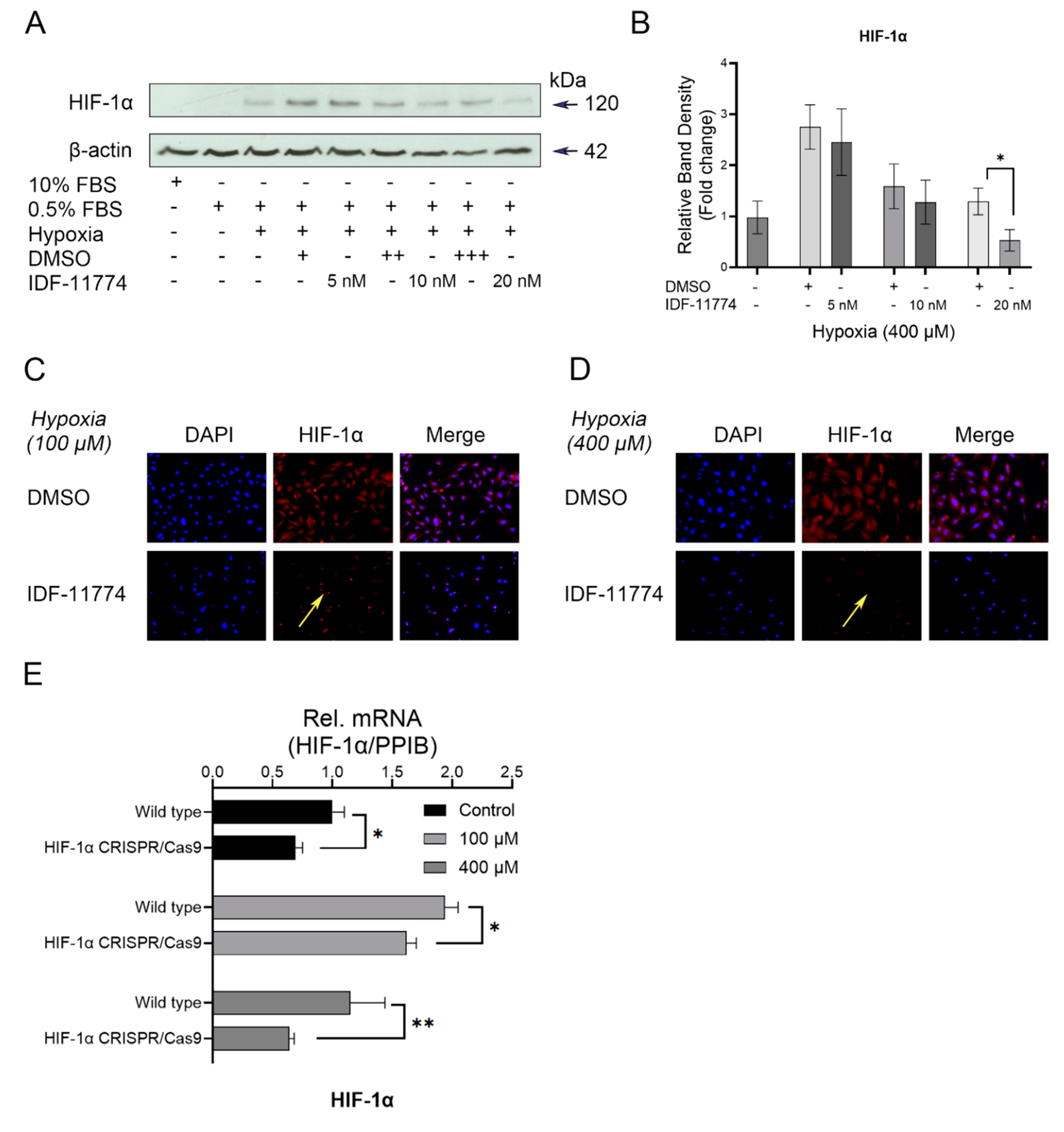
3.2. Increased Expression Levels of HIF-1α in Cementoblasts in Response to Hypoxia
3.3. Knockdown of HIF-1α Expression in Cementoblasts
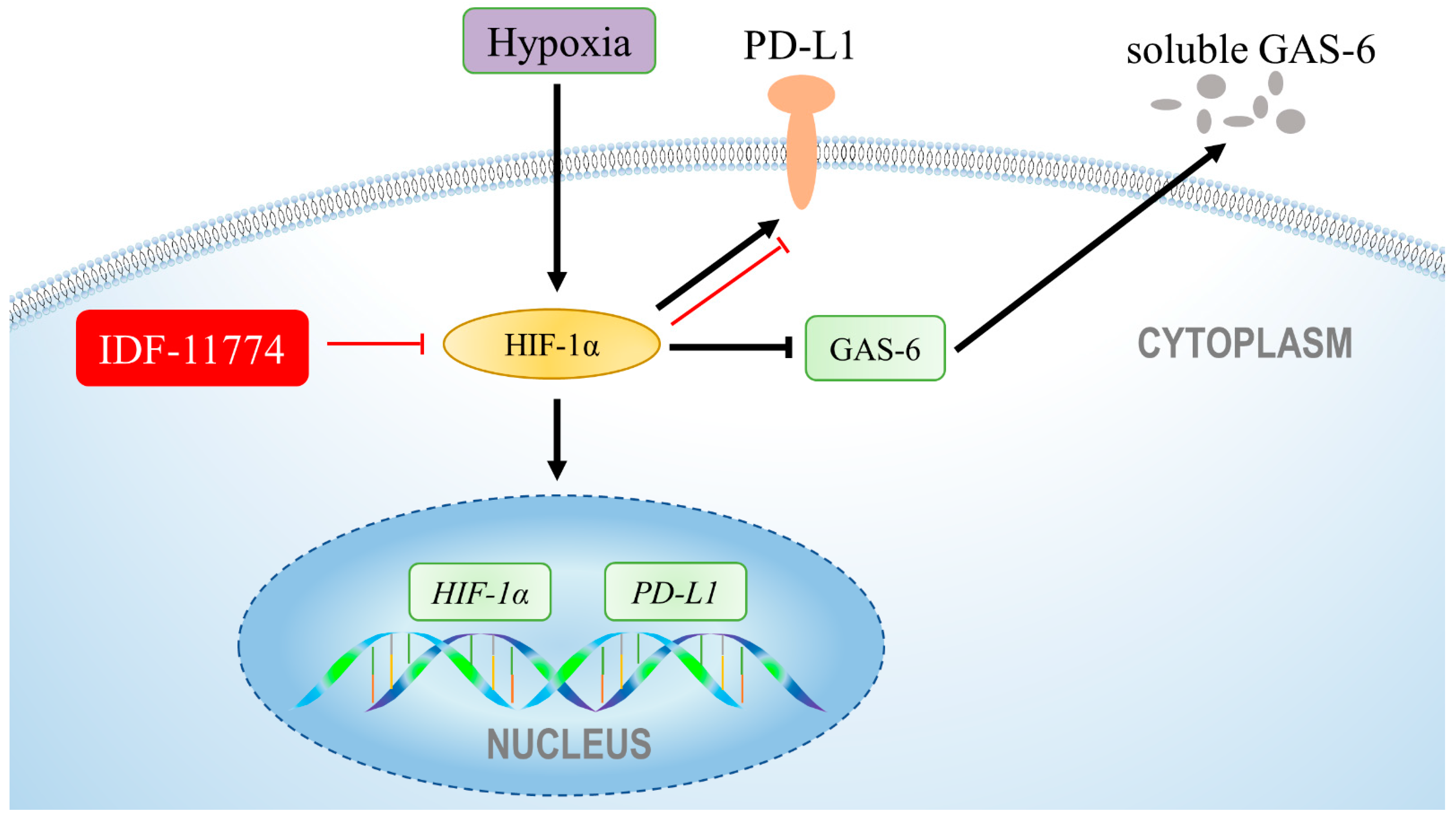
3.4. The Increased Expression of PD-L1 Is Directly Regulated by HIF-1α under Hypoxia in Cementoblasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yong, J.; Groeger, S.; Meyle, J.; Ruf, S. MAPK and β-Catenin signaling: Implication and interplay in orthodontic tooth movement. FBL 2022, 27, 54. [Google Scholar] [CrossRef] [PubMed]
- von Bremen, J.; Lorenz, N.; Ludwig, B.; Ruf, S. Increased BMI in children-an indicator for less compliance during orthodontic treatment with removable appliances. Eur. J. Orthod. 2018, 40, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.; von Bremen, J.; Ruiz-Heiland, G.; Ruf, S. Adiponectin as Well as Compressive Forces Regulate in vitro beta-Catenin Expression on Cementoblasts via Mitogen-Activated Protein Kinase Signaling Activation. Front. Cell Dev. Biol. 2021, 9, 645005. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.; Elisabeth Groeger, S.; Ruf, S.; Ruiz-Heiland, G. Influence of leptin and compression in GAS-6 mediated homeostasis of periodontal ligament cell. Oral Dis. 2021; Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Masella, R.S.; Meister, M. Current concepts in the biology of orthodontic tooth movement. Am. J. Orthod. Dentofacial Orthop. 2006, 129, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Linares, A.; Hartsfield, J.K., Jr. Cellular and Molecular Pathways Leading to External Root Resorption. J. Dent. Res. 2017, 96, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Brezniak, N.; Wasserstein, A. Orthodontically induced inflammatory root resorption. Part I: The basic science aspects. Angle Orthod. 2002, 72, 175–179. [Google Scholar] [PubMed]
- Yong, J.; von Bremen, J.; Ruiz-Heiland, G.; Ruf, S. Adiponectin Interacts In-Vitro With Cementoblasts Influencing Cell Migration, Proliferation and Cementogenesis Partly Through the MAPK Signaling Pathway. Front. Pharmacol. 2020, 11, 585346. [Google Scholar] [CrossRef] [PubMed]
