BMP Signaling Pathway in Dentin Development and Diseases
Abstract
1. Introduction
2. BMP Family
2.1. BMP Ligand Expression during Tooth Development and Mutations of BMPs Related to Dentin Defects
2.1.1. BMP-2
2.1.2. BMP-3
2.1.3. BMP-4
2.1.4. BMP-5
2.1.5. BMP-6
2.1.6. BMP-7
2.1.7. Activin-like Kinase-1 (ALK1, ACVRL1, or TSR-1) and ALK2 (ACVR1, ActR-IA)
2.1.8. ALK-3 (Bmpr-1A)
2.1.9. ALK-5 (TGFβR1 or NDSTN)
2.1.10. ALK-6 (BMPR-IB)
2.1.11. Type II Bmp Receptor (Bmpr-II)
2.2. Structure of BMPs and Their Partners
3. BMP Downstream Gene Expression during Tooth Development
3.1. Runt-Related Transcription Factor 2 (Runx2)
3.2. Osterix (Osx or Sp7)
3.3. Distal-Less (Dlx) Homeobox Gene 3 (Dlx3)
3.4. Msh (Muscle Segment Homeobox) Drosophila Homolog 1 (Msx1) and Msx2
3.5. Paired Box 9 (Pax9)
3.6. Dentin Matrix Protein 1 (DMP1)
3.7. Dentin Sialophosphoprotein (DSPP)
4. BMP Signaling during Tooth Development
4.1. Smad-Dependent Signaling in Dentin Development and Formation
4.2. Smad-Independent Signaling in Dentin Development and Formation
5. Negative Regulation of BMP Signaling in Odontoblast Differentiation and Dentinogenesis
6. Therapeutic Implications of BMPs for Endodontic Regeneration
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mitsiadis, T.A.; Orsini, G.; Jimenez-Rojo, L. Stem cell-based approaches in dentistry. Eur. Cell Mater. 2015, 30, 248–257. [Google Scholar] [CrossRef]
- Nanci, A. Ten Cate’s Oral Histology: Development, Structure, and Function, 8th ed.; Mosby: St. Louis, MI, USA, 2012; pp. 1–13. [Google Scholar]
- Zhang, Y.D.; Chen, Z.; Song, Y.Q.; Liu, C.; Chen, Y.P. Making a tooth: Growth factors, transcription factors, and stem cells. Cell Res. 2005, 15, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Thesleff, I. Epithelial-mesenchymal signaling regulating tooth morphogenesis. J. Cell Sci. 2003, 116, 1647–1648. [Google Scholar] [CrossRef]
- Chen, S.; Gluhak-Heinrich, J.; Martinez, M.; Li, T.; Wu, Y.; Chuang, H.H.; Chen, L.; Dong, J.; Gay, I.; MacDougall, M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J. Biol. Chem. 2008, 283, 19359–19370. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, W.; Xia, X.; Wang, F.; MacDougall, M.; Chen, S. Dentine sialophosphoprotein signal in dentineogenesis and dentine regeneration. Eur. Cells Mater. 2021, 42, 43–62. [Google Scholar] [CrossRef]
- Linde, A.; Goldberg, M. Dentinogenesis (review). Crit. Rev. Oral. Biol. Med. 1993, 4, 679–728. [Google Scholar] [CrossRef]
- MacDougall, M.; Simmons, D.; Luan, X.; Nydegger, J.; Feng, J.; Gu, T.T. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J. Biol. Chem. 1997, 272, 835–842. [Google Scholar] [CrossRef]
- Lopez-Cazaux, S.; Bluteau, G.; Magne, D.; Lieubeau, B.; Guicheux, J.; Alliot-Licht, B. Culture medium modulates the behavior of human dental pulpderived cells: Technical note. Eur. Cell Mater. 2006, 11, 35–42. [Google Scholar] [CrossRef]
- Orban, B.J. Orban’s Oral Histology and Embryology; Mosby: St. Louis, MI, USA, 1980. [Google Scholar]
- Li, J.; Parada, C.; Chai, Y. Cellular and molecular mechanisms of tooth root development. Development 2017, 144, 374–384. [Google Scholar] [CrossRef]
- Tucker, A.S.; Sharpe, P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 2004, 5, 499–508. [Google Scholar] [CrossRef]
- Chai, Y.; Maxson, R.E., Jr. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006, 235, 2353–2375. [Google Scholar] [CrossRef]
- Urist, M.R. Bone: Formation by autoinduction. Science 1965, 150, 893–899. [Google Scholar] [CrossRef]
- Wang, E.A.; Rosen, V.; Cordes, P.; Hewick, R.M.; Kriz, M.J.; Luxenberg, D.P.; Sibley, B.S.; Wozney, J.M. Purification and characterization of other distinct bone-inducing factors. Proc. Natl. Acad. Sci. USA 1988, 85, 9484–9488. [Google Scholar] [CrossRef]
- Wozney, J.M.; Rosen, V.; Celeste, A.J.; Mitsock, L.M.; Whitters, M.J.; Kriz, R.W.; Hewick, R.M.; Wang, E.A. Novel regulators of bone formation: Molecular clones and activities. Science 1988, 242, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Celeste, A.J.; Iannazzi, J.A.; Taylor, R.C.; Hewick, R.M.; Rosen, V.; Wang, E.A.; Wozney, J.M. Identification of transforming growth factor beta family members present in bone-inductive protein purified from bovine bone. Proc. Natl. Acad. Sci. USA 1990, 87, 9843–9847. [Google Scholar] [CrossRef]
- Ozkaynak, E.; Rueger, D.C.; Drier, E.A.; Corbett, C.; Ridge, R.J.; Sampath, T.K.; Oppermann, H. OP-1 cDNA encodes an osteogenic protein in the TGF-beta family. EMBO J. 1990, 9, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Sampath, T.K.; Coughlin, J.E.; Whetstone, R.M.; Banach, D.; Corbett, C.; Ridge, R.J.; Ozkaynak, E.; Oppermann, H.; Rueger, D.C. Bovine osteogenic protein is composed of dimers of OP-1 and BMP-2A, two members of the transforming growth factor-beta superfamily. J. Biol. Chem. 1990, 265, 13198–13205. [Google Scholar] [CrossRef]
- Sampath, T.K.; Reddi, A.H. Homology of bone-inductive proteins from human, monkey, bovine, and rat extracellular matrix. Proc. Natl. Acad. Sci. USA 1983, 80, 6591–6595. [Google Scholar] [CrossRef] [PubMed]
- Carreira, A.; Lojudice, F.; Halcsik, E.; Navarro, R.; Sogayar, M.; Granjeiro, J. Bone morphogenetic proteins: Facts, challenges, and future perspectives. J. Dent. Res. 2014, 93, 335–345. [Google Scholar] [CrossRef]
- Katagiri, T.; Watabe, T. Bone morphogenetic proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a021899. [Google Scholar] [CrossRef]
- Salazar, V.S.; Gamer, L.W.; Rosen, V. BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 2016, 12, 203–221. [Google Scholar] [CrossRef]
- Madaleno, C.D.S.; Jatzlau, J.; Knaus, P. BMP signalling in a mechanical context—Implications for bone biology. Bone 2020, 137, 115416. [Google Scholar] [CrossRef] [PubMed]
- Berg, T.; Wozney, J.; Thesleff, I. Expression patterns of bone morphogenetic proteins (bmps) in the developing mouse tooth suggest poles in morphogenesis and cell differentiation. Dev. Dyn. 1997, 210, 383–396. [Google Scholar]
- Nadiri, A.; Kuchler-Bopp, S.; Haikel, Y.; Lesot, H. Immunolocalization of BMP-2/-4, FGF-4, and WNT10b in the developing mouse first lower molar. J. Histochem. Cytochem. 2004, 52, 103–112. [Google Scholar] [CrossRef]
- Heikinheimo, K.; Bègue-Kirn, C.; Ritvos, O.; Tuuri, T.; Ruch, J.V. Activin and bone morphogenetic protein (BMP) signalling during tooth development. Eur. J. Oral Sci. 1998, 106, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Shen, B.; Ruan, N.; Guan, Z.; Zhang, Y.; Chen, Y.; Hu, X. Expression patterns of genes critical for BMP signaling pathway in developing human primary tooth germs. Histochem. Cell Biol. 2014, 142, 657–665. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, L.; Wang, F.; Zhang, C.; Wang, J.; He, J.; Wang, S. Expression of BMP2/4/7 during the odontogenesis of deciduous molars in miniature pig embryos. Histochem. J. 2018, 49, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, T.; Tummers, M.; Thesleff, I. Expression of bone morphogenetic proteins and MSX genes during root formation. J. Dent. Res. 2003, 82, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Kémoun, P.; Laurencin-Dalicieux, S.; Rue, J.; Vaysse, F.; Roméas, A.; Arzate, H.; Conte-Auriol, F.; Farges, J.; Salles, J.; Brunel, G. Localization of STRO-1, BMP-2/-3/-7, BMP receptors and phosphorylated Smad-1 during the formation of mouse periodontium. Tissue Cell 2007, 39, 257–266. [Google Scholar] [CrossRef]
- Casagrande, L.; Demarco, F.; Zhang, Z.; Araujo, F.; Shi, S.; Nör, J. Dentin-derived BMP-2 and odontoblast differentiation. J. Dent. Res. 2010, 89, 603–608. [Google Scholar] [CrossRef]
- Lu, Y.; Qian, Y.; Zhang, J.; Gong, M.; Wang, Y.; Gu, N.; Ma, L.; Xu, M.; Ma, J.; Zhang, W.; et al. Genetic variants of BMP2 and their association with the risk of non-syndromic tooth agenesis. PLoS ONE 2016, 11, e0158273. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Liu, H.C.; Han, D.; Feng, H.L. Detection and functional analysis of BMP2 gene mutation in patients with tooth agenesis. Beijing Da Xue Xue Bao Yi Xue Ban 2019, 51, 9–15. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bradley, A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 1996, 122, 2977–2986. [Google Scholar] [CrossRef]
- Yang, W.; Harris, M.; Cui, Y.; Mishina, Y.; Harris, S.; Gluhak-Heinrich, J. Bmp2 is required for odontoblast differentiation and pulp vasculogenesis. J. Dent. Res. 2011, 91, 58–64. [Google Scholar] [CrossRef]
