1. Introduction
Focal adhesions are multi-protein assemblies in which transmembrane communication occurs at cell–matrix junctions. They function as a mechanical link that connects actin in the cell to the extracellular substrate and a channel to transmit biochemical signals between the cell and extracellular matrix (ECM). Therefore, focal adhesion plays a vital role in maintaining cell homeostasis in response to external stimuli or internal changes in cells [
1,
2].
The central structure of focal adhesion, integrins, are heterodimers composed of α and β subunits and transmembrane glycoproteins located at focal adhesions. The extracellular domain of integrin binds to substrates, such as fibronectin or proteoglycan, and the cytoplasmic domain is linked to actin filaments. Integrins structurally attach cells to the ECM [
3]. Furthermore, the structures of the α and β subunits change after chemical or mechanical stimulation from inside or outside the cell, resulting in the transmission of biosignals in and out of the cell [
4]. By performing these two basic functions, integrin plays an essential role in various biological activities of the cell, including cell migration, proliferation, and division [
5,
6,
7]. Unlike growth factor receptors, integrin does not have intrinsic catalytic activity due to its lack of a tyrosine kinase domain in its structure. Therefore, it operates through collaboration with an extensive collection of cytoplasmic proteins called focal adhesion molecules [
1,
2]. Focal adhesion molecules are highly dynamic structures that attach to and detach from integrin, connect actin and integrin, and transmit signals from integrin into the cell. To date, the roles of various focal adhesion molecules, including vinculin, paxillin, talin, α-actinin, zyxin, vasodilator-stimulated phosphoprotein (VASP), focal adhesion kinase (FAK), and p130Cas, and their association with integrin have been gradually elucidated. However, further research is required to elucidate their complicated mechanisms [
8,
9].
Proliferative and differentiation potential are critical factors in the clinical application of stem cells. Human palatine tonsil-derived mesenchymal stem cells (TMSCs) have a higher proliferation rate and differentiation potential than those of bone-marrow-derived MSCs (BMSCs) or adipose-tissue-derived MSCs (AMSC) [
10,
11,
12]. In our previous study, we reported that the expression of tensin3 (
TNS3) was significantly higher in TMSCs than in BMSCs or AMSCs and that TNS3 influenced the high proliferative capacity and differentiation potential of TMSC by regulating integrin beta-1 (ITG β1) activity [
13].
The TNS family comprises four isoforms of focal adhesion proteins involved in cell adhesion and migration by competitively binding to the cytoplasmic tail of ITG β1 against other integrin adaptor proteins [
14]. However, studies on TNSs are limited compared to those on other focal adhesion molecules such as talin and kindlin. We revealed the role of TNS3 in focal adhesion and its relationship with integrin in a previous study [
13]. However, the roles of the remaining three TNS isoforms have not been investigated. In this study, we comprehensively investigated the role of the TNS family of proteins in the focal adhesion of TMSCs.
2. Materials and Methods
2.1. Generation of Palatine TMSCs
The tonsils of four patients with chronic tonsillitis were collected during tonsillectomy after obtaining informed consent. Immediately after surgery, the tonsils were washed several times with phosphate-buffered saline (PBS). The tissue was digested in 0.075% collagenase type I (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C to isolate the stem cells. Enzyme activity was inhibited by incubating the cells in alpha-modified Eagle’s medium (α-MEM) containing 10% fetal bovine serum (FBS) for 30 min, and the sample was centrifuged at 1200× g for 10 min. The pellet was filtered through a 100 μm nylon mesh to remove cell debris, and cell adhesion was confirmed by incubation for 1–2 days at 37 °C in a 5% CO2 atmosphere in α-MEM-containing 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. The cells adhered stably to the plate, and the plate was washed with PBS to remove residual non-adherent cells. The resulting cell population was further maintained. All study protocols were reviewed and approved by the Institutional Review Board.
2.2. Quantitative Reverse Transcription Polymerase Chain Reaction and RNA Interference
mRNA expression was confirmed using quantitative PCR. Primers for
TNS1,
TNS2,
TNS3, and
TNS4 were obtained from Bio-Rad (Hercules, CA, USA). The primer sequences for p16, p19, p21, Cyclin E,
CDC25, Oct-4,
Nanog, Sox-2, osteocalcin,
ALP (alkaline phosphatase), peroxisome proliferator-activated receptor-gamma (
PPARγ), and lipoprotein lipase (
LPL) are shown in
Table 1. All primer sequences were selected according to the established PubMed GenBank sequence.