- Klein, Y.; Fleissig, O.; Polak, D.; Barenholz, Y.; Mandelboim, O.; Chaushu, S. Immunorthodontics: In vivo gene expression of orthodontic tooth movement. Sci. Rep. 2020, 10, 8172. [Google Scholar] [CrossRef]
- Gianchecchi, E.; Delfino, D.V.; Fierabracci, A. Recent insights into the role of the PD-1/PD-L1 pathway in immunological tolerance and autoimmunity. Autoimmun. Rev. 2013, 12, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Ritprajak, P.; Azuma, M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015, 51, 221–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Ni, Z.; Liu, X.; Feng, S.; Dong, X.; Shi, X.; Zhai, J.; Mai, S.; Jiang, J.; Wang, Z.; et al. Beyond T Cells: Understanding the Role of PD-1/PD-L1 in Tumor-Associated Macrophages. J. Immunol. Res. 2019, 2019, 1919082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Q.; Qiu, X.; Zhang, Z.; Zhang, S.; Zhang, Y.; Liang, Y.; Guo, J.; Peng, H.; Chen, M.; Fu, Y.X.; et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat. Commun. 2020, 11, 4835. [Google Scholar] [CrossRef]
- Shen, J.K.; Cote, G.M.; Choy, E.; Yang, P.; Harmon, D.; Schwab, J.; Nielsen, G.P.; Chebib, I.; Ferrone, S.; Wang, X.; et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer Immunol. Res. 2014, 2, 690–698. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Gu, Y.; Liao, Y.; Bang, S.; Donnelly, C.R.; Chen, O.; Tao, X.; Mirando, A.J.; Hilton, M.J.; Ji, R.R. PD-1 blockade inhibits osteoclast formation and murine bone cancer pain. J. Clin. Investig. 2020, 130, 3603–3620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, C.M.; Zhang, P.; Wang, X.; Chen, J.; Yang, J.; Lu, W.; Zhou, W.; Yuan, W.; Feng, Y. Expression of programmed death 1 ligand 1 on periodontal tissue cells as a possible protective feedback mechanism against periodontal tissue destruction. Mol. Med. Rep. 2016, 13, 2423–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.; Sun, M.; Xia, Y.; Xie, Y.; Shu, R. LPS Stimulates Gingival Fibroblasts to Express PD-L1 via the p38 Pathway under Periodontal Inflammatory Conditions. Arch. Oral Biol. 2021, 129, 105161. [Google Scholar] [CrossRef] [PubMed]