- Yang, G.; Yuan, G.; MacDougall, M.; Zhi, C.; Chen, S. BMP-2 induced Dspp transcription is mediated by Dlx3/Osx signaling pathway in odontoblasts. Sci. Rep. 2017, 7, 10775. [Google Scholar] [CrossRef]
- Malik, Z.; Alexiou, M.; Hallgrimsson, B.; Economides, A.; Luder, H.; Graf, D. Bone morphogenetic protein 2 coordinates early tooth mineralization. J. Dent. Res. 2018, 97, 835–843. [Google Scholar] [CrossRef]
- Wu, L.; Wang, F.; Donly, K.J.; Wan, C.; Luo, D.; Harris, S.E.; MacDougall, M.; Chen, S. Establishment of immortalized mouse BMP2 knock-out dental papilla mesenchymal cells necessary for study of odontoblastic differentiation and odontogenesis. J. Cell. Physiol. 2015, 230, 2588–2595. [Google Scholar] [CrossRef]
- Takahashi, H.; Ikeda, T. Transcripts for two members of the transforming growth factor-beta superfamily BMP-3 and BMP-7 are expressed in developing rat embryos. Dev. Dyn. 1996, 207, 439–449. [Google Scholar] [CrossRef]
- Thomadakis, G.; Ramoshebi, L.N.; Crooks, J.; Rueger, D.C.; Ripamonti, U. Immunolocalization of bone morphogenetic protein-2 and -3 and osteogenic protein-1 during murine tooth root morphogenesis and in other craniofacial structures. Eur. J. Oral Sci. 1999, 107, 368–377. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, M.; Tian, X.; Wang, M.; Zhang, F. Experimental animal study on BMP-3 expression in periodontal tissues in the process of orthodontic tooth movement. Exp. Ther. Med. 2018, 17, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lu, Y.; Mues, G.; Wang, S.; Bonds, J.; D’Souza, R. Functional evaluation of a novel tooth agenesis-associated bone morphogenetic protein 4 prodomain mutation. Eur. J. Oral Sci. 2013, 121, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, H.; Fan, Z.; Xie, C.; Liu, H.; Liu, Y.; Han, D.; Wong, S.-W.; Feng, H. BMP4 mutations in tooth agenesis and low bone mass. Arch. Oral Biol. 2019, 103, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B.L. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995, 9, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Gluhak-Heinrich, J.; Guo, D.; Yang, W.; Harris, M.; Lichtler, A.; Kream, B.; Zhang, J.; Feng, J.; Smith, L.; Dechow, P.; et al. New roles and mechanism of action of BMP4 in postnatal tooth cytodifferentiation. Bone 2010, 46, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Jani, P.; Liu, C.; Zhang, H.; Younes, K.; Benson, M.D.; Qin, C. The role of bone morphogenetic proteins 2 and 4 in mouse dentinogenesis. Arch. Oral Biol. 2018, 90, 33–39. [Google Scholar] [CrossRef]
- Pathi, S.; Rutenberg, J.B.; Johnson, R.L.; Vortkamp, A. Interaction of Ihh and BMP/Noggin signaling during cartilage differentiation. Dev. Biol. 1999, 209, 239–253. [Google Scholar] [CrossRef]
- Kettunen, P.; Nie, X.; Kvinnsland, I.H.; Luukko, K. Histological development and dynamic expression of Bmp2–6 mRNAs in the embryonic and postnatal mousecranial base. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2006, 288, 1250–1258. [Google Scholar] [CrossRef]
- Mailhot, G.; Yang, M.; Mason-Savas, A.; MacKay, C.A.; Leav, I.; Odgren, P.R. BMP-5 expression increases during chondrocyte differentiation in vivo and in vitro and promotes proliferation and cartilage matrix synthesis in primary chondrocyte cultures. J. Cell. Physiol. 2007, 214, 56–64. [Google Scholar] [CrossRef]
- Kingsley, D.M.; Bland, A.E.; Grubber, J.M.; Marker, P.C.; Russell, L.B.; Copeland, N.G.; Jenkins, N.A. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell 1992, 71, 399–410. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Q.; Guo, Y.; Lin, L.; Yang, Q.; Jiang, H. Whole-genome sequencing identifies two novel rare mutations in BMP5 and BMP2 in monozygotic twins with microtia. J. Craniofacial Surg. 2021, 33, e212–e217. [Google Scholar] [CrossRef]
- Wilkins, J.M.; Southam, L.; Mustafa, Z.; Chapman, K.; Loughlin, J. Association of a functional microsatellite within intron 1 of the BMP5 gene with susceptibility to osteoarthritis. BMC Med. Genet. 2009, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Gao, B.; Xu, G.; Weng, D.; Xie, M.; Qian, Y. Association between single nucleotide polymorphisms of asporin (ASPN) and BMP5 with the risk of knee osteoarthritis in a Chinese Han population. Cell Biophys. 2014, 70, 1603–1608. [Google Scholar] [CrossRef]
- Shih, H.-Y.; Hsu, S.-Y.; Ouyang, P.; Lin, S.-J.; Chou, T.-Y.; Chiang, M.-C.; Cheng, Y.-C. Bmp5 regulates neural crest cell survival and proliferation via two different signaling pathways. Stem Cells 2016, 35, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Heikinheimo, K. Stage-specific expression of decapentaplegic-vg-related genes 2, 4, and 6 (bone morphogenetic proteins 2, 4, and 6) during human tooth morphogenesis. J. Dent. Res. 1994, 73, 590–597. [Google Scholar] [CrossRef]
- Oralová, V.; Chlastáková, I.; Radlanski, R.J.; Matalová, E. Distribution of BMP6 in the alveolar bone during mouse mandibular molar eruption. Connect. Tissue Res. 2014, 55, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Meynard, D.; Kautz, L.; Darnaud, V.; Canonne-Hergaux, F.; Coppin, H.; Roth, M.-P. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat. Genet. 2009, 41, 478–481. [Google Scholar] [CrossRef]
- Daher, R.; Kannengiesser, C.; Houamel, D.; Lefebvre, T.; Bardou-Jacquet, E.; Ducrot, N.; de Kerguenec, C.; Jouanolle, A.-M.; Robreau, A.-M.; Oudin, C.; et al. Heterozygous mutations in BMP6 pro-peptide lead to inappropriate hepcidin synthesis and moderate iron overload in humans. Gastroenterology 2016, 150, 672–683.e4. [Google Scholar] [CrossRef]
- Piubelli, C.; Castagna, A.; Marchi, G.; Rizzi, M.; Busti, F.; Badar, S.; Marchetti, M.; De Gobbi, M.; Roetto, A.; Xumerle, L.; et al. Identification of new BMP6 pro-peptide mutations in patients with iron overload. Am. J. Hematol. 2017, 92, 562–568. [Google Scholar] [CrossRef]
- Solloway, M.J.; Dudley, A.T.; Bikoff, E.K.; Lyons, K.M.; Hogan, B.L.; Robertson, E.J. Mice lacking Bmp6 function. Dev. Genet. 1998, 22, 321–339. [Google Scholar] [CrossRef]
- Perry, M.J.; McDougall, K.E.; Hou, S.-C.; Tobias, J. Impaired growth plate function in bmp-6 null mice. Bone 2008, 42, 216–225. [Google Scholar] [CrossRef]
- Cleves, P.A.; Hart, J.C.; Agoglia, R.M.; Jimenez, M.T.; Erickson, P.A.; Gai, L.; Miller, C.T. An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLoS Genet. 2018, 14, e1007449. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Rutherford, B.; Upholt, W.B.; Mina, M. Effects of BMP-7 on mouse tooth mesenchyme and chick mandibular mesenchyme. Dev. Dyn. 1999, 216, 320–335. [Google Scholar] [CrossRef]
- Helder, M.; Karg, H.; Bervoets, T.; Vukicevic, S.; Burger, E.; D’Souza, R.; Wöltgens, J.; Karsenty, G.; Bronckers, A. Bone morphogenetic protein-7 (osteogenic protein-1, OP-1) and tooth development. J. Dent. Res. 1998, 77, 545–554. [Google Scholar] [CrossRef]
- Godin, R.E.; Takaesu, N.T.; Robertson, E.J.; Dudley, A.T. Regulation of BMP7 expression during kidney development. Development 1998, 125, 3473–3482. [Google Scholar] [CrossRef]
- Morgan, E.A.; Nguyen, S.B.; Scott, V.; Stadler, H.S. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development 2003, 130, 3095–3109. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhao, S.; Jolly, A.; Wang, L.; Pan, H.; Yuan, J.; Chen, S.; Koch, A.; Ma, C.; Tian, W.; et al. Perturbations of genes essential for Müllerian duct and Wölffian duct development in Mayer-Rokitansky-Küster-Hauser syndrome. Am. J. Hum. Genet. 2021, 108, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Jeremias, F.; Pierri, R.A.; Souza, J.F.; Fragelli, C.M.B.; Restrepo, M.; Finoti, L.S.; Bussaneli, D.G.; Cordeiro, R.C.; Secolin, R.; Maurer-Morelli, C.V.; et al. Family-based genetic association for molar-incisor hypomineralization. Caries Res. 2016, 50, 310–318. [Google Scholar] [CrossRef]
- Dudley, A.T.; Lyons, K.M.; Robertson, E.J. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995, 9, 2795–2807. [Google Scholar] [CrossRef]
- Luo, G.; Hofmann, C.; Bronckers, A.L.; Sohocki, M.; Bradley, A.; Karsenty, G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 1995, 9, 2808–2820. [Google Scholar] [CrossRef]
- Saito, K.; Takahashi, K.; Asahara, M.; Kiso, H.; Togo, Y.; Tsukamoto, H.; Huang, B.; Sugai, M.; Shimizu, A.; Motokawa, M.; et al. Effects of Usag-1 and Bmp7 deficiencies on murine tooth morphogenesis. BMC Dev. Biol. 2016, 16, 14. [Google Scholar] [CrossRef]