GAPDH was used as an internal control. Small interfering RNA (siRNA) oligonucleotide duplexes targeting
TNS1,
2,
3, and
4 mRNA were purchased from ON-TARGETplus SMARTpool (Dharmacon, Thermo Fisher Scientific, Waltham, MA, USA; a nontargeted oligonucleotide duplex was also used). Transfection was performed according to the instructions of the Dharma FECT Transfection Reagent. Real-time quantification was conducted on LightCycler (Roche, Basel, Switzerland) using a fluorescent SYBR Green I PCR mixture. PCR kinetics and quantitative data were analyzed using LightCycler version 3.3 software.
2.3. Western Blotting
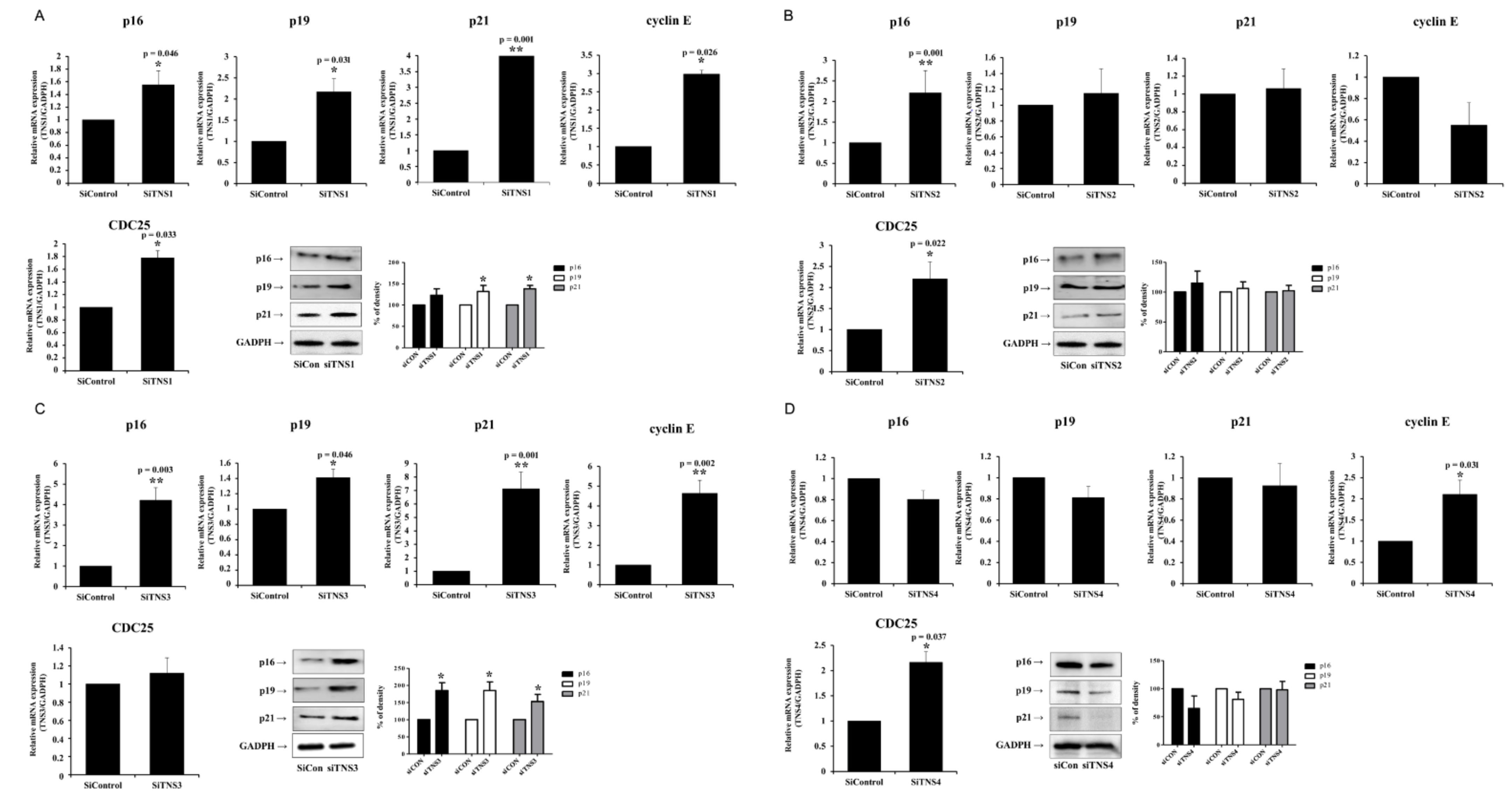
Protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific). Total protein equivalents for each sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 10% acrylamide gels; the proteins were transferred to polyvinylidene fluoride membranes. The membrane was immediately placed into a blocking buffer containing 5% bovine serum albumin for 1 h. The membrane was incubated with anti-p16, 19, 21, SOX-2, talin1, and kindlin 1 antibodies (1:2000; Santa Cruz Biotechnology, Dallas, TX, USA and Cell Signaling Technology, Danvers, MA, USA) at 4 °C overnight. Anti-GAPDH antibody (1:2000; Santa Cruz Biotechnology) was used as an internal control. After three 10 min washes, the membranes were incubated with anti-mouse and anti-rabbit peroxidase-conjugated secondary antibodies (1:10,000 dilution) for 1 h at 21 °C. Antibody labeling was detected using West-zol Plus and chemiluminescence FluorchemTMSP (Alpha Innotech Corporation, San Leandro, CA, USA).
