VMP1 Regulated by chi-miR-124a Effects Goat Myoblast Proliferation, Autophagy, and Apoptosis through the PI3K/ULK1/mTOR Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Sample Collection
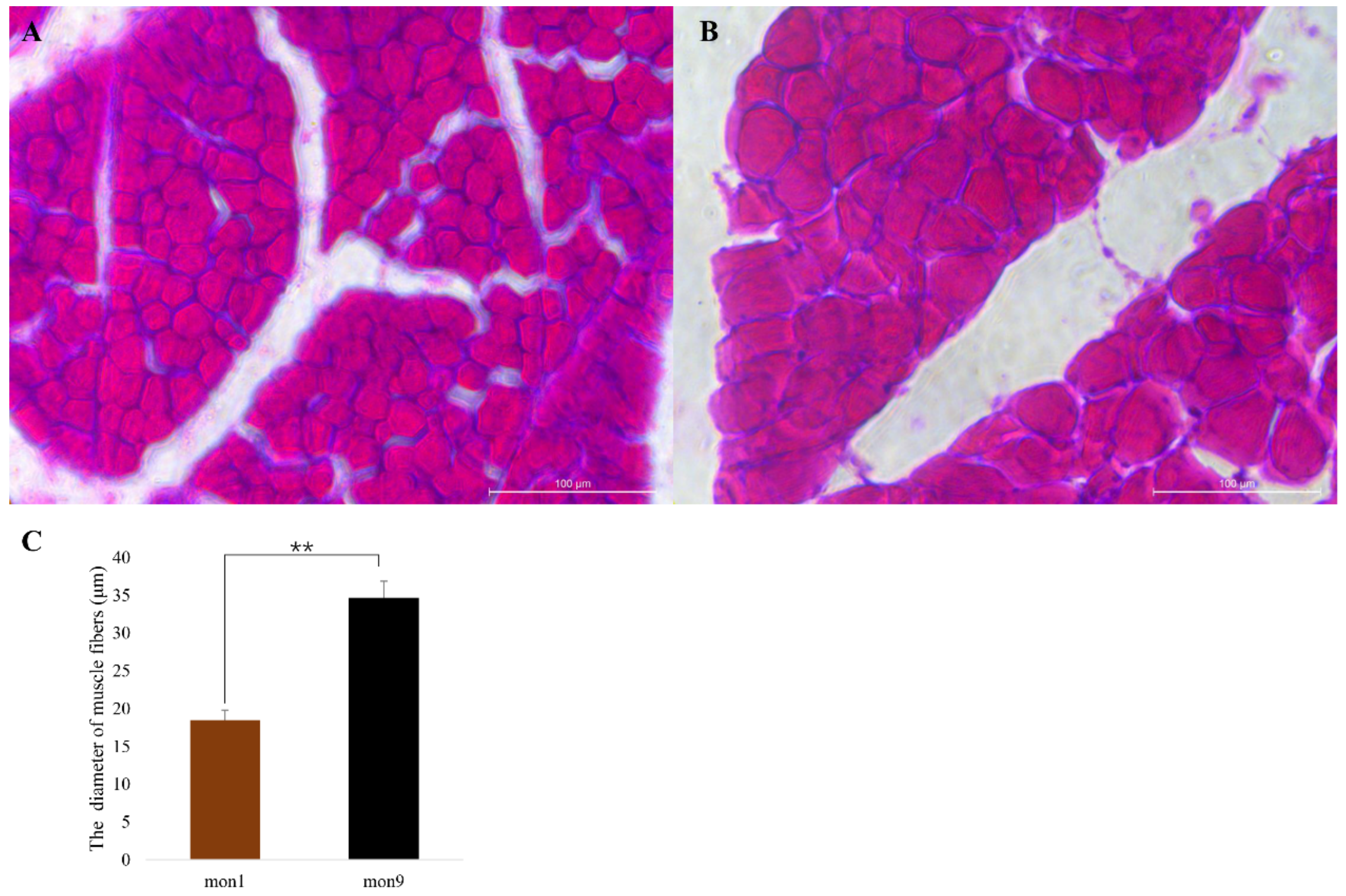
2.3. Hematoxylin–Eosin Staining
2.4. RNA Extraction, Library Construction, and Differential Expression Analysis
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses
2.6. Reverse-Transcription (RT)-qPCR Verification
2.7. miRNA Prediction for VMP1 Targeting
2.8. Cell Culture
2.9. Plasmid Construction and Transfection
2.10. Dual-Luciferase Reporter Assay
2.11. Western Blot Analysis
2.12. Cell Proliferation Assay
2.13. Ethynyldeoxyuridine Incorporation Assay
2.14. Cell Apoptosis Analysis
2.15. PI3K/ULK1/mTOR Pathway Analysis
2.16. Confocal Microscopy
2.17. Statistical Analysis
3. Results
3.1. RNA-Seq Analysis
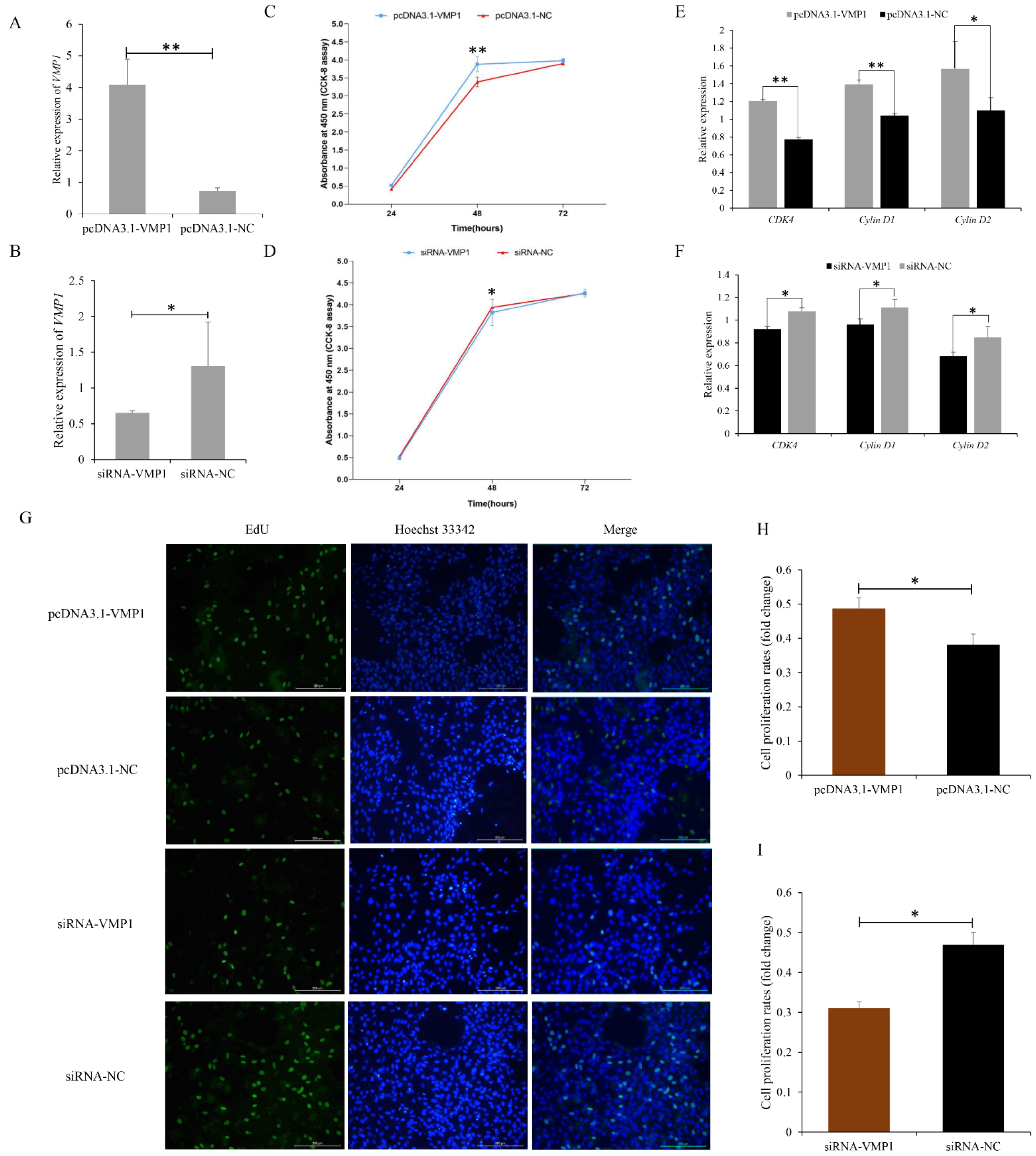
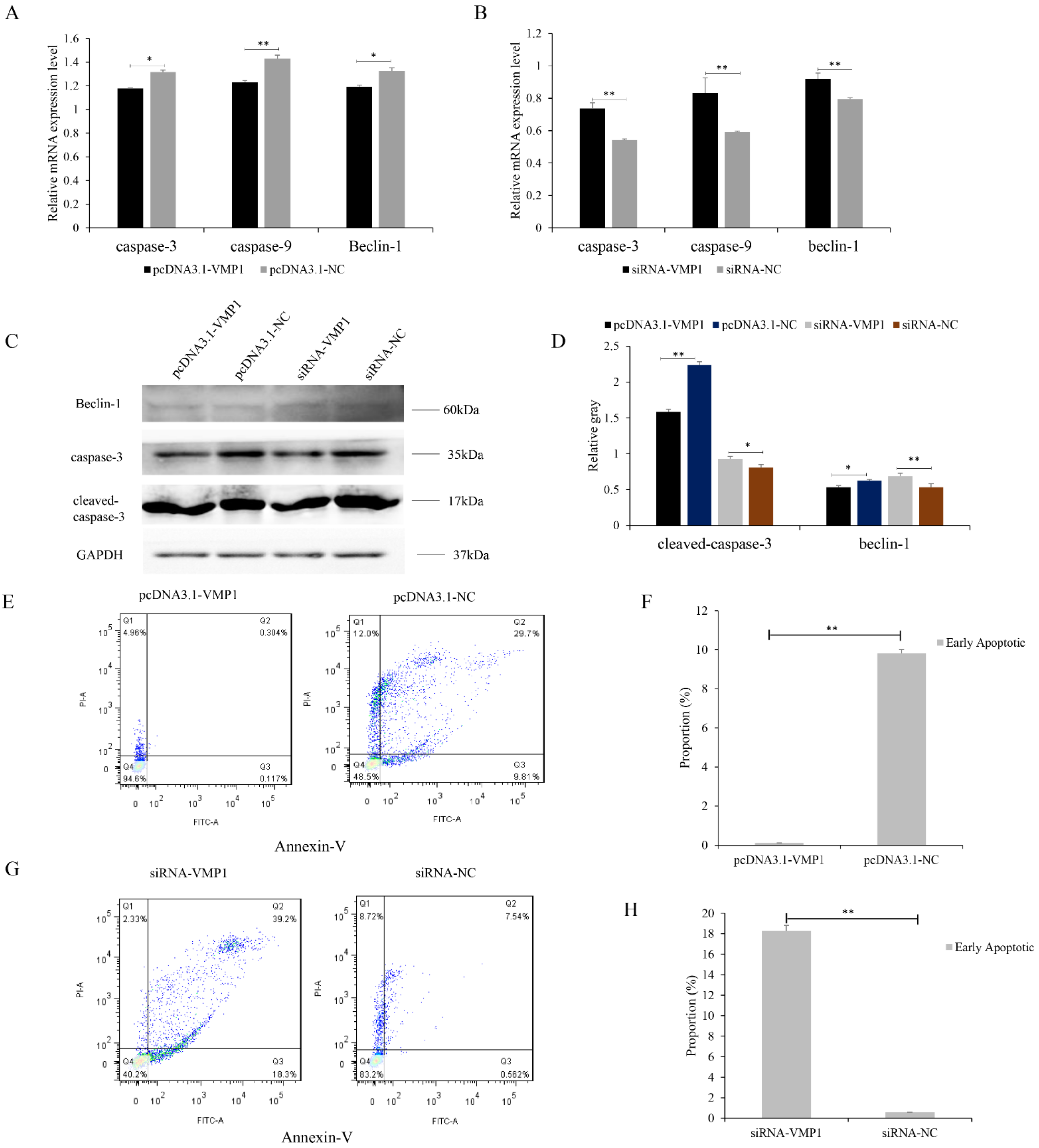
3.2. Function of VMP1 in Cell Proliferation and Apoptosis of Goat Myoblasts
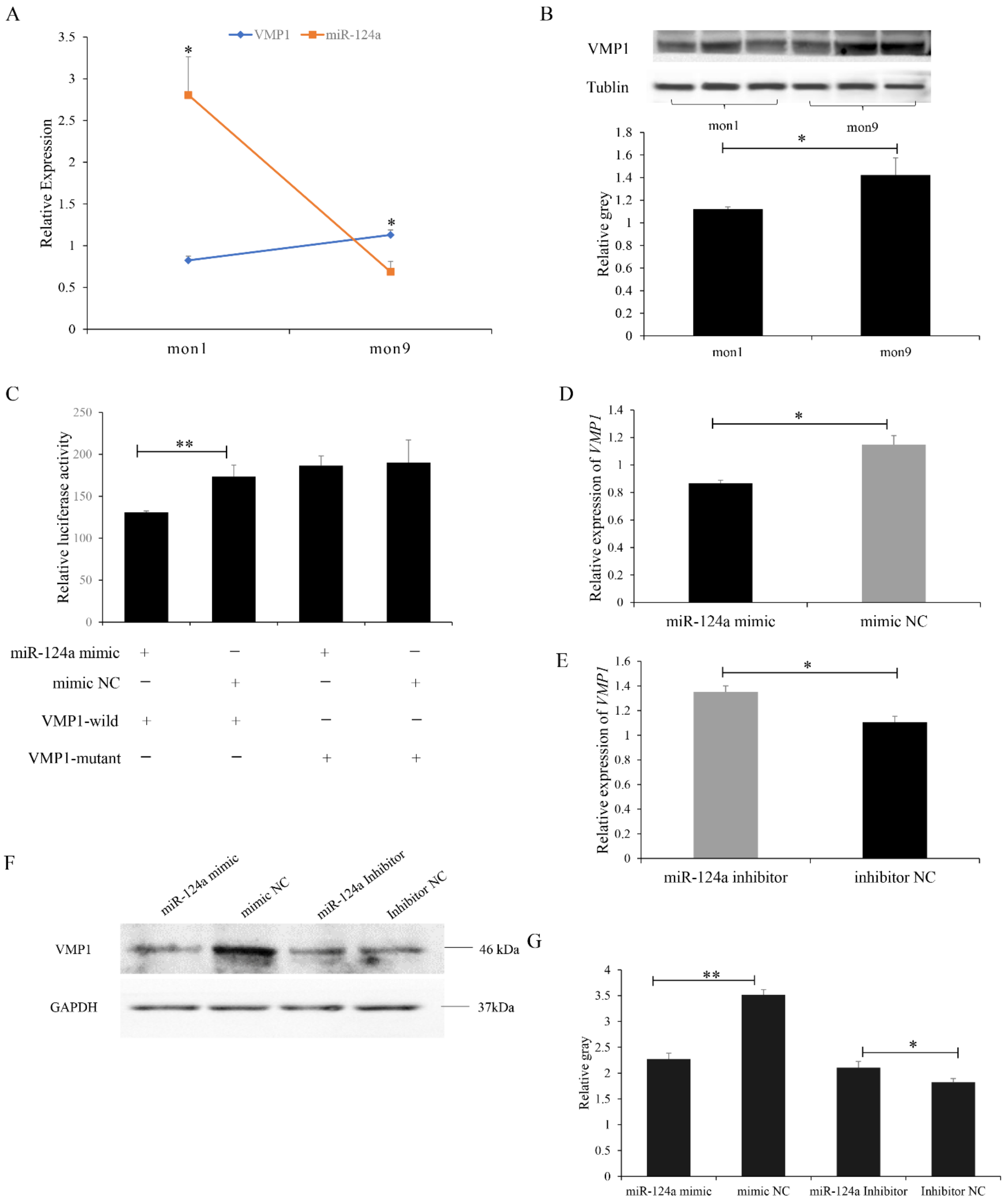
3.3. miR-124a Targets VMP1
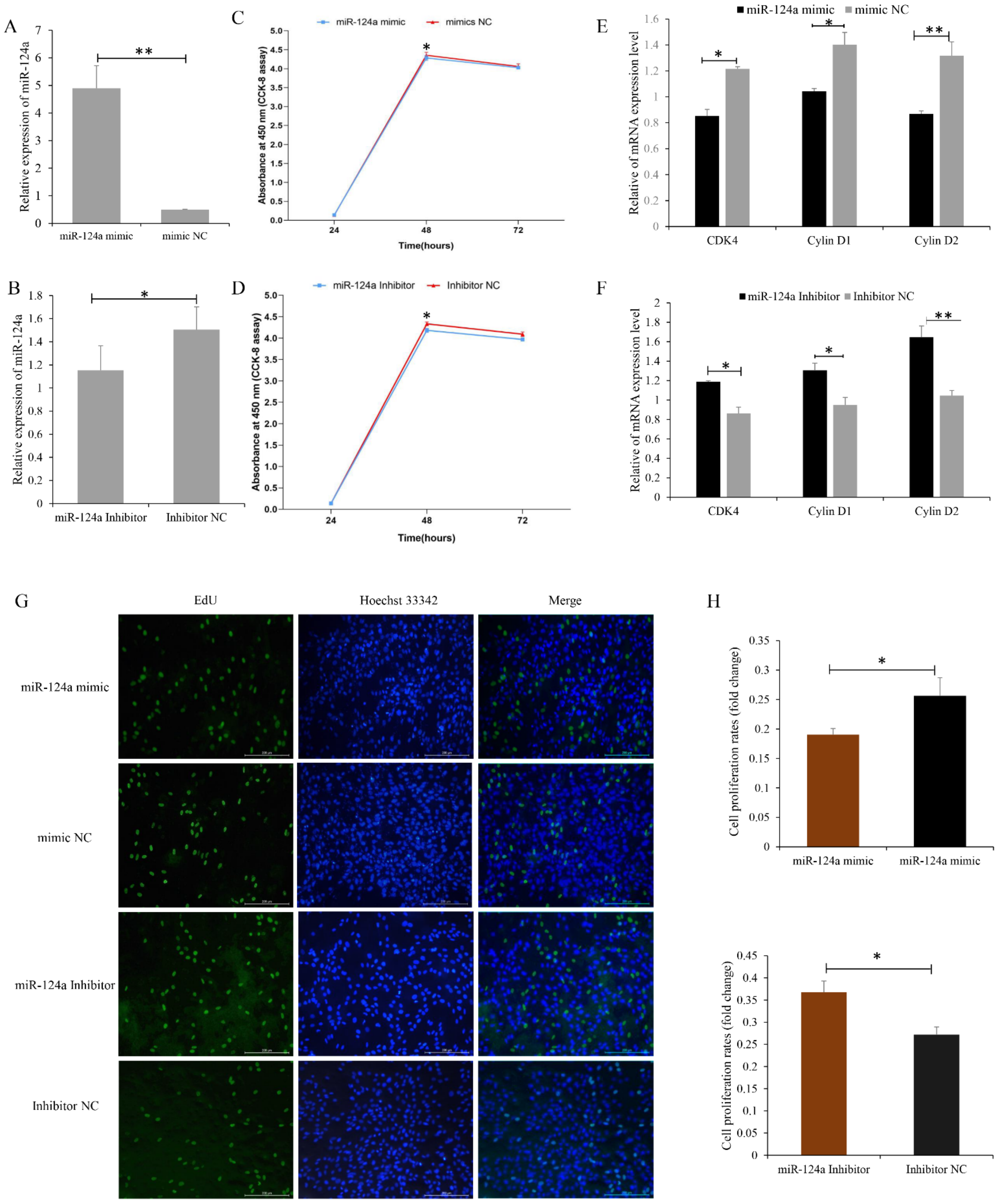
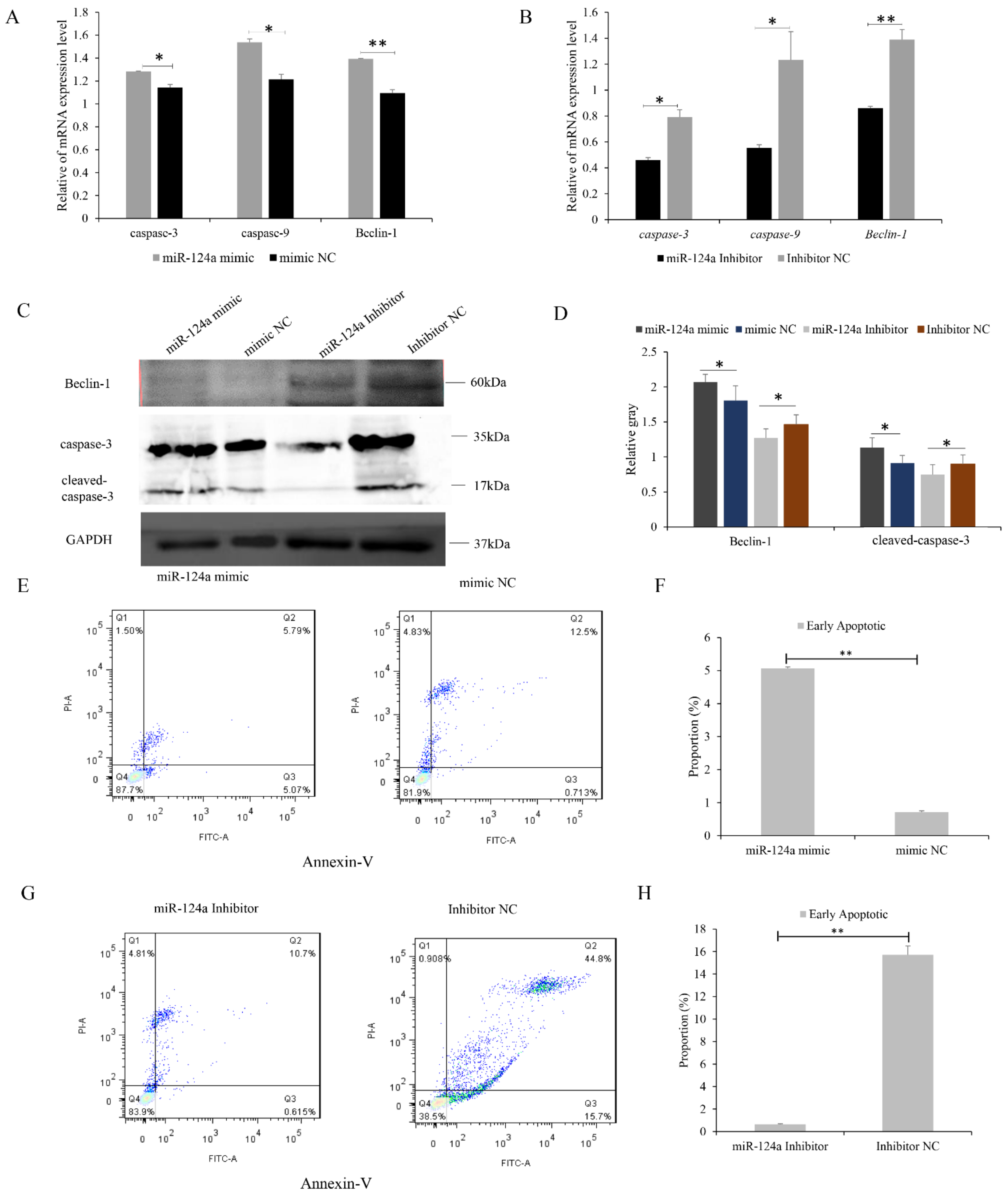
3.4. miR-124a Inhibits Proliferation of, and Induces Cell Apoptosis in, Goat Myoblasts
3.5. VMP1 Promotes Goat Myoblast Proliferation via the PI3K/Akt/mTOR Pathway
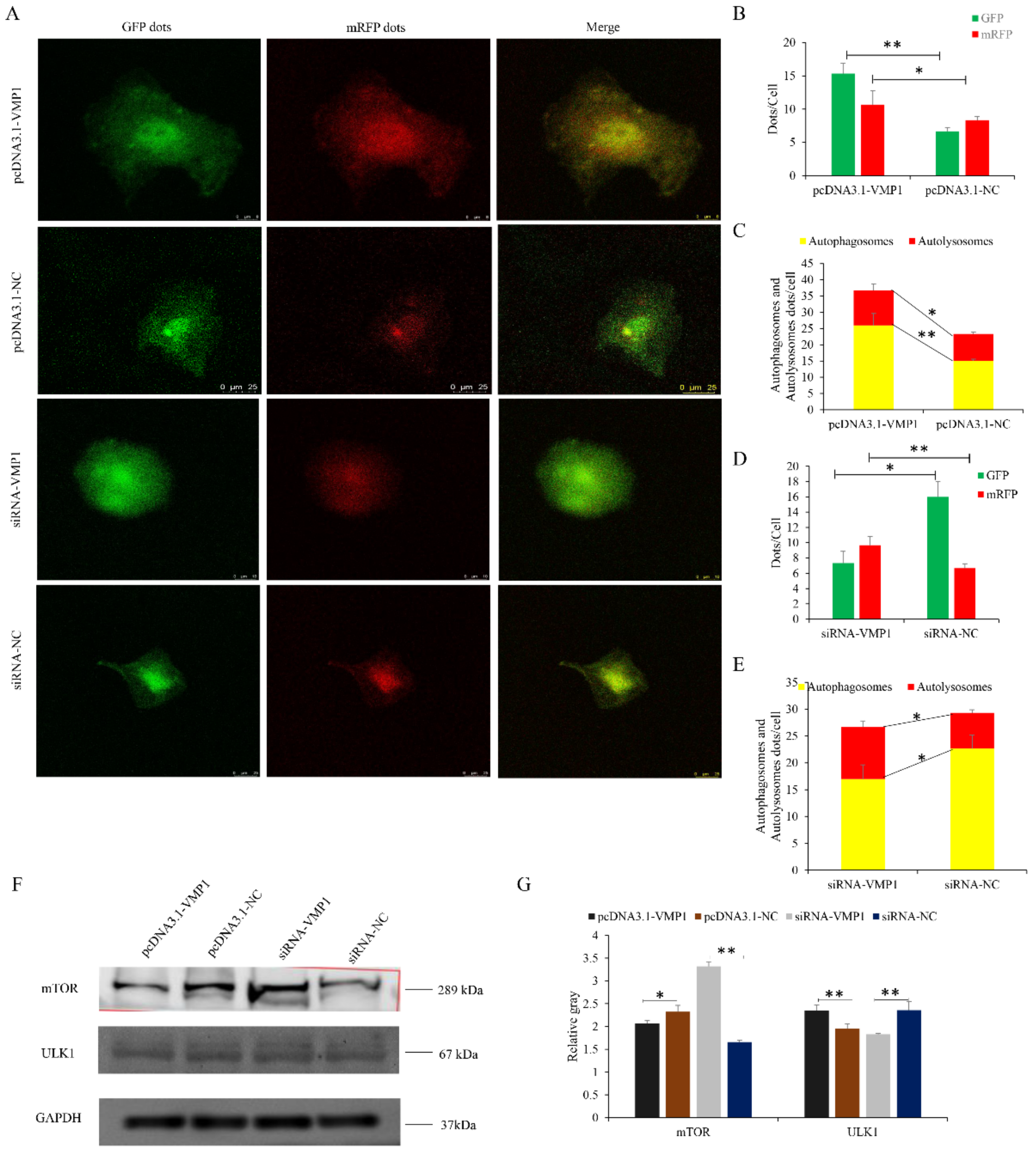
3.6. VMP1 Promotes Myoblast Autophagy
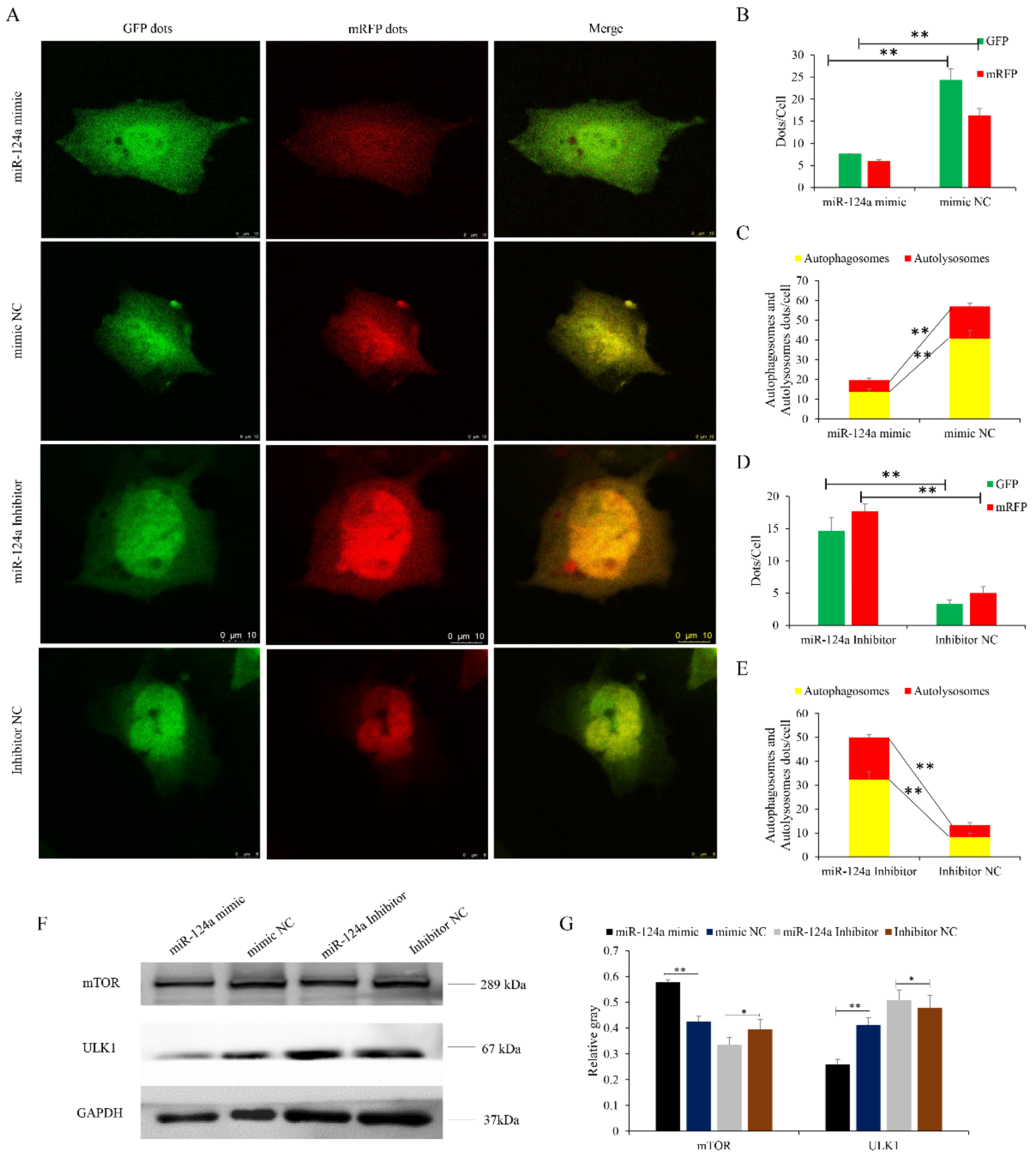
3.7. miR-124a Regulates Myoblast Autophagy by Targeting VMP1
3.8. miR-124a Impedes the VMP1/ULK1/mTOR Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deans, C.; Wigmore, S.J. Systemic inflammation, cachexia, and prognosis in patients with cancer. Curr. Opin. Clin. Nutr. Metab. Care 2005, 8, 265–269. [Google Scholar] [CrossRef]
- Teixeira, A.; Silva, S.; Rodrigues, S. Advances in sheep and goat meat products research. Adv. Food Nutr. Res. 2019, 87, 305–370. [Google Scholar]