- Zurowski, C.; Jamniczky, H.; Graf, D.; Theodor, J. Deletion/loss of bone morphogenetic protein 7 changes tooth morphology and function in Mus musculus: Implications for dental evolution in mammals. R. Soc. Open Sci. 2018, 5, 170761. [Google Scholar] [CrossRef] [PubMed]
- Malik, Z.; Roth, D.M.; Eaton, F.; Theodor, J.M.; Graf, D. Mesenchymal Bmp7 controls onset of tooth mineralization: A novel way to regulate molar cusp shape. Front. Physiol. 2020, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Verschueren, K.; Dewulf, N.; Goumans, M.J.; Lonnoy, O.; Feijen, A.; Grimsby, S.; Vande Spiegle, K.; ten Dijke, P.; Morén, A.; Vanscheeuwijck, P.; et al. Expression of type I and type IB receptors for activin in midgestation mouse embryos suggests distinct functions in organogenesis. Mech. Dev. 1995, 52, 109–123. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Q.; Zhao, H.; Hu, Y.; Liu, C.; Yan, G.; Li, D.; Mishina, Y.; Shi, C.; Sun, H. ACVR1 is essential for periodontium development and promotes alveolar bone formation. Arch. Oral Biol. 2018, 95, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.; Smoke, R.H.; Rutherford, R. Expression of genes for bone morphogenetic proteins and receptors in human dental pulp. Arch. Oral Biol. 1996, 41, 919–923. [Google Scholar] [CrossRef]
- Toyono, T.; Nakashima, M.; Kuhara, S.; Akamine, A. Temporal changes in expression of transforming growth factor-β superfamily members and their receptors during bovine preodontoblast differentiation in vitro. Arch. Oral Biol. 1997, 42, 481–488. [Google Scholar] [CrossRef]
- Toyono, T.; Nakashima, M.; Kuhara, S.; Akamine, A. Expression of TGF-beta superfamily receptors in dental pulp. J. Dent. Res. 1997, 76, 1555–1560. [Google Scholar] [CrossRef]
- Cheifetz, S. BMP receptors in limb and tooth formation. Crit. Rev. Oral Biol. Med. 1999, 10, 182–198. [Google Scholar] [CrossRef]
- Chien, H.-H.; Lin, W.-L.; Cho, M.-I. Expression of TGF-beta isoforms and their receptors during mineralized nodule formation by rat periodontal ligament cells in vitro. J. Periodontal Res. 1999, 34, 301–309. [Google Scholar] [CrossRef]
- McDonald, J.E.; Miller, F.J.; Hallam, S.E.; Nelson, L.; Marchuk, D.A.; Ward, K.J. Clinical manifestations in a large hereditary hemorrhagic telangiectasia (HHT) type 2 kindred. Am. J. Med. Genet. 2000, 93, 320–327. [Google Scholar] [CrossRef]
- Wehner, L.-E.; Folz, B.J.; Argyriou, L.; Twelkemeyer, S.; Teske, U.; Geisthoff, U.W.; Werner, J.A.; Engel, W.; Nayernia, K. Mutation analysis in hereditary haemorrhagic telangiectasia in Germany reveals 11 novel ENG and 12 novel ACVRL1/ALK1 mutations. Clin. Genet. 2006, 69, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Furtado, J.; Poulet, M.; Chung, M.; Yun, S.; Lee, S.; Sessa, W.C.; Franco, C.A.; Schwartz, M.A.; Eichmann, A. Defective flow-migration coupling causes arteriovenous malformations in hereditary hemorrhagic telangiectasia. Circulation 2021, 144, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, C.; Zhao, H.; Zhou, Y.; Hu, Y.; Yan, G.; Liu, C.; Li, D.; Hao, X.; Mishina, Y.; et al. Distinctive role of ACVR1 in dentin formation: Requirement for dentin thickness in molars and prevention of osteodentin formation in incisors of mice. Histochem. J. 2018, 50, 43–61. [Google Scholar] [CrossRef]
- Fiori, J.L.; Billings, P.C.; De La Peña, L.S.; Kaplan, F.S.; Shore, E.M. Dysregulation of the BMP-p38 MAPK signaling pathway in cells from patients with Fibrodysplasia Ossificans Progressiva (FOP). J. Bone Miner. Res. 2006, 21, 902–909. [Google Scholar] [CrossRef]
- Kaplan, F.S.; Kobori, J.A.; Orellana, C.; Calvo, I.; Rosello, M.; Martinez, F.; Lopez, B.; Xu, M.; Pignolo, R.J.; Shore, E.M.; et al. Multi-system involvement in a severe variant of fibrodysplasia ossificans progressiva (ACVR1 c.772G > A.; R258G): A report of two patients. Am. J. Med. Genet. A 2015, 167, 2265–2271. [Google Scholar] [CrossRef]
- Schoenmaker, T.; Mokry, M.; Micha, D.; Netelenbos, C.; Bravenboer, N.; Gilijamse, M.; Eekhoff, E.; de Vries, T. Activin-A induces early differential gene expression exclusively in periodontal ligament fibroblasts from Fibrodysplasia Ossificans Progressiva Patients. Biomedicines 2021, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Mishina, Y.; Suzuki, A.; Ueno, N.; Behringer, R.R. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995, 9, 3027–3037. [Google Scholar] [CrossRef]
- Andl, T.; Ahn, K.; Kairo, A.; Chu, E.Y.; Wine-Lee, L.; Reddy, S.T.; Croft, N.J.; Cebra-Thomas, J.A.; Metzger, D.; Chambon, P.; et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development 2004, 131, 2257–2268. [Google Scholar] [CrossRef]
- Li, L.; Lin, M.; Wang, Y.; Cserjesi, P.; Chen, Z.; Chen, Y. BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev. Biol. 2011, 349, 451–461. [Google Scholar] [CrossRef]
- Omi, M.; Kulkarni, A.K.; Raichur, A.; Fox, M.; Uptergrove, A.; Zhang, H.; Mishina, Y. BMP-Smad signaling regulates postnatal crown Dentinogenesis in mouse molar. JBMR Plus 2019, 4, e10249. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Lin, M.; Yuan, G.; Yang, G.; Zheng, Y.; Chen, Y. Augmented BMPRIA-mediated BMP signaling in cranial neural crest lineage leads to cleft palate formation and delayed tooth differentiation. PLoS ONE 2013, 8, e66107. [Google Scholar] [CrossRef] [PubMed]
- Iseki, S.; Osumi-Yamashita, N.; Miyazono, K.; Franzén, P.; Ichijo, H.; Ohtani, H.; Hayashi, Y.; Eto, K. Localization of transforming growth factor-β type I and type II receptors in mouse development. Exp. Cell Res. 1995, 219, 339–347. [Google Scholar] [CrossRef]
- Zhang, H.; Zhan, Y.; Zhang, Y.; Yuan, G.; Yang, G. Dual roles of TGF-β signaling in the regulation of dental epithelial cell proliferation. Histochem. J. 2020, 52, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Fan, Z.; Wang, S.; Du, J. ALK5 is essential for tooth germ differentiation during tooth development. Biotech. Histochem. 2019, 94, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, D.; Han, T.; Sun, Y.; Zhang, J. TGF-beta1 and TGFBR1 are expressed in ameloblasts and promote MMP20 expression. Anat. Rec. 2009, 292, 885–890. [Google Scholar] [CrossRef]
- Sloan, A.; Matthews, J.; Smith, A. TGF-β receptor expression in human odontoblasts and pulpal cells. Histochem. J. 1999, 31, 565–569. [Google Scholar] [CrossRef]
- Sloan, A.; Couble, M.-L.; Bleicher, F.; Magloire, H.; Smith, A.; Farges, J.-C. Expression of TGF-β receptors I and II in the human dental pulp by in situ hybridization. Adv. Dent. Res. 2001, 15, 63–67. [Google Scholar] [CrossRef]
- Hosoya, A.; Kim, J.-Y.; Cho, S.-W.; Jung, H.-S. BMP4 signaling regulates formation of Hertwig’s epithelial root sheath during tooth root development. Cell Tissue Res. 2008, 333, 503–509. [Google Scholar] [CrossRef]
- Yi, S.; Daluiski, A.; Pederson, R.; Rosen, V.; Lyons, K. The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development 2000, 127, 621–630. [Google Scholar] [CrossRef]
- Shi, C.; Iura, A.; Terajima, M.; Liu, F.; Lyons, K.; Pan, H.; Zhang, H.; Yamauchi, M.; Mishina, Y.; Sun, H. Deletion of BMP receptor type IB decreased bone mass in association with compromised osteoblastic differentiation of bone marrow mesenchymal progenitors. Sci. Rep. 2016, 6, 24256. [Google Scholar] [CrossRef]
- Lee, H.-K.; Park, J.-T.; Cho, Y.-S.; Bae, H.-S.; Cho, M.-I.; Park, J.-C. Odontogenic ameloblasts-associated protein (ODAM), via phosphorylation by bone morphogenetic protein receptor type IB (BMPR-IB), is implicated in ameloblast differentiation. J. Cell. Biochem. 2011, 113, 1754–1765. [Google Scholar] [CrossRef] [PubMed]
- Hinck, A.P.; Mueller, T.D.; Springer, T.A. Structural biology and evolution of the TGF-β family. Cold Spring Harb. Perspect. Biol. 2016, 8, a022103. [Google Scholar] [CrossRef] [PubMed]
- Yadin, D.; Knaus, P.; Mueller, T.D. Structural insights into BMP receptors: Specificity, activation and inhibition. Cytokine Growth Factor Rev. 2016, 27, 13–34. [Google Scholar] [CrossRef]
- Sengle, G.; Charbonneau, N.L.; Ono, R.N.; Sasaki, T.; Alvarez, J.; Keene, D.R.; Bächinger, H.P.; Sakai, L.Y. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J. Biol. Chem. 2008, 283, 13874–13888. [Google Scholar] [CrossRef] [PubMed]