- Groeger, S.; Domann, E.; Gonzales, J.R.; Chakraborty, T.; Meyle, J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology 2011, 216, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Francisco, L.M.; Sharpe, A.H. PD-1 and its ligands in T-cell immunity. Curr. Opin. Immunol. 2007, 19, 309–314. [Google Scholar] [CrossRef]
- Niklas, A.; Proff, P.; Gosau, M.; Romer, P. The role of hypoxia in orthodontic tooth movement. Int. J. Dent. 2013, 2013, 841840. [Google Scholar] [CrossRef]
- Pacary, E.; Tixier, E.; Coulet, F.; Roussel, S.; Petit, E.; Bernaudin, M. Crosstalk between HIF-1 and ROCK pathways in neuronal differentiation of mesenchymal stem cells, neurospheres and in PC12 neurite outgrowth. Mol. Cell Neurosci. 2007, 35, 409–423. [Google Scholar] [CrossRef]
- Yong, J.; von Bremen, J.; Groeger, S.; Ruiz-Heiland, G.; Ruf, S. Hypoxia-inducible factor 1-alpha acts as a bridge factor for crosstalk between ERK1/2 and caspases in hypoxia-induced apoptosis of cementoblasts. J. Cell Mol. Med. 2021, 25, 9710–9723. [Google Scholar] [CrossRef]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef]
- Kyrkanides, S. Hypoxia-inducible factor-1alpha may be the first host response in orthodontic tooth movement. Am. J. Orthod. Dentofacial Orthop. 2021, 160, 163–164. [Google Scholar] [CrossRef] [PubMed]
- Kirschneck, C.; Strassmair, N.; Cieplik, F.; Paddenberg, E.; Jantsch, J.; Proff, P.; Schroder, A. Myeloid HIF1alpha Is Involved in the Extent of Orthodontically Induced Tooth Movement. Biomedicines 2021, 9, 796. [Google Scholar]
- Kirschneck, C.; Thuy, M.; Leikam, A.; Memmert, S.; Deschner, J.; Damanaki, A.; Spanier, G.; Proff, P.; Jantsch, J.; Schroder, A. Role and Regulation of Mechanotransductive HIF-1alpha Stabilisation in Periodontal Ligament Fibroblasts. Int. J. Mol. Sci. 2020, 21, 9530. [Google Scholar]