2.4. Cell Proliferation
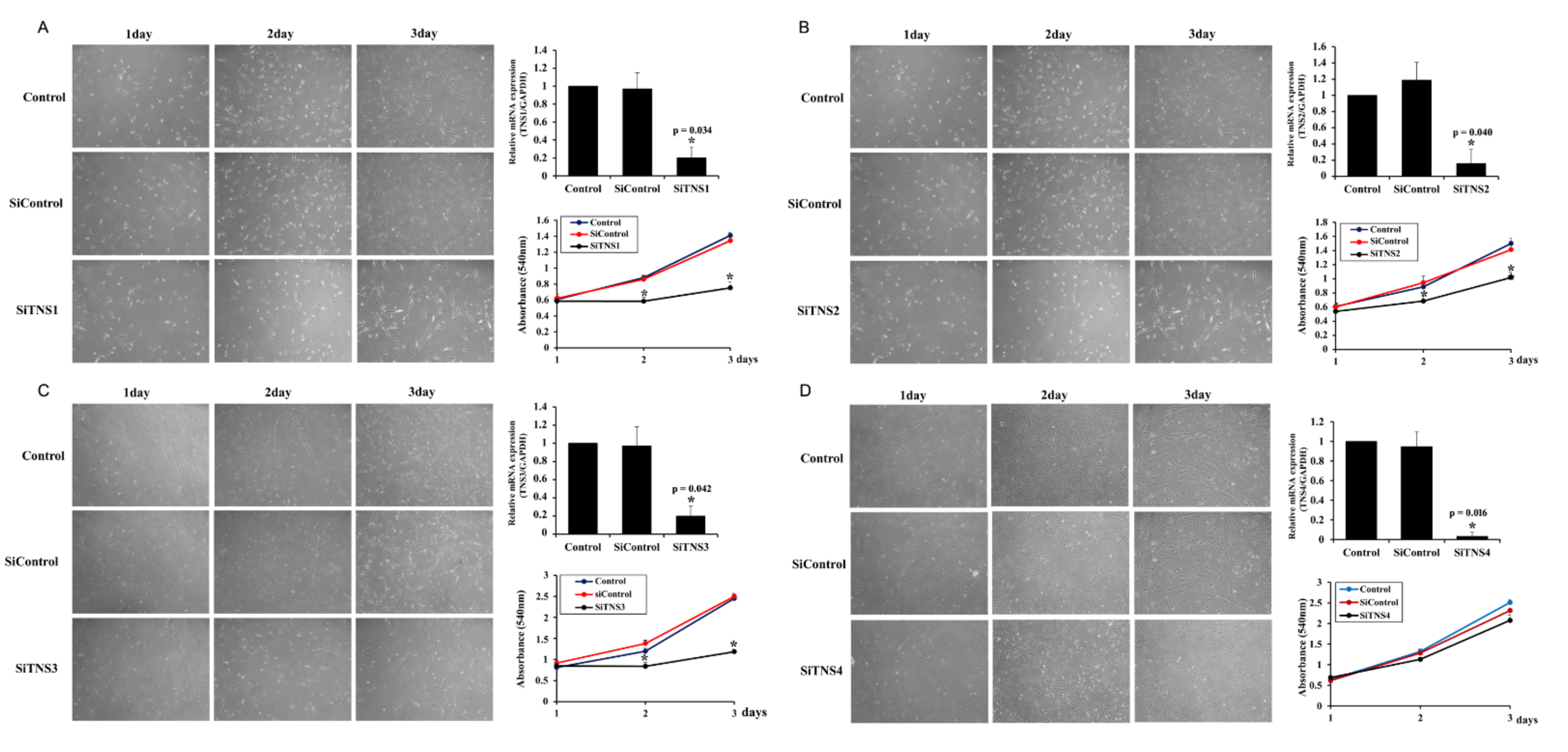
Cell proliferation was measured using the cell counting kit-8 (CCK8) assay. After culturing the cells in a medium containing CCK8 solution (Dojindo Laboratories, Kumamoto, Japan) at 37 °C for 1 h, absorbance at 450 nm was measured with a spectrophotometer. This experiment was repeated four times.