- Mueller, T.D.; Nickel, J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012, 586, 1846–1859. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhu, J.; Wang, R.; Chen, X.; Mi, L.; Walz, T.; Springer, T.A. Latent TGF-β structure and activation. Nature 2011, 474, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Nelsen, S.M.; Christian, J.L. Site-specific cleavage of BMP4 by Furin, PC6, and PC7. J. Biol. Chem. 2009, 284, 27157–27166. [Google Scholar] [CrossRef]
- Bragdon, B.; Moseychuk, O.; Saldanha, S.; King, D.; Julian, J.; Nohe, A. Bone morphogenetic proteins: A critical review. Cell. Signal. 2011, 23, 609–620. [Google Scholar] [CrossRef]
- Saremba, S.; Nickel, J.; Seher, A.; Kotzsch, A.; Sebald, W.; Mueller, T.D. Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J. 2007, 275, 172–183. [Google Scholar] [CrossRef]
- Garrigue-Antar, L.; Hartigan, N.; Kadler, K.E. Post-translational modification of bone morphogenetic protein-1 is required for secretion and stability of the protein. J. Biol. Chem. 2002, 277, 43327–43334. [Google Scholar] [CrossRef]
- Hashimoto, O.; Moore, R.K.; Shimasaki, S. Posttranslational processing of mouse and human BMP-15: Potential implication in the determination of ovulation quota. Proc. Natl. Acad. Sci. USA 2005, 102, 5426–5431. [Google Scholar] [CrossRef] [PubMed]
- Little, S.C.; Mullins, M.C. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat. Cell Biol. 2009, 11, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Sieber, C.; Kopf, J.; Hiepen, C.; Knaus, P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009, 20, 343–355. [Google Scholar] [CrossRef]
- Brazil, D.P.; Church, R.H.; Surae, S.; Godson, C.; Martin, F. BMP signalling: Agony and antagony in the family. Trends Cell Biol. 2015, 25, 249–264. [Google Scholar] [CrossRef]
- Ulloa, L.; Doody, J.; Massagué, J. Inhibition of transforming growth factor-β/SMAD signalling by the interferon-gamma/STAT pathway. Nature 1999, 397, 710–713. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ren, W.; Warburton, R.; Toksoz, D.; Fanburg, B.L. Serotonin induces Rho/ROCKdependent activation of Smads 1/5/8 in pulmonary artery smooth muscle cells. FASEB J. 2009, 23, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Miralles, I.; Schisler, J.C.; Patterson, C. New insights into bone morphogenetic protein signaling: Focus on angiogenesis. Curr. Opin. Hematol. 2009, 16, 195–201. [Google Scholar] [CrossRef]
- Israel, D.I.; Nove, J.; Kerns, K.M.; Kaufman, R.J.; Rosen, V.; Cox, K.A.; Wozney, J.M. Heterodimeric bone morphogenetic proteins show enhanced activity in vitro and in vivo. Growth Factors 1996, 13, 291–300. [Google Scholar] [CrossRef]
- Guo, J.; Wu, G. The signaling and functions of heterodimeric bone morphogenetic proteins. Cytokine Growth Factor Rev. 2012, 23, 61–67. [Google Scholar] [CrossRef]
- Suzuki, A.; Kaneko, E.; Maeda, J.; Ueno, N. Mesoderm induction by BMP-4 and -7 heterodimers. Biochem. Biophys. Res. Commun. 1997, 232, 153–156. [Google Scholar] [CrossRef]
- Yuan, S.; Pan, Q.; Liu, W.; Wu, B.; Han, X.; Bi, Z. Recombinant BMP 4/7 fusion protein induces differentiation of bone marrow stem cells. J. Cell Biochem. 2011, 112, 3054–3060. [Google Scholar] [CrossRef]
- Griffith, D.L.; Keck, P.C.; Sampath, T.K.; Rueger, D.C.; Carlson, W.D. Three-dimensional structure of recombinant human osteogenic protein 1: Structural paradigm for the transforming growth factor b superfamily. Proc. Natl. Acad. Sci. USA 1996, 93, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Scheufler, C.; Sebald, W.; Hülsmeyer, M. Crystal structure of human bone morphogenetic protein-2 at 2.7 Å resolution. J. Mol. Biol. 1999, 287, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Zhao, Q.; Baker, K.A.; Naik, C.; Chen, C.; Pukac, L.; Singh, M.; Tsareva, T.; Parice, Y.; Mahoney, A.; et al. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J. Biol. Chem. 2005, 280, 25111–25118. [Google Scholar] [CrossRef] [PubMed]
- Schreuder, H.; Liesum, A.; Pohl, J.; Kruse, M.; Koyama, M. Crystal structure of recombinant human growth and differentiation factor 5: Evidence for interaction of the type I and type II receptor-binding sites. Biochem. Biophys. Res. Commun. 2005, 329, 1076–1086. [Google Scholar] [CrossRef]
- Allendorph, G.P.; Isaacs, M.J.; Kawakami, Y.; Belmonte, J.C.I.; Choe, S. BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors. Biochemistry 2007, 46, 12238–12247. [Google Scholar] [CrossRef]
- Kirsch, T.; Sebald, W.; Dreyer, M.K. Crystal structure of the BMP-2–BRIA ectodomain complex. Nat. Genet. 2000, 7, 492–496. [Google Scholar] [CrossRef]
- Groppe, J.; Greenwald, J.; Wiater, E.; Rodriguez-Leon, J.M.; Economides, A.N.; Kwiatkowski, W.; Affolter, M.; Vale, W.W.; Belmonte, J.C.I.; Choe, S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature 2002, 420, 636–642. [Google Scholar] [CrossRef]
- Greenwald, J.; Groppe, J.; Gray, P.; Wiater, E.; Kwiatkowski, W.; Vale, W.; Choe, S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol. Cell. 2003, 11, 605–617. [Google Scholar] [CrossRef]
- Keller, S.L.; Nickel, J.; Zhang, J.-L.; Sebald, W.; Mueller, T.D. Molecular recognition of BMP-2 and BMP receptor IA. Nat. Struct. Mol. Biol. 2004, 11, 481–488. [Google Scholar] [CrossRef]
- Allendorph, G.P.; Vale, W.W.; Choe, S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc. Natl. Acad. Sci. USA 2006, 103, 88. [Google Scholar] [CrossRef]
- Healey, E.G.; Bishop, B.; Elegheert, J.; Bell, C.H.; Padilla-Parra, S.; Siebold, C. Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat. Struct. Mol. Biol. 2015, 22, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Lee, B.; Thirunavukkarasu, K.; Zhou, L.; Pastore, L.; Baldini, A.; Hecht, J.; Geoffroy, V.; Ducy, P.; Karsenty, G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 1997, 16, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Mundlos, S.; Otto, F.; Mundlos, C.; Mulliken, J.; Aylsworth, A.; Albright, S.; Lindhout, D.; Cole, W.; Henn, W.; Knoll, J.; et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997, 89, 773–779. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Quack, I.; Vonderstrass, B.; Stock, M.; Aylsworth, A.; Becker, A.; Brueton, L.; Lee, P.; Majewski, F.; Mulliken, J.; Suri, M.; et al. Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am. J. Hum. Genet. 1999, 65, 1268–1278. [Google Scholar] [CrossRef]
- Zhou, G.; Chen, Y.; Zhou, L.; Thirunavukkarasu, K.; Hecht, J.; Chitayat, D.; Gelb, B.D.; Pirinen, S.; Berry, S.A.; Greenberg, C.R.; et al. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum. Mol. Genet. 1999, 8, 2311–2316. [Google Scholar] [CrossRef]
- D’Souza, R.; Aberg, T.; Gaikwad, J.; Cavender, A.; Owen, M.; Karsenty, G.; Thesleff, I. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development 1999, 126, 2911–2920. [Google Scholar] [CrossRef]
- Chen, S.; Gluhak-Heinrich, J.; Wang, Y.H.; Wu, Y.M.; Chuang, H.H.; Chen, L.; Yuan, G.H.; Dong, J.; Gay, I.; MacDougall, M. Runx2, OSX, and DSPP in tooth development. J. Dent. Res. 2009, 88, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, S.; McDonald, F. Runx2 and dental development. Eur. J. Oral Sci. 2006, 114, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Sodek, J.; Karsenty, G.; Thomas, H.; Ranly, D.; Chen, J. Expression of core binding factor Osf2/Cbfa-1 and bone sialoprotein in tooth development. Mech. Dev. 1999, 81, 169–173. [Google Scholar] [CrossRef]
- Hirata, A.; Sugahara, T.; Nakamura, H. Localization of runx2, osterix, and osteopontin in tooth root formation in rat molars. J. Histochem. Cytochem. 2008, 57, 397–403. [Google Scholar] [CrossRef]
- Han, J.; He, H. Expression and function of osteogenic genes runt-related transcription factor 2 and osterix in orthodontic tooth movement in rats. Int. J. Clin. Exp. Pathol. 2015, 8, 11895–11900. [Google Scholar]
- Chen, S.; Rani, S.; Wu, Y.; Unterbrink, A.; Gu, T.T.; Gluhak-Heinrich, J.; Chuang, H.-H.; MacDougall, M. Differential regulation of dentin sialophosphoprotein expression by runx2 during odontoblast cytodifferentiation. J. Biol. Chem. 2005, 280, 29717–29727. [Google Scholar] [CrossRef]
- James, M.J.; Järvinen, E.; Wang, X.-P.; Thesleff, I. Different roles of runx2 during early neural crest-derived bone and tooth development. J. Bone Miner. Res. 2006, 21, 1034–1044. [Google Scholar] [CrossRef]
- Bufalino, A.; Paranaíba, L.; Gouvêa, A.; Gueiros, L.A.; Martelli-Júnior, H.; Junior, J.; Lopes, M.; Graner, E.; De Almeida, O.; Vargas, P.; et al. Cleidocranial dysplasia: Oral features and genetic analysis of 11 patients. Oral Dis. 2011, 18, 184–190. [Google Scholar] [CrossRef]