- Shen, J.; Hao, Z.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Ke, N.; Song, Y.; Lu, Y.; Hu, L.; et al. Comparative transcriptome profile analysis of longissimus dorsi muscle tissues from two goat breeds with different meat production performance using RNA-Seq. Front. Genet. 2021, 11, 619399. [Google Scholar] [CrossRef]
- Hawke, T.J.; Garry, D.J. Myogenic satellite cells: Physiology to molecular biology. J. Appl. Physiol. 2001, 91, 534–551. [Google Scholar] [CrossRef]
- Merrell, A.J.; Kardon, G. Development of the diaphragm–A skeletal muscle essential for mammalian respiration. FEBS J. 2013, 280, 4026–4035. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.; Kardon, G. Origin of vertebrate limb muscle: The role of progenitor and myoblast populations. Curr. Top Dev. Biol. 2011, 96, 1–32. [Google Scholar]
- Noden, D.M.; Francis-West, P. The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 2006, 235, 1194–1218. [Google Scholar] [CrossRef]
- McNally, E.M.; Pytel, P. Muscle diseases: The muscular dystrophies. Annu. Rev. Pathol. 2007, 2, 87–109. [Google Scholar] [CrossRef] [Green Version]
- Hughes, S.M.; Blau, H.M. Migration of myoblasts across basal lamina during skeletal muscle development. Nature 1990, 345, 350–353. [Google Scholar] [CrossRef]
- Ovilo, C.; Benítez, R.; Fernández, A.; Núñez, Y.; Ayuso, M.; Fernández, A.I.; Rodríguez, C.; Isabel, B.; Rey, A.I.; López-Bote, C.; et al. Longissimus dorsi transcriptome analysis of purebred and crossbred Iberian pigs differing in muscle characteristics. BMC Genom. 2014, 15, 413. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wang, C.; Jin, E.; Gu, Y.; Li, S.; Li, Q. Identification of differentially expressed genes in longissimus dorsi muscle between Wei and Yorkshire pigs using RNA sequencing. Genes Genom. 2018, 40, 413–421. [Google Scholar] [CrossRef]