2.5. Cell-Cycle Analysis
For cell-cycle analysis, 6-well plates were seeded with 5 × 104 cells/well and treated with recombinant human fibroblast growth factor 5 (FGF5) at 37 °C for 48 h. The cells were harvested using 0.05% trypsin solution and centrifuged at 10,000× g for 15 min. The pellet was washed twice with Hank’s Balanced Salt Solution buffer and fixed in 70% ethanol overnight at −20 °C. The next day, the ethanol was removed, and the cells were resuspended in 500 mL of PBS containing 1 mg/mL propidium iodide and 100 µg RNase/mL; after incubation for 20 min, flow cytometry analysis was performed using a FACS Calibur (Becton Dickinson, San Jose, CA, USA).
2.6. Cell Migration Assay
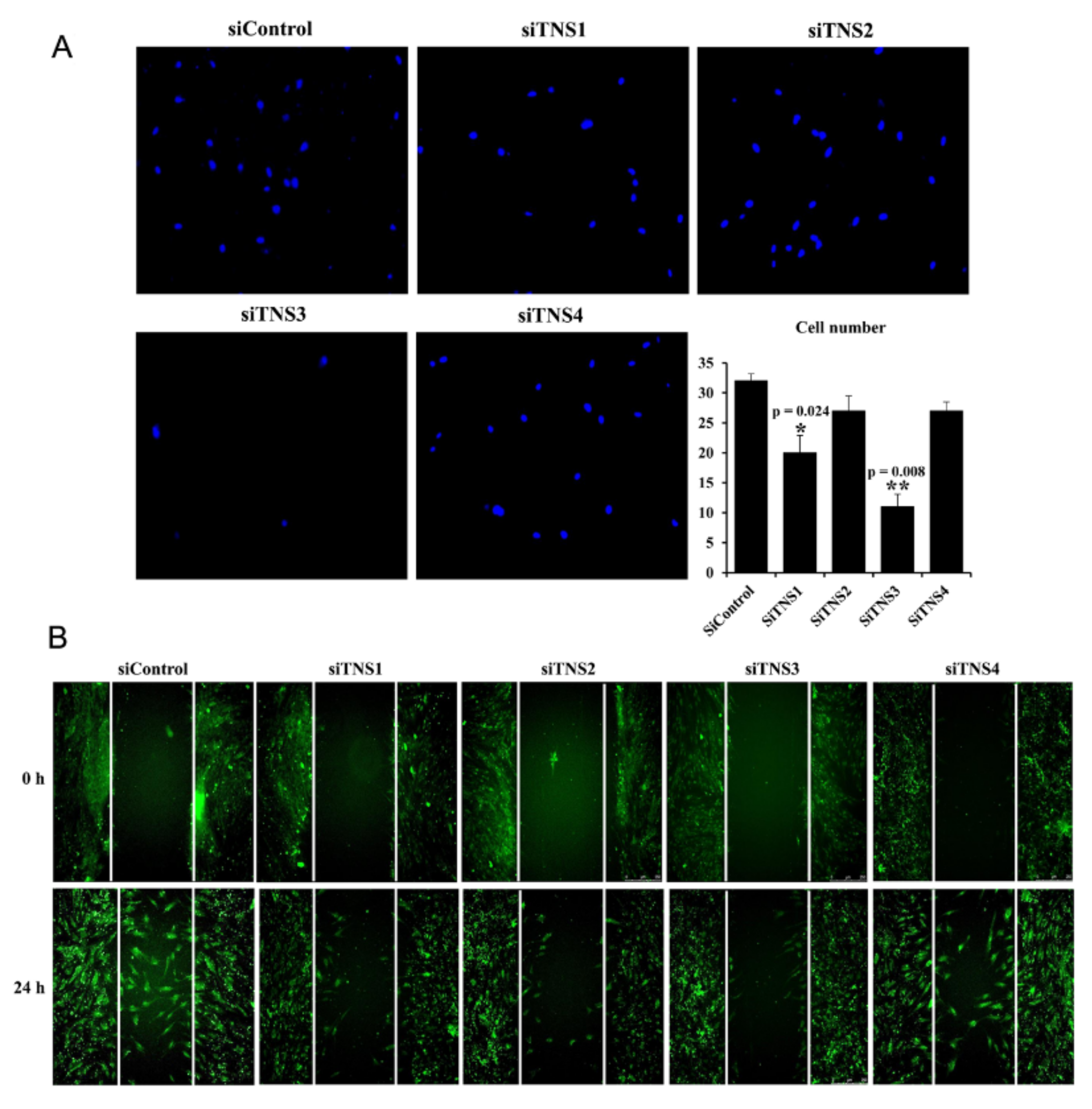
Cell migration was analyzed using a transwell chamber with a pore size of 8 μm. Cells were seeded into the upper chamber at 4 × 105/well, and IFN’- and TNF’-treated medium was dispensed into the lower chamber. The cells were incubated at 37 °C for 24 h to allow migration between the transwells by inflammatory cytokines. Cells that migrated and adhered to the lower chamber were washed, fixed with 10% formaldehyde, and stained with DAPI to measure cell migration. Stained cells were counted using a fluorescence microscope (Leica Microsystems, Wetzlar, Germany).