- Gaikwad, J.; Hoffmann, M.; Cavender, A.; Bronckers, A.; D’Souza, R. Molecular insights into the lineage-specific determination of odontoblasts: The role of cbfa1. Adv. Dent. Res. 2001, 15, 19–24. [Google Scholar] [CrossRef]
- Miyazaki, T.; Kanatani, N.; Rokutanda, S.; Yoshida, C.; Toyosawa, S.; Nakamura, R.; Takada, S.; Komori, T. Inhibition of the terminal differentiation of odontoblasts and their transdifferentiation into osteoblasts in Runx2 transgenic mice. Arch. Histol. Cytol. 2008, 71, 131–146. [Google Scholar] [CrossRef]
- Lee, M.-H.; Kim, Y.-J.; Yoon, W.-J.; Kim, J.-I.; Kim, B.-G.; Hwang, Y.-S.; Wozney, J.M.; Chi, X.-Z.; Bae, S.-C.; Choi, K.-Y.; et al. Dlx5 Specifically regulates runx2 type II expression by binding to homeodomain-response elements in the runx2 distal promoter. J. Biol. Chem. 2005, 280, 35579–35587. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Bae, J.-S.; Afzal, F.; Gutierrez, S.; Pratap, J.; Zaidi, S.K.; Lou, Y.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Structural coupling of Smad and runx2 for execution of the BMP2 osteogenic signal. J. Biol. Chem. 2008, 283, 8412–8422. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Chen, M.; Wang, Y.-J.; Kaneki, H.; Xing, L.; O’Keefe, R.J.; Chen, D. Smad6 interacts with runx2 and mediates smad ubiquitin regulatory factor 1-induced runx2 degradation. J. Biol. Chem. 2006, 281, 3569–3576. [Google Scholar] [CrossRef] [PubMed]
- Jani, P.; Zhang, H.; Benson, M.D.; Qin, C. Noggin inhibition of mouse dentinogenesis. J. Oral Biosci. 2019, 62, 72–79. [Google Scholar] [CrossRef]
- Cho, Y.-D.; Yoon, W.-J.; Woo, K.-M.; Baek, J.-H.; Park, J.-C.; Ryoo, H.-M. The canonical BMP signaling pathway plays a crucial part in stimulation of dentin sialophosphoprotein expression by BMP-2. J. Biol. Chem. 2010, 285, 36369–36376. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, W.; Xu, H.; Ning, Y.; Fang, W.; Liao, W.; Zou, J.; Yang, Y.; Shao, N. Effects of altered CXCL12/CXCR4 axis on BMP2/Smad/Runx2/Osterix axis and osteogenic gene expressions during osteogenic differentiation of MSCs. Am. J. Transl. Res. 2017, 9, 1680–1693. [Google Scholar]
- Xiao, M.; Yao, B.; Zhang, B.-D.; Bai, Y.; Sui, W.; Wang, W.; Yu, Q. Stromal-derived Factor-1α signaling is involved in bone morphogenetic protein-2-induced odontogenic differentiation of stem cells from apical papilla via the Smad and Erk signaling pathways. Exp. Cell Res. 2019, 381, 39–49. [Google Scholar] [CrossRef]
- Wang, X.; Liao, X.; Zhang, Y.; Wei, L.; Pang, Y. Schisandrin B regulates MC3T3-E1 subclone 14 cells proliferation and differentiation through BMP2-SMADs-RUNX2-SP7 signaling axis. Sci. Rep. 2020, 10, 14476. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Tomazelli, K.B.; Modolo, F.; Trentin, A.G.; Garcez, R.C.; Biz, M.T. Temporo-spatial analysis of Osterix, HNK1 and Sox10 during odontogenesis and maxillaries osteogenesis. Tissue Cell 2015, 47, 465–470. [Google Scholar] [CrossRef]
- Takahashi, A.; Ono, N.; Ono, W. The fate of Osterix-expressing mesenchymal cells in dental root formation and maintenance. Orthod. Craniofacial Res. 2017, 20 (Suppl. S1), 39–43. [Google Scholar] [CrossRef]
- Lee, D.S.; Roh, S.Y.; Park, J.-C. The Nfic-osterix pathway regulates ameloblast differentiation and enamel formation. Cell Tissue Res. 2018, 374, 531–540. [Google Scholar] [CrossRef]
- Hosoya, A.; Yukita, A.; Ninomiya, T.; Hiraga, T.; Yoshiba, K.; Yoshiba, N.; Kasahara, E.; Nakamura, H. Localization of SUMOylation factors and Osterix in odontoblast lineage cells during dentin formation and regeneration. Histochem. Cell Biol. 2013, 140, 201–211. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Kim, B.-I.; Jue, S.-S.; Park, J.H.; Shin, J.-W. Localization of osteopontin and osterix in periodontal tissue during orthodontic tooth movement in rats. Angle Orthod. 2011, 82, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Bae, C.; Lee, J.; Kim, J.; Yang, X.; de Crombrugghe, B.; Cho, E. Osterix regulates tooth root formation in a site-specific manner. J. Dent. Res. 2015, 94, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, Y.; Qin, C.; Liu, Y.; Ho, S.P.; Feng, J.Q. Essential role of Osterix for tooth root but not crown dentin formation. J. Bone Miner. Res. 2014, 30, 742–746. [Google Scholar] [CrossRef] [PubMed]
- He, Y.D.; Sui, B.D.; Li, M.; Huang, J.; Chen, S.; Wu, L.A. Site-specific function and regulation of Osterix in tooth root formation. Int. Endod. J. 2015, 49, 1124–1131. [Google Scholar] [CrossRef]
- Fiscaletti, M.; Biggin, A.; Bennetts, B.; Wong, K.; Briody, J.; Pacey, V.; Birman, C.; Munns, C.F. Novel variant in Sp7/Osx associated with recessive osteogenesis imperfecta with bone fragility and hearing impairment. Bone 2018, 110, 66–75. [Google Scholar] [CrossRef]
- Lapunzina, P.; Aglan, M.; Temtamy, S.; Caparros-Martin, J.A.; Valencia, M.; Letón, R.; Martínez-Glez, V.; Elhossini, R.; Amr, K.; Vilaboa, N.; et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010, 87, 110–114. [Google Scholar] [CrossRef]
- Yang, G.; Li, X.; Yuan, G.; Liu, P.; Fan, M. The effects of Osterix on the proliferation and odontoblastic differentiation of human dental papilla cells. J. Endod. 2014, 40, 1771–1777. [Google Scholar] [CrossRef]
- Bae, J.-M.; Clarke, J.C.; Rashid, H.; Adhami, M.D.; McCullough, K.; Scott, J.S.; Chen, H.; Sinha, K.M.; De Crombrugghe, B.; Javed, A. Specificity protein 7 is required for proliferation and differentiation of ameloblasts and odontoblasts. J. Bone Miner. Res. 2018, 33, 1126–1140. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Jiang, Z.; Wang, Y.; Xi, Y.; Yang, G. Molecular mechanisms for short root anomaly. Oral Dis. 2019, 27, 142–150. [Google Scholar] [CrossRef]
- Liu, H.; Lin, H.; Zhang, L.; Sun, Q.; Yuan, G.; Zhang, L.; Chen, S.; Chen, Z. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J. Biol. Chem. 2013, 288, 9261–9271. [Google Scholar] [CrossRef] [PubMed]
- Celil, A.B.; Hollinger, J.O.; Campbell, P.G. Osx transcriptional regulation is mediated by additional pathways to BMP2/Smad signaling. J. Cell. Biochem. 2005, 95, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.C.; Drissi, H.; Choudhury, G.G.; Ghosh-Choudhury, N. Integration of phosphatidylinositol 3-kinase, akt kinase, and Smad signaling pathway in BMP-2-induced Osterix expression. Calcif. Tissue Res. 2010, 87, 533–540. [Google Scholar] [CrossRef]
- Lee, M.-H.; Kwon, T.-G.; Park, H.-S.; Wozney, J.M.; Ryoo, H.-M. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 2003, 309, 689–694. [Google Scholar] [CrossRef]
- Matsubara, T.; Kida, K.; Yamaguchi, A.; Hata, K.; Ichida, F.; Meguro, H.; Aburatani, H.; Nishimura, R.; Yoneda, T. BMP2 Regulates Osterix through Msx2 and Runx2 during Osteoblast differentiation. J. Biol. Chem. 2008, 283, 29119–29125. [Google Scholar] [CrossRef]
- Tao, H.; Lin, H.; Sun, Z.; Pei, F.; Zhang, J.; Chen, S.; Liu, H.; Chen, Z. Klf4 Promotes Dentinogenesis and odontoblastic differentiation via modulation of TGF-β signaling pathway and interaction with histone acetylation. J. Bone Miner. Res. 2019, 34, 1502–1516. [Google Scholar] [CrossRef]
- Lee, D.-S.; Park, J.-T.; Kim, H.-M.; Ko, J.S.; Son, H.-H.; Gronostajski, R.M.; Cho, M.-I.; Choung, P.-H.; Park, J.-C. Nuclear factor I-C is essential for odontogenic cell proliferation and odontoblast differentiation during tooth root development. J. Biol. Chem. 2009, 284, 17293–17303. [Google Scholar] [CrossRef]
- Morasso, M.I.; Grinberg, A.; Robinson, G.; Sargent, T.D.; Mahon, K.A. Placental failure in mice lacking the homeobox gene Dlx3. Proc. Natl. Acad. Sci. USA 1999, 96, 162–167. [Google Scholar] [CrossRef]
- Merlo, G.R.; Zerega, B.; Paleari, L.; Trombino, S.; Mantero, S.; Levi, G. Multiple functions of Dlx genes. Int. J. Dev. Biol. 2000, 44, 619–626. [Google Scholar] [PubMed]
- Qiu, M.; Bulfone, A.; Ghattas, I.; Meneses, J.J.; Christensen, L.; Sharpe, P.T.; Presley, R.; Pedersen, R.A.; Rubenstein, J.L.R. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 andDlx-2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 1997, 185, 165–184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Stock, D.W.; Buchanan, A.V.; Weiss, K.M. Expression of Dlx genes during the development of the murine dentition. Dev. Genes Evol. 2000, 210, 270. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.W.; Mahon, K.A. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mech. Dev. 1994, 48, 199–215. [Google Scholar] [CrossRef]