- Silva-Vignato, B.; Coutinho, L.L.; Cesar, A.S.M.; Poleti, M.D.; Regitano, L.C.A.; Balieiro, J.C.C. Comparative muscle transcriptome associated with carcass traits of Nellore cattle. BMC Genom. 2017, 18, 506. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.M.; Xia, H.L.; Jiang, H.R.; Mao, Y.J.; Qu, K.X.; Huang, B.Z.; Gong, Y.C.; Yang, Z.P. Longissimus dorsi muscle transcriptomic analysis of Yunling and Chinese simmental cattle differing in intramuscular fat content and fatty acid composition. Genome 2018, 61, 549–558. [Google Scholar] [CrossRef]
- Sun, L.; Bai, M.; Xiang, L.; Zhang, G.; Ma, W.; Jiang, H. Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep. Sci. Rep. 2016, 6, 33586. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Wang, X.; Zhang, Q.; He, Y.; Zhang, X.; Yang, L.; Shi, J. Comparative transcriptome analysis identifying the different molecular genetic markers related to production performance and meat quality in longissimus dorsi tissues of MG × STH and STH sheep. Genes 2020, 11, 183. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Zhang, C.L.; Plath, M.; Fang, X.T.; Lan, X.Y.; Zhou, Y.; Chen, H. Global transcriptional profiling of longissimus thoracis muscle tissue in fetal and juvenile domestic goat using RNA sequencing. Anim. Genet. 2015, 46, 655–665. [Google Scholar] [CrossRef]
- Lin, Y.; Zhu, J.; Wang, Y.; Li, Q.; Lin, S. Identification of differentially expressed genes through RNA sequencing in goats (Capra hircus) at different postnatal stages. PLoS ONE 2017, 12, e0182602. [Google Scholar] [CrossRef] [Green Version]
- Zhong, T.; Jin, P.F.; Dong, E.N.; Li, L.; Wang, L.J.; Zhang, H.P. Caprine sex affects skeletal muscle profile and MRFs expression during postnatal development. Anim. Sci. J. 2013, 84, 442–448. [Google Scholar] [CrossRef]
- Zhan, S.Y.; Chen, L.; Li, L.; Wang, L.J.; Zhong, T.; Zhang, H.P. Molecular characterization and expression patterns of insulin-like growth factor-binding protein genes in postnatal Nanjiang brown goats. Genet. Mol. Res. 2015, 14, 12547–12560. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Yang, W.; Cui, M.; Dai, B.; Dong, Y.; Yang, J.; Zhang, X.; Liu, D.; Liang, H.; et al. Comparison of gene editing efficiencies of CRISPR/Cas9 and TALEN for generation of MSTN knock-out cashmere goats. Theriogenology 2019, 132, 1–11. [Google Scholar] [CrossRef]