2.7. Adipogenic and Osteogenic Differentiation
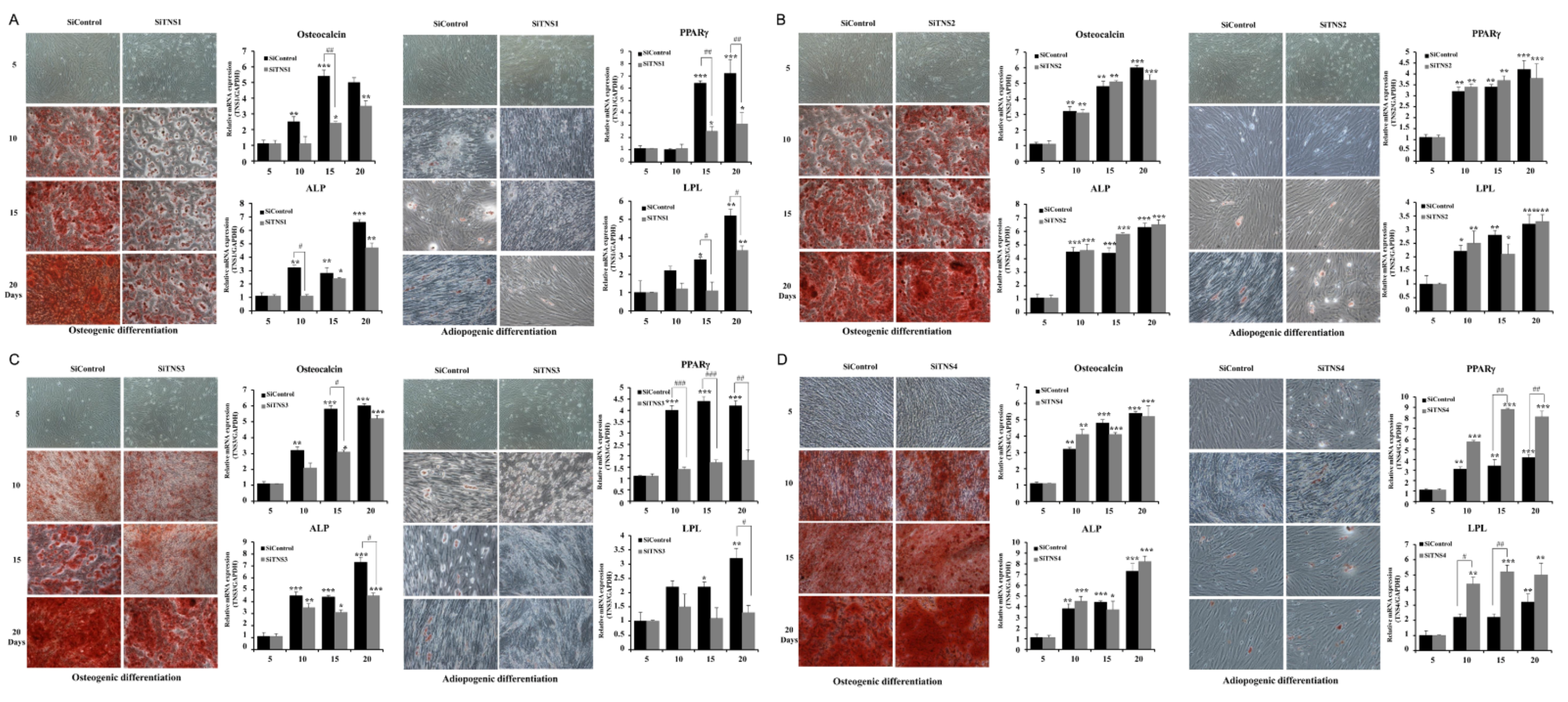
To confirm that the TMSC cells separated from the tonic chamber were stem cells, we examined their ability to differentiate into bone formation and adipogenesis systems. TMSCs were cultured in medium containing lipogenesis-induced reagents (10% FBS, and 1 μM dexamethasone (Sigma-Aldrich, St. Louis, MO, USA) α-MEM, 100 μg/mL 3-isobutyl-L-methylxanthine, 5 μg/mL insulin, and 60 μM) to induce lipocyte differentiation. Oil Red O (Sigma-Aldrich) staining was performed after treatment with indomethacin for three weeks to evaluate lipid accumulation in cells. The cells were fixed to 70% ethanol at room temperature for 15 min and then treated with 2% Oil RedO reagent at room temperature for 1 h. To visualize lipid droplets, the cells were washed with 70% ethanol, and the excess stains were removed through washing with distilled water. In addition, TMSCs were cultured in a medium treated with osteogenesis-induced reagents (10% FBS, 0.1 mM dexamethasone, 10 μM glycerophosphate, and α-MEM supplemented with 50 μg/mL ascorbic acid) to induce osteocyte differentiation. After fixing the cells in 70% ethanol and washing with distilled water, the cells were incubated at room temperature for 15 min in 2% Alizarin Red S solution and then washed several times with distilled water.
2.8. Flow Cytometry
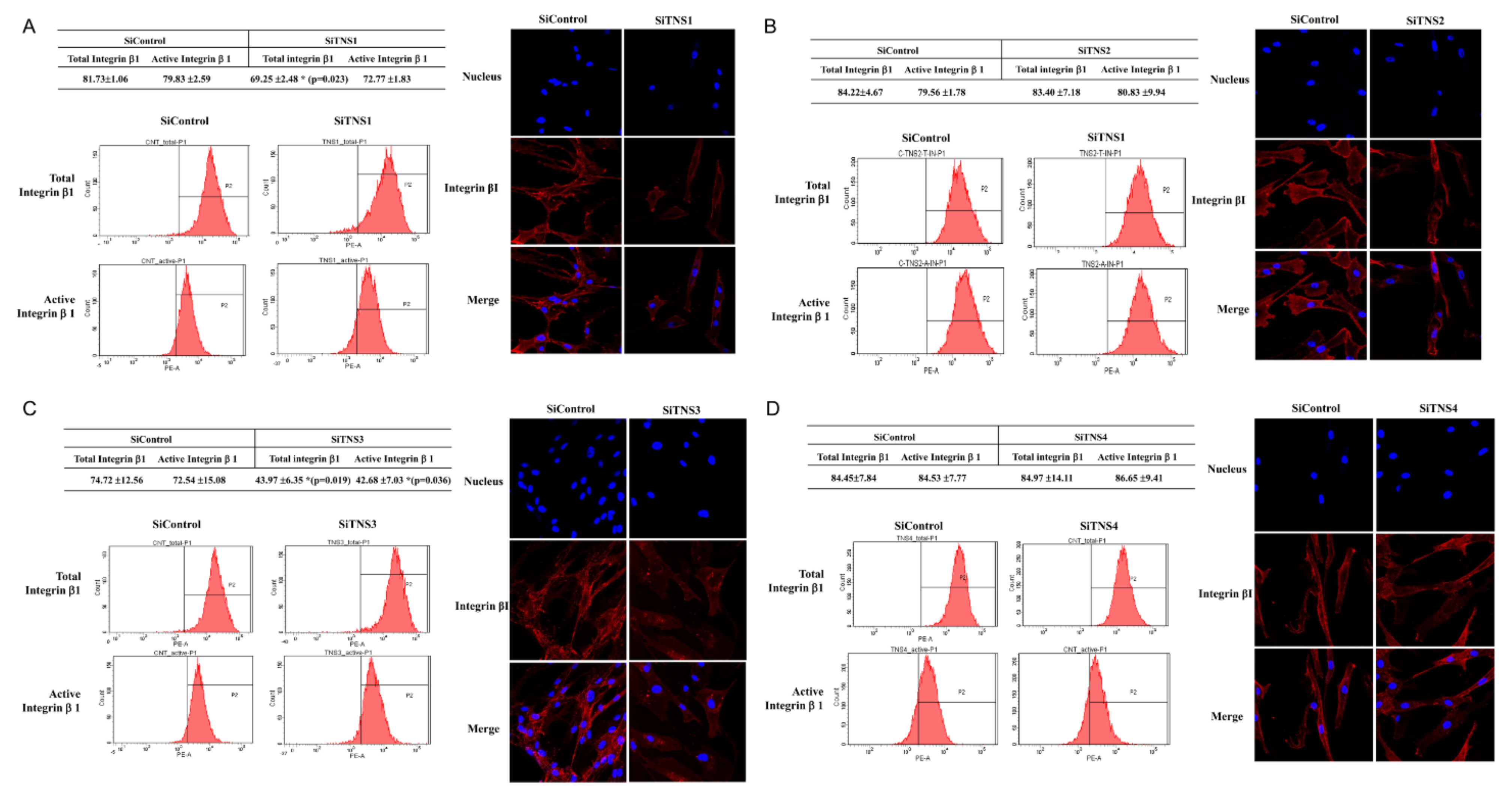