- Weiss, K.M.; Bollekens, J.; Ruddle, F.H.; Takashita, K. Distal-less and other homeobox genes in the development of the dentition. J. Exp. Zool. 1994, 270, 273–284. [Google Scholar] [CrossRef]
- Lézot, F.; Thomas, B.; Greene, S.R.; Hotton, D.; Yuan, Z.; Castaneda, B.; Bolaños, A.; Depew, M.; Sharpe, P.; Gibson, C.W.; et al. Physiological implications of DLX homeoproteins in enamel formation. J. Cell. Physiol. 2008, 216, 688–697. [Google Scholar] [CrossRef]
- Ghoul-Mazgar, S.; Hotton, D.; Lézot, F.; Blin-Wakkach, C.; Asselin, A.; Sautier, J.-M.; Berdal, A. Expression pattern of Dlx3 during cell differentiation in mineralized tissues. Bone 2005, 37, 799–809. [Google Scholar] [CrossRef]
- Dong, J.; Amor, D.; Aldred, M.J.; Gu, T.; Escamilla, M.; MacDougall, M. DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. Am. J. Med. Genet. Part A 2005, 133, 138–141. [Google Scholar] [CrossRef]
- Lee, S.K.; Lee, Z.H.; Lee, S.J.; Ahn, B.D.; Kim, Y.J.; Lee, S.H.; Kim, J.W. DLX3 mutation in a new family and its phenotypic variations. J. Dent. Res. 2008, 87, 354–357. [Google Scholar] [CrossRef]
- Wright, J.T.; Hong, S.P.; Simmons, D.; Daly, B.; Uebelhart, D.; Luder, H.U. DLX3 c.561_562delCT mutation causes attenuated phenotype of tricho-dento-osseous syndrome. Am. J. Med. Genet. Part A 2008, 146, 343–349. [Google Scholar] [CrossRef]
- Price, J.A.; Wright, J.T.; Kula, K.; Bowden, D.W.; Hart, T.C. A common DLX3 gene mutation is responsible for tricho-dento-osseous syndrome in Virginia and North Carolina families. J. Med. Genet. 1998, 35, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, P.; Lukinmaa, P.-L.; Alapulli, H.; Methuen, M.; Suojärvi, T.; Kivirikko, S.; Peltola, J.; Asikainen, M.; Alaluusua, S. DLX3 homeodomain mutations cause Tricho-Dento-osseous syndrome with novel phenotypes. Cells Tissues Organs 2011, 194, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, D.; Zhang, H.; Liu, H.; Wong, S.; Zhao, N.; Feng, H. Morphological analyses and a novel de novo DLX3 mutation associated with tricho-dento-osseous syndrome in a Chinese family. Eur. J. Oral Sci. 2015, 123, 228–234. [Google Scholar] [CrossRef]
- Choi, S.; Song, I.; Feng, J.; Gao, T.; Haruyama, N.; Gautam, P.; Robey, P.; Hart, T.C. Mutant DLX 3 disrupts odontoblast polarization and dentin formation. Dev. Biol. 2010, 344, 682–692. [Google Scholar] [CrossRef][Green Version]
- Duverger, O.; Zah, A.; Isaac, J.; Sun, H.-W.; Bartels, A.K.; Lian, J.B.; Berdal, A.; Hwang, J.; Morasso, M.I. Neural crest deletion of Dlx3 leads to major dentin defects through down-regulation of Dspp. J. Biol. Chem. 2012, 287, 12230–12240. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Fu, J.; Chen, Z.; Yang, G.; Yuan, G. Dlx3 ubiquitination by nuclear Mdm2 is essential for dentinogenesis in mice. J. Dent. Res. 2022, 26, 1–11. [Google Scholar] [CrossRef]
- Viale-Bouroncle, S.; Felthaus, O.; Schmalz, G.; Brockhoff, G.; Reichert, T.E.; Morsczeck, C. The transcription factor DLX3 regulates the osteogenic differentiation of human dental follicle precursor cells. Stem Cells Dev. 2012, 21, 1936–1947. [Google Scholar] [CrossRef]
- Mackenzie, A.; Leeming, G.; Jowett, A.; Ferguson, M.; Sharpe, P. The homeobox gene Hox 7.1 has specific regional and temporal expression patterns during early murine craniofacial embryogenesis, especially tooth development in vivo and in vitro. Development 1991, 111, 269–285. [Google Scholar] [CrossRef]
- Keränen, S.V.E.; Åberg, T.; Kettunen, P.; Thesleff, I.; Jernvall, J. Association of developmental regulatory genes with the development of different molar tooth shapes in two species of rodents. Dev. Genes Evol. 1998, 208, 477–486. [Google Scholar] [CrossRef]
- Tucker, A.S.; Al Khamis, A.; Sharpe, P.T. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 1998, 212, 533–539. [Google Scholar] [CrossRef]
- van den Boogaard, M.J.; Dorland, M.; Beemer, F.A.; van Amstel, H.K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000, 24, 342–343. [Google Scholar] [CrossRef]
- Lidral, A.; Reising, B. The role of MSX1 in human tooth agenesis. J. Dent. Res. 2002, 81, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Satokata, I.; Maas, R.L. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 1994, 6, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Vastardis, H.; Karimbux, N.; Guthua, S.W.; Seidman, J.; Seidman, C.E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 1996, 13, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kong, H.; Mues, G.; D’Souza, R. Msx1 mutations: How do they cause tooth agenesis? J. Dent. Res. 2011, 90, 311–316. [Google Scholar] [CrossRef]
- Bei, M.; Maas, R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development 1998, 125, 4325–4333. [Google Scholar] [CrossRef]
- Xin, T.; Zhang, T.; Li, Q.; Yu, T.; Zhu, Y.; Yang, R.; Zhou, Y. A novel mutation of MSX1 in oligodontia inhibits odontogenesis of dental pulp stem cells via the ERK pathway. Stem Cell Res. Ther. 2018, 9, 221. [Google Scholar] [CrossRef]
- Feng, X.-Y.; Zhao, Y.-M.; Wang, W.-J.; Ge, L.-H. Msx1 regulates proliferation and differentiation of mouse dental mesenchymal cells in culture. Eur. J. Oral Sci. 2013, 121, 412–420. [Google Scholar] [CrossRef]
- Feng, X.-Y.; Wu, X.-S.; Wang, J.-S.; Zhang, C.-M.; Wang, S.-L. Homeobox protein MSX-1 inhibits expression of bone morphogenetic protein 2, bone morphogenetic protein 4, and lymphoid enhancer-binding factor 1 via Wnt/β-catenin signaling to prevent differentiation of dental mesenchymal cells during the late bell stage. Eur. J. Oral Sci. 2017, 126, 1–12. [Google Scholar] [CrossRef]
- Yang, G.; Yuan, G.; Ye, W.; Cho, K.W.Y.; Chen, Y. An atypical canonical bone morphogenetic protein (BMP) signaling pathway regulates Msh homeobox 1 (Msx1) expression during odontogenesis. J. Biol. Chem. 2014, 289, 31492–31502. [Google Scholar] [CrossRef]
- Hu, X.; Lin, C.; Ruan, N.; Huang, Z.; Zhang, Y.; Hu, X. Operation of the atypical canonical bone morphogenetic protein signaling pathway during early human odontogenesis. Front. Physiol. 2022, 13, 823275. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, A.; Ferguson, M.; Sharpe, P. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development 1992, 115, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Jowett, A.; Vainio, S.; Ferguson, M.; Sharpe, P.; Thesleff, I. Epithelial-mesenchymal interactions are required for msx 1 and msx 2 gene expression in the developing murine molar tooth. Development 1993, 117, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Keränen, S.V.E.; Kettunen, P.; Åberg, T.; Thesleff, I.; Jernvall, J. Gene expression patterns associated with suppression of odontogenesis in mouse and vole diastema regions. Dev. Genes Evol. 1999, 209, 495–506. [Google Scholar] [CrossRef]
- Babajko, S.; de La Dure-Molla, M.; Jedeon, K.; Berdal, A. MSX2 in ameloblast cell fate and activity. Front. Physiol. 2015, 5, 00510. [Google Scholar] [CrossRef]
- Bidder, M.; Latifi, T.; Towler, D.A. Reciprocal temporospatial patterns of Msx2 and osteocalcin gene expression during murine odontogenesis. J. Bone Miner. Res. 1998, 13, 609–619. [Google Scholar] [CrossRef]
- Yamamoto, H.; Cho, S.-W.; Kim, E.-J.; Kim, J.-Y.; Fujiwara, N.; Jung, H.-S. Developmental properties of the hertwig’s epithelial root sheath in mice. J. Dent. Res. 2004, 83, 688–692. [Google Scholar] [CrossRef]
- Molla, M.; Descroix, V.; Aïoub, M.; Simon, S.; Castañeda, B.; Hotton, D.; Bolaños, A.; Simon, Y.; Lezot, F.; Goubin, G.; et al. Enamel protein regulation and dental and periodontal physiopathology in Msx2 mutant mice. Am. J. Pathol. 2010, 177, 2516–2526. [Google Scholar] [CrossRef]
- Jabs, E.W.; Müller, U.; Li, X.; Ma, L.; Luo, W.; Haworth, I.S.; Klisak, I.; Sparkes, R.; Warman, M.L.; Mulliken, J.B.; et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell 1993, 75, 443–450. [Google Scholar] [CrossRef]
- Ma, L.; Golden, S.; Wu, L.; Maxson, R. The molecular basis of Boston-type craniosynostosis: The Pro148-->His mutation in the N-terminal arm of the MSX2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum. Mol. Genet. 1996, 5, 1915–1920. [Google Scholar] [CrossRef]