- Xia, Q.; Huang, X.; Huang, J.; Zheng, Y.; March, M.E.; Li, J.; Wei, Y. The role of autophagy in skeletal muscle diseases. Front Physiol. 2021, 12, 638983. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.G.; Chen, Y.; Miao, G.; Zhao, H.; Qu, W.; Li, D.; Wang, Z.; Liu, N.; Li, L.; Chen, S.; et al. The ER-Localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 2017, 67, 974–989.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, K.; Hama, Y.; Izume, T.; Tamura, N.; Ueno, T.; Yamashita, Y.; Sakamaki, Y.; Mimura, K.; Morishita, H.; Shihoya, W.; et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 2018, 217, 3817–3828. [Google Scholar] [CrossRef] [Green Version]
- Morita, K.; Hama, Y.; Mizushima, N. TMEM41B functions with VMP1 in autophagosome formation. Autophagy 2019, 15, 922–923. [Google Scholar] [CrossRef]
- Ropolo, A.; Grasso, D.; Pardo, R.; Sacchetti, M.L.; Archange, C.; Lo Re, A.; Seux, M.; Nowak, J.; Gonzalez, C.D.; Iovanna, J.L.; et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 2007, 282, 37124–37133. [Google Scholar] [CrossRef] [Green Version]
- Guardiola, O.; Andolfi, G.; Tirone, M.; Iavarone, F.; Brunelli, S.; Minchiotti, G. Induction of acute skeletal muscle regeneration by cardiotoxin injection. J. Vis. Exp. 2017, 119, 54515. [Google Scholar] [CrossRef]
- Jia, B.Y.; Ba, H.X.; Wang, G.W.; Yang, Y.; Cui, X.Z.; Peng, Y.H.; Zheng, J.J.; Xing, X.M.; Yang, F.H. Transcriptome analysis of sika deer in China. Mol. Genet. Genom. 2016, 291, 1941–1953. [Google Scholar] [CrossRef]
- Law, C.W.; Alhamdoosh, M.; Su, S.A.; Dong, X.Y.; Tian, L.Y.; Smyth, G.K.; Ritchie, M.E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. Version 3. F1000Research 2016, 5, ISCB Comm J-1408. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Tibshirani, R. Statistical significance for genome wide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.W.; Bai, Y.; Zhang, Q.Y.; Zhang, H.; Fan, Y.K.; Han, H.Y.; Liu, Y.F. Associations of CALM1 and DRD1 polymorphisms, and their expression levels, with Taihang chicken egg-production traits. Anim. Biotechnol. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fauconneau, B.; Paboeuf, G. Effect of fasting and refeeding on in vitro muscle cell proliferation in rainbow trout (Oncorhynchus mykiss). Cell Tissue Res. 2000, 301, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Duran, B.O.S.; Góes, G.A.; Zanella, B.T.T.; Freire, P.P.; Valente, J.S.; Salomão, R.A.S.; Fernandes, A.; Mareco, E.A.; Carvalho, R.F.; Dal-Pai-Silva, M. Ascorbic acid stimulates the in vitro myoblast proliferation and migration of pacu (Piaractus mesopotamicus). Sci. Rep. 2019, 9, 2229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Chen, K.; Tang, Y.X.; Luan, X.R.; Zheng, X.X.; Lu, X.M.; Mao, J.Y.; Hu, L.Q.; Zhang, S.F.; Zhang, X.N.; et al. LncRNA-HOTAIR activates autophagy and promotes the imatinib resistance of gastrointestinal stromal tumor cells through a mechanism involving the miR-130a/ATG2B pathway. Cell Death Dis. 2021, 12, 367. [Google Scholar] [CrossRef]
- Alarcin, E.; Bal-Öztürk, A.; Avci, H.; Ghorbanpoor, H.; Dogan Guzel, F.; Akpek, A.; Yesiltas, G.; Canak-Ipek, T.; Avci-Adali, M. Current strategies for the regeneration of skeletal muscle tissue. Int. J. Mol. Sci. 2021, 22, 5929. [Google Scholar] [CrossRef]
- Folkerts, H.; Wierenga, A.T.; van den Heuvel, F.A.; Woldhuis, R.R.; Kluit, D.S.; Jaques, J.; Schuringa, J.J.; Vellenga, E. Elevated VMP1 expression in acute myeloid leukemia amplifies autophagy and is protective against venetoclax-induced apoptosis. Cell Death Dis. 2019, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Ropolo, A.; Catrinacio, C.; Renna, F.J.; Boggio, V.; Orquera, T.; Gonzalez, C.D.; Vaccaro, M.I. A novel E2F1-EP300-VMP1 pathway mediates gemcitabine-induced autophagy in pancreatic cancer cells carrying oncogenic KRAS. Front. Endocrinol. 2020, 11, 411. [Google Scholar] [CrossRef]