- D’Errico, J.A.; Berry, J.E.; Ouyang, H.; Strayhorn, C.L.; Windle, J.J.; Somerman, M.J. Employing a transgenic animal model to obtain cementoblasts in vitro. J. Periodontol. 2000, 71, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Han, X.; Guo, Y.; Wu, H.; Ren, J.; Li, J.; Ai, D.; Wang, L.; Bai, D. Response of immortalized murine cementoblast cells to hypoxia in vitro. Arch. Oral Biol. 2013, 58, 1718–1725. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Yong, J.; Groeger, S.; Ruiz-Heiland, G.; Ruf, S. Selection and validation of reference gene for RT-qPCR studies in co-culture system of mouse cementoblasts and periodontal ligament cells. BMC Res. Notes 2022, 15, 57. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, T.; Akiba, H.; Iwai, H.; Matsuda, H.; Aoki, M.; Tanno, Y.; Shin, T.; Tsuchiya, H.; Pardoll, D.M.; Okumura, K.; et al. Expression of programmed death 1 ligands by murine T cells and APC. J. Immunol. 2002, 169, 5538–5545. [Google Scholar] [CrossRef] [Green Version]
- Groeger, S.; Denter, F.; Lochnit, G.; Schmitz, M.L.; Meyle, J. Porphyromonas gingivalis Cell Wall Components Induce Programmed Death Ligand 1 (PD-L1) Expression on Human Oral Carcinoma Cells by a Receptor-Interacting Protein Kinase 2 (RIP2)-Dependent Mechanism. Infect. Immun. 2020, 88, e00051-20. [Google Scholar] [CrossRef]
- Groeger, S.; Howaldt, H.P.; Raifer, H.; Gattenloehner, S.; Chakraborty, T.; Meyle, J. Oral Squamous Carcinoma Cells Express B7-H1 and B7-DC Receptors in Vivo. Pathol. Oncol. Res. 2017, 23, 99–110. [Google Scholar] [CrossRef]
- Mazanet, M.M.; Hughes, C.C. B7-H1 is expressed by human endothelial cells and suppresses T cell cytokine synthesis. J. Immunol. 2002, 169, 3581–3588. [Google Scholar] [CrossRef] [Green Version]
- van Duijn, A.; Willemsen, K.J.; van Uden, N.O.P.; Hoyng, L.; Erades, S.; Koster, J.; Luiten, R.M.; Bakker, W.J. A secondary role for hypoxia and HIF1 in the regulation of (IFNgamma-induced) PD-L1 expression in melanoma. Cancer Immunol. Immunother. 2021, 71, 529–540. [Google Scholar]
- Ruf, M.; Moch, H.; Schraml, P. PD-L1 expression is regulated by hypoxia inducible factor in clear cell renal cell carcinoma. Int. J. Cancer 2016, 139, 396–403. [Google Scholar] [CrossRef] [Green Version]