- Wilkie, A.O.; Tang, Z.; Elanko, N.; Walsh, S.; Twigg, S.; Hurst, J.A.; Wall, S.A.; Chrzanowska, K.; Maxson, R.E. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat. Genet. 2000, 24, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Amri, N.; Djolé, S.X.; Petit, S.; Babajko, S.; Coudert, A.E.; Castaneda, B.; Simon, S.; Berdal, A. Distorted patterns of dentinogenesis and eruption in Msx2 null mutants: Involvement of sost/sclerostin. Am. J. Pathol. 2016, 186, 2577–2587. [Google Scholar] [CrossRef][Green Version]
- Aïoub, M.; Lézot, F.; Molla, M.; Castaneda, B.; Robert, B.; Goubin, G.; Néfussi, J.; Berdal, A. Msx2−/− transgenic mice develop compound amelogenesis imperfecta, dentinogenesis imperfecta and periodental osteopetrosis. Bone 2007, 41, 851–859. [Google Scholar] [CrossRef]
- Hur, S.-W.; Oh, S.-H.; Jeong, B.-C.; Choi, H.; Kim, J.-W.; Lee, K.-N.; Hwang, Y.-C.; Ryu, J.-H.; Kim, S.-H.; Koh, J.-T. COUP-TFII Stimulates dentin sialophosphoprotein expression and mineralization in odontoblasts. J. Dent. Res. 2015, 94, 1135–1142. [Google Scholar] [CrossRef]
- Zhou, Y.L.; Lei, Y.; Snead, M.L. Functional antagonism between Msx2 and CCAAT/enhancer-binding protein α in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J. Biol. Chem. 2000, 275, 29066–29075. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Satoh, K.; Hirota, M.; Kimura, K.; Kanno, A.; Masamune, A.; Shimosegawa, T. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J. Cell. Physiol. 2007, 213, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Valdimarsdottir, L.; Hrafnkelsdottir, H.E.; Runarsson, J.F.; Omarsdottir, A.R.; Oostwaard, D.W.-V.; Mummery, C.; Valdimarsdottir, G. BMP4 Promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells 2013, 32, 636–648. [Google Scholar] [CrossRef]
- Hussein, S.M.; Duff, E.K.; Sirard, C. Smad4 and β-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J. Biol. Chem. 2003, 278, 48805–48814. [Google Scholar] [CrossRef]
- Rath, B.; Nam, J.; Deschner, J.; Schaumburger, J.; Tingart, M.; Grässel, S.; Grifka, J.; Agarwal, S. Biomechanical forces exert anabolic effects on osteoblasts by activation of SMAD 1/5/8 through type 1 BMP receptor. Biorheology 2011, 48, 37–48. [Google Scholar] [CrossRef]
- Rodríguez-Carballo, E.; Ulsamer, A.; Susperregui, A.R.; Manzanares-Céspedes, C.; Sánchez-García, E.; Bartrons, R.; Rosa, J.L.; Ventura, F. Conserved regulatory motifs in osteogenic gene promoters integrate cooperative effects of canonical Wnt and BMP pathways. J. Bone Miner. Res. 2010, 26, 718–729. [Google Scholar] [CrossRef]
- Neubüser, A.; Peters, H.; Balling, R.; Martin, G.R. Antagonistic interactions between FGF and BMP signaling pathways: A mechanism for positioning the sites of tooth formation. Cell 1997, 90, 247–255. [Google Scholar] [CrossRef]
- Peters, H.; Neubüser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Huang, Y.; He, F.; Gu, S.; Zhang, G.; Chen, Y.; Zhang, Y. Expression survey of genes critical for tooth development in the human embryonic tooth germ. Dev. Dyn. 2007, 236, 1307–1312. [Google Scholar] [CrossRef] [PubMed]
- Stockton, D.W.; Das, P.; Goldenberg, M.; D’Souza, R.; Patel, P. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 2000, 24, 18–19. [Google Scholar] [CrossRef]
- Bonczek, O.; Balcar, V.; Šerý, O. PAX9 gene mutations and tooth agenesis: A review. Clin. Genet. 2017, 92, 467–476. [Google Scholar] [CrossRef]
- Wong, S.-W.; Han, D.; Zhang, H.; Liu, Y.; Zhang, X.; Miao, M.; Wang, Y.; Zhao, N.; Zeng, L.; Bai, B.; et al. Nine novel PAX9 mutations and a distinct tooth agenesis genotype-phenotype. J. Dent. Res. 2017, 97, 155–162. [Google Scholar] [CrossRef]
- Blake, J.A.; Ziman, M. Pax genes: Regulators of lineage specification and progenitor cell maintenance. Development 2014, 141, 737–751. [Google Scholar] [CrossRef]
- Yin, W.; Bian, Z. The gene network underlying hypodontia. J. Dent. Res. 2015, 94, 878–885. [Google Scholar] [CrossRef]
- Kapadia, H.; Mues, G.; D’Souza, R. Genes affecting tooth morphogenesis. Orthod. Craniofacial Res. 2007, 10, 105–113. [Google Scholar] [CrossRef]
- Kong, H.; Wang, Y.; Patel, M.; Mues, G.; D’Souza, R.N. Regulation of Bmp4 expression in odontogenic mesenchyme: From simple to complex. Cells Tissues Organs 2011, 194, 156–160. [Google Scholar] [CrossRef]
- Zhou, J.; Gao, Y.; Lan, Y.; Jia, S.; Jiang, R. Pax9 regulates a molecular network involving Bmp4, Fgf10, Shh signaling and the Osr2 transcription factor to control palate morphogenesis. Development 2013, 140, 4709–4718. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, Z.; Yu, Q.; Weng, M.; Chen, Z. The function and regulatory network of Pax9 gene in palate development. J. Dent. Res. 2019, 98, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Seki, D.; Takeshita, N.; Oyanagi, T.; Sasaki, S.; Takano, I.; Hasegawa, M.; Takano-Yamamoto, T. Differentiation of odontoblast-like cells from mouse induced pluripotent stem cells by Pax9 and Bmp4 transfection. STEM CELLS Transl. Med. 2015, 4, 993–997. [Google Scholar] [CrossRef]
- Feng, J.; Jing, J.; Li, J.; Zhao, H.; Punj, V.; Zhang, T.; Xu, J.; Chai, Y. BMP signaling orchestrates a transcriptional network to control the fate of mesenchymal stem cells in mice. Development 2017, 144, 2560–2569. [Google Scholar] [CrossRef]
- George, A.; Sabsay, B.; Simonian, P.A.; Veis, A. Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J. Biol. Chem. 1993, 268, 12624–12630. [Google Scholar] [CrossRef]
- MacDougall, M.; Gu, T.T.; Luan, X.; Simmons, D.; Chen, J. Identification of a novel isoform of mouse dentin matrix protein 1: Spatial expression in mineralized tissues. J. Bone Miner. Res. 1998, 13, 422–431. [Google Scholar] [CrossRef]
- D’Souza, R.N.; Cavender, A.; Sunavala, G.; Alvarez, J.; Ohshima, T.; Kulkarni, A.B.; MacDougall, M. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J. Bone Miner. Res. 1997, 12, 2040–2049. [Google Scholar] [CrossRef]
- Feng, J.; Huang, H.; Lu, Y.; Ye, L.; Xie, Y.; Tsutsui, T.; Kunieda, T.; Castranio, T.; Scott, G.; Bonewald, L.; et al. The Dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. J. Dent. Res. 2003, 82, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Baba, O.; Qin, C.; Brunn, J.C.; Wygant, J.N.; McIntyre, B.W.; Butler, W.T. Colocalization of dentin matrix protein 1 and dentin sialoprotein at late stages of rat molar development. Matrix Biol. 2004, 23, 371–379. [Google Scholar] [CrossRef]
- Hao, J.; Zou, B.; Narayanan, K.; George, A. Differential expression patterns of the dentin matrix proteins during mineralized tissue formation. Bone 2004, 34, 921–932. [Google Scholar] [CrossRef]
- Martinez, E.F.; da Silva, L.A.H.; Furuse, C.; de Araújo, N.S.; de Araújo, V.C. Dentin matrix protein 1 (DMP1) expression in developing human teeth. Braz. Dent. J. 2009, 20, 365–369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Toyosawa, S.; Okabayashi, K.; Komori, T.; Ijuhin, N. mRNA expression and protein localization of dentin matrix protein 1 during dental root formation. Bone 2004, 34, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; MacDougall, M.; Zhang, S.; Xie, Y.; Zhang, J.; Li, Z.; Lu, Y.; Mishina, Y.; Feng, J.Q. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J. Biol. Chem. 2004, 279, 19141–19148. [Google Scholar] [CrossRef]
- Feng, J.Q.; Ward, L.M.; Liu, S.; Lu, Y.; Xie, Y.; Yuan, B.; Yu, X.; Rauch, F.; Davis, S.I.; Zhang, S.; et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat. Genet. 2006, 38, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Lorenz-Depiereux, B.; Bastepe, M.; Benet-Pagès, A.; Amyere, M.; Wagenstaller, J.; Müller-Barth, U.; Badenhoop, K.; Kaiser, S.M.; Rittmaster, R.S.; Shlossberg, A.H.; et al. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat. Genet. 2006, 38, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Turan, S.; Aydin, C.; Bereket, A.; Akcay, T.; Güran, T.; Yaralioglu, B.A.; Bastepe, M.; Jüppner, H. Identification of a novel dentin matrix protein-1 (DMP-1) mutation and dental anomalies in a kindred with autosomal recessive hypophosphatemia. Bone 2010, 46, 402–409. [Google Scholar] [CrossRef]
- Liu, T.; Wang, J.; Xie, X.; Wang, K.; Sui, T.; Liu, D.; Lai, L.; Zhao, H.; Li, Z.; Feng, J.Q. DMP1 Ablation in the rabbit results in mineralization defects and abnormalities in haversian canal/osteon microarchitecture. J. Bone Miner. Res. 2019, 34, 1115–1128. [Google Scholar] [CrossRef]
- Yamamoto, R.; Oida, S.; Yamakoshi, Y. Dentin Sialophosphoprotein-derived proteins in the dental pulp. J. Dent. Res. 2015, 94, 1120–1127. [Google Scholar] [CrossRef]