- Biswas, A.K.; Acharyya, S. Understanding cachexia in the context of metastatic progression. Nat. Rev. Cancer 2020, 20, 274–284. [Google Scholar] [CrossRef]
- Van Linthout, S.; Miteva, K.; Tschöpe, C. Crosstalk between fibroblasts and inflammatory cells. Cardiovasc. Res. 2014, 102, 258–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, D.; Xu, J.; Dirain, M.L.; Leeuwenburgh, C. Calorie restriction combined with resveratrol induces autophagy and protects 26-month-old rat hearts from doxorubicin-induced toxicity. Free Radic. Biol. Med. 2014, 74, 252–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.S. Role of mitochondrial function in cell death and body metabolism. Front. Biosci. 2016, 21, 1233–1244. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Song, C.; Cao, X.; Li, H.; Cai, H.; Ma, Y.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; et al. MiR-208b regulates cell cycle and promotes skeletal muscle cell proliferation by targeting CDKN1A. J. Cell Physiol. 2019, 234, 3720–3729. [Google Scholar] [CrossRef]
- Zheng, L.F.; Chen, P.J.; Xiao, W.H. Signaling pathways controlling skeletal muscle mass. Sheng Li Xue Bao 2019, 71, 671–679. [Google Scholar] [PubMed]
- Andrade, G.M.; da Silveira, J.C.; Perrini, C.; Del Collado, M.; Gebremedhn, S.; Tesfaye, D.; Meirelles, F.V.; Perecin, F. The role of the PI3K-Akt signaling pathway in the developmental competence of bovine oocytes. PLoS ONE 2017, 12, e0185045. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Sun, Y.; Qiu, X.; Huang, Q.; Kong, L.; Lu, J.J. VMP1, a novel prognostic biomarker, contributes to glioma development by regulating autophagy. J. Neuroinflamm. 2021, 18, 165. [Google Scholar] [CrossRef]
- Loncle, C.; Molejon, M.I.; Lac, S.; Tellechea, J.I.; Lomberk, G.; Gramatica, L.; Fernandez Zapico, M.F.; Dusetti, N.; Urrutia, R.; Iovanna, J.L. The pancreatitis-associated protein VMP1, a key regulator of inducible autophagy, promotes Kras(G12D)-mediated pancreatic cancer initiation. Cell Death Dis. 2016, 7, e2295. [Google Scholar] [CrossRef] [Green Version]