We used flow cytometric analysis to measure the overall and active cell surface ITG11 levels of TMSCs. The cells (5 × 104) were incubated with phycoerythrin-conjugated monoclonal antibodies against human guns and active ITG11 in 100 μL of PBS containing 0.5% bovine serum albumin and 2 mmol/LEDTA. The labeled cells were analyzed by cell analysis using a FACS Caliber leucocyte analyzer equipped with CellQuest Pro software (BD Biosciences).
2.9. Immunocytochemical Staining
The cells were fixed in 4% paraformaldehyde for 10 min, followed by incubation in 0.5% Tween 20 for 10 min to ensure permeability. The cells were incubated in a solution containing 1% bovine serum albumin for 1 h at room temperature, and then stained with purified rabbit anti-human ITG β11 at 4 °C for 2 h, followed by incubation with phycoerythrin-conjugated chlorine anti-rabbit IgG (e-Bioscience, San Diego, CA, USA) and FITC-conjugated anti-mouse IgG (e-Bioscience) for 1 h at 21 °C. The cells were stained with 4′,6-diamidino-2-phenylindole in VECTASHIELD (Vector Laboratories, Burlingame, CA, USA) mounting medium and examined using a confocal laser scanning microscope.
2.10. Statistical Analysis
The data are shown as the mean ± standard deviations for all experiments. One-way analysis of variance (SPSS version 18.0 software, SPSS, Inc., Chicago, IL, USA) and Scheffé’s tests were used to detect significant differences between the groups. A p-value < 0.05 was considered to indicate statistically significant results.
4. Discussion
The role of focal adhesion molecules, such as talin and kindlin, and their relationship with integrins in stem cells have been actively studied in recent years [
2,
9,