- Merceron, C.; Ranganathan, K.; Wang, E.; Tata, Z.; Makkapati, S.; Khan, M.P.; Mangiavini, L.; Yao, A.Q.; Castellini, L.; Levi, B.; et al. Hypoxia-inducible factor 2alpha is a negative regulator of osteoblastogenesis and bone mass accrual. Bone Res. 2019, 7, 7. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, F.; Kou, X.; Liu, D.; Yang, R.; Wang, X.; Song, Y.; He, D.; Gan, Y.; Zhou, Y. T Cells Are Required for Orthodontic Tooth Movement. J. Dent. Res. 2015, 94, 1463–1470. [Google Scholar] [CrossRef]
- Shelby, A.; Pendleton, C.; Thayer, E.; Johnson, G.K.; Xie, X.J.; Brogden, K.A. PD-L1 correlates with chemokines and cytokines in gingival crevicular fluid from healthy and diseased sites in subjects with periodontitis. BMC Res. Notes 2020, 13, 532. [Google Scholar] [CrossRef]
- Wongtim, K.; Ikeda, E.; Ohno, T.; Nagai, S.; Okuhara, S.; Kure, K.; Azuma, M. Overexpression of PD-L1 in gingival basal keratinocytes reduces periodontal inflammation in a ligature-induced periodontitis model. J. Periodontol. 2021, 93, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Philips, E.A.; Garcia-Espana, A.; Tocheva, A.S.; Ahearn, I.M.; Adam, K.R.; Pan, R.; Mor, A.; Kong, X.P. The structural features that distinguish PD-L2 from PD-L1 emerged in placental mammals. J. Biol. Chem. 2020, 295, 4372–4380. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yong, J.; Gröger, S.; von Bremen, J.; Meyle, J.; Ruf, S. Immunorthodontics: PD-L1, a Novel Immunomodulator in Cementoblasts, Is Regulated by HIF-1α under Hypoxia. Cells 2022, 11, 2350. https://doi.org/10.3390/cells11152350
Yong J, Gröger S, von Bremen J, Meyle J, Ruf S. Immunorthodontics: PD-L1, a Novel Immunomodulator in Cementoblasts, Is Regulated by HIF-1α under Hypoxia. Cells. 2022; 11(15):2350. https://doi.org/10.3390/cells11152350
Chicago/Turabian StyleYong, Jiawen, Sabine Gröger, Julia von Bremen, Joerg Meyle, and Sabine Ruf. 2022. "Immunorthodontics: PD-L1, a Novel Immunomodulator in Cementoblasts, Is Regulated by HIF-1α under Hypoxia" Cells 11, no. 15: 2350. https://doi.org/10.3390/cells11152350
APA StyleYong, J., Gröger, S., von Bremen, J., Meyle, J., & Ruf, S. (2022). Immunorthodontics: PD-L1, a Novel Immunomodulator in Cementoblasts, Is Regulated by HIF-1α under Hypoxia. Cells, 11(15), 2350. https://doi.org/10.3390/cells11152350