- Gudmundsdottir, E.T.; Freysteinsdottir, E.S.; Olafsdottir, K.A.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B.; Reynisdottir, I. High expression of the vacuole membrane protein 1 (VMP1) is a potential marker of poor prognosis in HER2 positive breast cancer. PLoS ONE 2019, 14, e0221413. [Google Scholar]
- Wang, C.; Peng, R.; Zeng, M.; Zhang, Z.; Liu, S.; Jiang, D.; Lu, Y.; Zou, F. An autoregulatory feedback loop of miR-21/VMP1 is responsible for the abnormal expression of miR-21 in colorectal cancer cells. Cell Death Dis. 2020, 11, 1067. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, L.; Zhang, X.; Zhan, J.; Chen, J. TMEM49-related apoptosis and metastasis in ovarian cancer and regulated cell death. Mol. Cell Biochem. 2016, 416, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tabara, L.C.; Escalante, R. VMP1 establishes ER-microdomains that regulate membrane contact sites and autophagy. PLoS ONE 2016, 11, e0166499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo-Garrido, J.; Carilla-Latorre, S.; Lazaro-Dieguez, F.; Egea, G.; Escalante, R. Vacuole membrane protein 1 is an endoplasmic reticulum protein required for organelle biogenesis, protein secretion, and development. Mol. Biol. Cell. 2008, 19, 3442–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Vaccaro, M.I.; Ropolo, A.; Grasso, D.; Iovanna, J.L. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy 2008, 4, 388–390. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Hummel, E.; Osterrieder, A.; Meyer, A.J.; Frigerio, L.; Sparkes, I.; Hawes, C. KMS1 and KMS2, two plant endoplasmic reticulum proteins involved in the early secretory pathway. Plant J. 2011, 66, 613–628. [Google Scholar] [CrossRef]
- Tamargo-Gómez, I.; Mariño, G. AMPK: Regulation of metabolic dynamics in the context of autophagy. Int. J. Mol. Sci. 2018, 19, 3812. [Google Scholar] [CrossRef] [Green Version]