15]. However, relatively little is known about the role of TNS in focal adhesions. Here, we demonstrated that the TNS family regulates biological processes, including proliferation and differentiation of TMSCs, through integrins at focal adhesions. The two most critical factors for the clinical value of stem cells are self-renewal and pluripotency. We previously showed that TMSCs are significantly superior in proliferation and differentiation potential compared to conventional AMSCs or BMSCs [
11,
13]. In addition, we demonstrated significantly higher TNS3 in TMSCs than in AMSCs and BMSCs, and that TNS3 affects the proliferation and differentiation of TMSCs [
13].
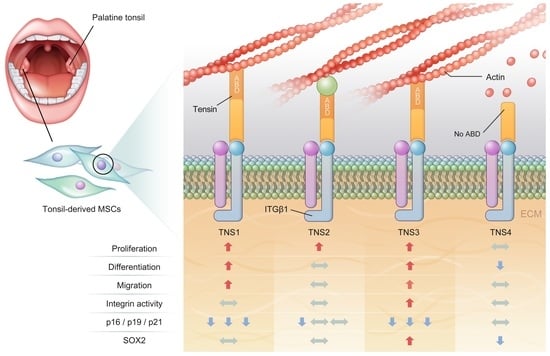
This study investigated the effect of the entire TNS family, including TNS1, 2, 3, and 4, on TMSC biology. TNS1, 2, and 3 showed similar results, but TNS4 showed varying results. When TNS1, 2, and 3 were inhibited, TMSC proliferation was significantly reduced. However, TNS4 did not have a discernable effect on proliferation. Similarly, no significant change was observed in the cell cycle or pluripotency with TNS4 inhibition. Unlike TNS1 and 3, which affected ITGβ1 activity, TNS4 did not alter ITGβ1 activity. The suppression of TNS1 and 3 decreased osteogenic and adipogenic differentiation, whereas the inhibition of TNS4 increased adipogenic differentiation. We investigated the changes in Wnt signaling to reveal the pathway by which TNS4 is involved in adipogenesis. However, we did not observe any change in the expression and nuclear translation of β-catenin. In siTNS4, Akt phosphorylation remained unaltered, whereas ERK phosphorylation and AMPK were decreased, which are thought to increase adipogenic differentiation.
TNS plays a role in transferring structural and chemical changes in cells through integrin to the inside and outside of cells during focal adhesions. To perform this role, it has a multi-domain protein that can bind to various signaling molecules present in focal adhesion, as well as the cytoplasmic tail of ITGβ1 [
14]. TNS is structurally divided into three regions. The N-terminus contains an actin-binding domain (ABD) that interacts with actin filaments. The opposite C-terminus contains the Src homology 2 (SH2) domain that can bind to tyrosine-phosphorylated proteins such as FAK, PI3 kinase, MET, and p130Cas [
16]. C-terminus also has a phosphotyrosine-binding (PTB) domain, where it binds to the cytoplasmic tail of ITGβ1 [
17]. In addition, focal adhesion-binding (FAB) sites are present in both the N- and C-terminal regions. Except for the protein kinase C (C1) domain of TNS2, the structures of the C-terminus and N-terminus of TNS1, 2, and 3 are almost identical [
18,
19]. However, TNS4, also called a COOH-terminus TNS-like molecule (CTEN), does not have an N-terminal region, unlike the other three TNSs. Therefore, TNS4 does not bind to actin due to the absence of ABD, and it plays a functionally different role from that of TNS1, 2, and 3 because of its conformational specificity [
20,
21]. TNS1, 2, and 3 play a role in the maturation of focal adhesions and progression to fibrillar adhesions by stably linking actin and integrin [
16,
22,
23]. On the other hand, TNS4 are known to promote cell migration by uncoupling between integrin and actin cytoskeleton. TNS4 may also have a different relationship with the tyrosine-phosphorylated proteins, which interact through the SH2 domain, unlike other TNS families. TNS4 directly interacts with MET through the SH2 domain and regulates MET-dependent cell migration. The downregulation of TNS4 reduces the level of cell-surface expression of MET and affects MET downstream signaling, but the silencing of TNS3 did not significantly affect MET [
20].
TNS3 had the most significant influence on most biological processes, including the proliferation and differentiation of TMSCs. TNS3 treatment also led to the most substantial change in ITGβ1 activity. TNS1 exhibited results similar to those of TNS3 in almost all experiments. However, the range of the change was smaller than that of TNS3. TNS2 did not affect migration or differentiation, except for TMSC proliferation. The N- and C-terminal regions of TNS1, 2, and 3 were almost identical, with differences observed in the middle region [
14]. Further investigation is required to ascertain whether the different roles of TNS1, 2, and 3 are due to the divergence in this middle region or the high expression rate of TNS3 in TMSCs.
Various biological functions of TMSCs are likely achieved through the cooperation among TNS families with a similar structure but play slightly different roles in focal adhesion. For example, TNS3 plays a vital role in focal adhesion, and a stable integrin–TNS–actin complex is formed through the ABD and PTB domains on both sides of TNS3. However, due to the downregulation of TNS3 and upregulation of TNS4 caused by external stimuli, such as the epidermal growth factor, TNS3 is replaced with TNS4 without ABD. Consequently, the complex with actin stress fiber, which maintains the cell shape, is disrupted, and cells begin to migrate. This “reciprocal TNS3-TNS4 switch” is thought to play a critical role in various biological processes, including cell migration at focal adhesions [
21,
24].
This study had several limitations. First, only the effects of TNS on integrin and focal adhesion kinases were investigated, and the reciprocal regulation loop between TNS and tyrosine-phosphorylated proteins was not investigated. There we could not determine how TNS4 exhibits different actions from other TNSs. Furthermore, to examine the effect of TNS on the biological process of TMSCs, only the inhibition by siRNA was investigated, and overexpression-mediated changes were not estimated. Nonetheless, we comprehensively investigated the effects of four TNSs on the performance of TMSC functions, including proliferation, cell cycle regulation, maintenance of pluripotency, and differentiation for the first time. We further established that TNS3 regulates integrin activity, whereas TNS4 does not. In particular, we revealed that TNS3 and TNS4 have opposing actions during adipogenic and osterogenic differentiation. In order to more precisely control the differentiation of TMSCs, further studies on the mechanisms of action of TNS3 and TNS4 in focal adhesion during differentiation will be conducted, and we expect that this study will serve as a foundation for future therapeutic applications of TMSC.