- Calvo-Garrido, J.; King, J.S.; Muñoz-Braceras, S.; Escalante, R. Vmp1 regulates PtdIns3P signaling during autophagosome formation in Dictyostelium discoideum. Traffic 2014, 15, 1235–1246. [Google Scholar] [CrossRef] [Green Version]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal 2014, 20, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Morishita, H.; Zhao, Y.G.; Tamura, N.; Nishimura, T.; Kanda, Y.; Sakamaki, Y.; Okazaki, M.; Li, D.; Mizushima, N. A critical role of VMP1 in lipoprotein secretion. eLife 2019, 8, e48834. [Google Scholar] [CrossRef]
- Xiong, X.; Wu, M.; Zhang, H.; Li, J.; Lu, B.; Guo, Y.; Zhou, T.; Guo, H.; Peng, R.; Li, X.; et al. Atg5 siRNA inhibits autophagy and enhances norcantharidin-induced apoptosis in hepatocellular carcinoma. Int. J. Oncol. 2015, 47, 1321–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhou, Z.; Li, K.; Wang, P.; Chen, Y.; Deng, S.; Li, W.; Yu, K.; Wang, K. VMP1 Regulated by chi-miR-124a Effects Goat Myoblast Proliferation, Autophagy, and Apoptosis through the PI3K/ULK1/mTOR Signaling Pathway. Cells 2022, 11, 2227. https://doi.org/10.3390/cells11142227
Liu Y, Zhou Z, Li K, Wang P, Chen Y, Deng S, Li W, Yu K, Wang K. VMP1 Regulated by chi-miR-124a Effects Goat Myoblast Proliferation, Autophagy, and Apoptosis through the PI3K/ULK1/mTOR Signaling Pathway. Cells. 2022; 11(14):2227. https://doi.org/10.3390/cells11142227
Chicago/Turabian StyleLiu, Yufang, Zuyang Zhou, Kunyu Li, Peng Wang, Yulin Chen, Shoulong Deng, Wenting Li, Kun Yu, and Kejun Wang. 2022. "VMP1 Regulated by chi-miR-124a Effects Goat Myoblast Proliferation, Autophagy, and Apoptosis through the PI3K/ULK1/mTOR Signaling Pathway" Cells 11, no. 14: 2227. https://doi.org/10.3390/cells11142227
APA StyleLiu, Y., Zhou, Z., Li, K., Wang, P., Chen, Y., Deng, S., Li, W., Yu, K., & Wang, K. (2022). VMP1 Regulated by chi-miR-124a Effects Goat Myoblast Proliferation, Autophagy, and Apoptosis through the PI3K/ULK1/mTOR Signaling Pathway. Cells, 11(14), 2227. https://doi.org/10.3390/cells11142227