- Ritchie, H.H.; Berry, J.E.; Somerman, M.J.; Hanks, C.T.; Bronckers, A.L.J.J.; Hotton, D.; Papagerakis, P.; Berdal, A.; Butler, W.T. Dentin sialoprotein (DSP) transcripts: Developmentally-sustained expression in odontoblasts and transient expression in pre-ameloblasts. Eur. J. Oral Sci. 1997, 105, 405–413. [Google Scholar] [CrossRef]
- Hou, C.; Liu, Z.X.; Tang, K.L.; Wang, M.G.; Sun, J.; Wang, J.; Li, S. Developmental changes and regional localization of Dspp, Mepe, Mimecan and Versican in postnatal developing mouse teeth. Histochem. J. 2011, 43, 9–16. [Google Scholar] [CrossRef]
- Bleicher, F.; Couble, M.; Farges, J.; Couble, P.; Magloire, H. Sequential expression of matrix protein genes in developing rat teeth. Matrix Biol. 1999, 18, 133–143. [Google Scholar] [CrossRef]
- Vijaykumar, A.; Ghassem-Zadeh, S.; Vidovic-Zdrilic, I.; Komitas, K.; Adameyko, I.; Krivanek, J.; Fu, Y.; Maye, P.; Mina, M. Generation and characterization of DSPP-Cerulean/DMP1-Cherry reporter mice. Genesis 2019, 57, e23324. [Google Scholar] [CrossRef]
- Qin, C.; Brunn, J.; Cadena, E.; Ridall, A.; Tsujigiwa, H.; Nagatsuka, H.; Nagai, N.; Butler, W. The expression of dentin sialophosphoprotein gene in bone. J. Dent. Res. 2002, 81, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Chaplet, M.; Waltregny, D.; Detry, C.; Fisher, L.W.; Castronovo, V.; Bellahcène, A. Expression of dentin sialophosphoprotein in human prostate cancer and its correlation with tumor aggressiveness. Int. J. Cancer. 2006, 118, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Wang, Y.; Gluhak-Heinrich, J.; Yang, G.; Chen, L.; Li, T.; Wu, L.-A.; Chen, Z.; MacDougall, M.; Chen, S. Tissue-specific expression of dentin sialophosphoprotein (DSPP) and its polymorphisms in mouse tissues. Cell Biol. Int. 2009, 33, 816–829. [Google Scholar] [CrossRef] [PubMed]
- Ogbureke, K.; Abdelsayed, R.A.; Kushner, H.; Li, L.; Fisher, L.W. Two members of the SIBLING family of proteins, DSPP and BSP, may predict the transition of oral epithelial dysplasia to oral squamous cell carcinoma. Cancer 2010, 116, 1709–1717. [Google Scholar] [CrossRef]
- Xiao, S.; Yu, C.; Chou, X.; Yuan, W.; Wang, Y.; Bu, L.; Fu, G.; Qian, M.; Yang, J.; Shi, Y.; et al. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet. 2001, 27, 201–204. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, J.; Li, C.; Gao, S.; Qiu, C.; Liu, P.; Wu, G.; Qiang, B.; Lo, W.H.; Shen, Y. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat. Genet. 2001, 27, 151–152. [Google Scholar] [CrossRef]
- Rajpar, M.H.; Koch, M.J.; Davies, R.M.; Mellody, K.T.; Kielty, C.M.; Dixon, M.J. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum. Mol. Genet. 2002, 11, 2559–2565. [Google Scholar] [CrossRef]
- Kim, J.-W.; Simmer, J.P. Hereditary dentin defects. J. Dent. Res. 2007, 86, 392–399. [Google Scholar] [CrossRef]
- Simmer, J.P.; Zhang, H.; Moon, S.J.H.; Donnelly, L.A.-J.; Lee, Y.-L.; Seymen, F.; Koruyucu, M.; Chan, H.-C.; Lee, K.Y.; Wu, S.; et al. The modified shields classification and 12 families with defined DSPP mutations. Genes 2022, 13, 858. [Google Scholar] [CrossRef] [PubMed]
- Hursey, R.J.; Witkop, C.J.; Miklashek, D.; Sackett, L.M. Dentinogenesis imperfecta in a racial isolate with multiple hereditary defects. Oral Surg. Oral Med. Oral Pathol. 1956, 9, 641–658. [Google Scholar] [CrossRef]
- Sreenath, T.; Thyagarajan, T.; Hall, B.; Longenecker, G.; D’Souza, R.; Hong, S.; Wright, J.T.; MacDougall, M.; Sauk, J.; Kulkarni, A.B. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J. Biol. Chem. 2003, 278, 24874–24880. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H.; Miyazono, K.; ten Dijke, P. TGF-b signaling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390, 465–471. [Google Scholar] [CrossRef]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef]
- Miyazono, K.; Maeda, S.; Imamura, T. BMP receptor signaling: Transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005, 16, 251–263. [Google Scholar] [CrossRef]
- Sánchez-Duffhues, G.; García de Vinuesa, A.; ten Dijke, P. Bone Morphogenetic Proteins Signal Transduction in Vascular Diseases; Nova Science Publishers: New York, NY, USA, 2012. [Google Scholar]
- Sánchez-Duffhues, G.; Hiepen, C.; Knaus, P.; Dijke, P.T. Bone morphogenetic protein signaling in bone homeostasis. Bone 2015, 80, 43–59. [Google Scholar] [CrossRef]
- Goumans, M.-J.; Zwijsen, A.; Ten Dijke, P.; Bailly, S. Bone morphogenetic proteins in vascular homeostasis and disease. Cold Spring Harb. Perspect. Biol. 2018, 10, a031989. [Google Scholar] [CrossRef]
- Yu, P.B.; Beppu, H.; Kawai, N.; Li, E.; Bloch, K.D. BMP type II receptor deletion reveals BMP ligand-specific gain of signaling in pulmonary artery smooth muscle cells. J. Biol. Chem. 2005, 280, 24443–24450. [Google Scholar] [CrossRef]
- ten Dijke, P.; Yamashita, H.; Sampath, T.K.; Reddi, A.H.; Estevez, M.; Riddle, D.L.; Ichijo, H.; Heldin, C.H.; Miyazono, K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J. Biol. Chem. 1994, 269, 16985–16988. [Google Scholar] [CrossRef]
- Ebisawa, T.; Tada, K.; Kitajima, I.; Tojo, K.; Sampath, T.; Kawabata, M.; Miyazono, K.; Imamura, T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J. Cell Sci. 1999, 112, 3519–3527. [Google Scholar] [CrossRef] [PubMed]
- Nishitoh, H.; Ichijo, H.; Kimura, M.; Matsumoto, T.; Makishima, F.; Yamaguchi, A.; Yamashita, H.; Enomoto, S.; Miyazono, K. Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J. Biol. Chem. 1996, 271, 21345–21352. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Mallet, C.; Mazerbourg, S.; Feige, J.-J.; Bailly, S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 2006, 109, 1953–1961. [Google Scholar] [CrossRef]
- Daluiski, A.; Engstrand, T.; Bahamonde, M.E.; Gamer, L.W.; Agius, E.; Stevenson, S.L.; Cox, K.; Rosen, V.; Lyons, K.M. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat. Genet. 2001, 27, 84–88. [Google Scholar] [CrossRef]
- Mazerbourg, S.; Klein, C.; Roh, J.; Kaivo-Oja, N.; Mottershead, D.G.; Korchynskyi, O.; Ritvos, O.; Hsueh, A.J.W. Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5. Mol. Endocrinol. 2004, 18, 653–665. [Google Scholar] [CrossRef]
- Kokabu, S.; Gamer, L.; Cox, K.; Lowery, J.; Tsuji, K.; Raz, R.; Economides, A.; Katagiri, T.; Rosen, V. BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol. Endocrinol. 2012, 26, 87–94. [Google Scholar] [CrossRef]
- Wu, M.; Chen, G.; Li, Y.-P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef]
- Bhatt, S.; Diaz, R.; Trainor, P. Signals and switches in mammalian neural crest cell differentiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a008326. [Google Scholar] [CrossRef]
- Graf, D.; Malik, Z.; Hayano, S.; Mishina, Y. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 2015, 27, 129–139. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smadindependent pathways in TGF-b family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Sammar, M.; Stricker, S.; Schwabe, G.C.; Sieber, C.; Hartung, A.; Hanke, M.; Oishi, I.; Pohl, J.; Minami, Y.; Sebald, W.; et al. Modulation of GDF5/BRI-b signalling through interaction with the tyrosine kinase receptor Ror2. Genes Cells 2004, 9, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Endo, M.; Hata, K.; Higuchi, C.; Takaoka, K.; Yoshikawa, H.; Yamashita, T. Neogenin, a receptor for bone morphogenetic proteins. J. Biol. Chem. 2011, 286, 5157–5165. [Google Scholar] [CrossRef] [PubMed]
- Nickel, J.; Dijke, P.T.; Mueller, T.D. TGF-β family co-receptor function and signaling. Acta Biochim. Biophys. Sin. 2018, 50, 12–36. [Google Scholar] [CrossRef] [PubMed]
- Gipson, G.R.; Goebel, E.J.; Hart, K.N.; Kappes, E.C.; Kattamuri, C.; McCoy, J.C.; Thompson, T.B. Structural perspective of BMP ligands and signaling. Bone 2020, 140, 115549. [Google Scholar] [CrossRef] [PubMed]
- Zouvelou, V.; Luder, H.-U.; Mitsiadis, T.A.; Graf, D. Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J. Exp. Zool. Part B Mol. Dev. Evol. 2009, 312, 361–374. [Google Scholar] [CrossRef]
- Feng, J.; Yang, G.; Yuan, G.; Gluhak-Heinrich, J.; Yang, W.; Wang, L.; Chen, Z.; McDaniel, J.S.; Donly, K.J.; Harris, S.E.; et al. Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs 2011, 194, 216–221. [Google Scholar] [CrossRef]
- Guo, F.; Feng, J.; Wang, F.; Li, W.; Gao, Q.; Chen, Z.; Shoff, L.; Donly, K.J.; Gluhak-Heinrich, J.; Chun, Y.H.P.; et al. Bmp2 deletion causes an amelogenesis imperfecta phenotype via regulating enamel gene expression. J. Cell. Physiol. 2014, 230, 1871–1882. [Google Scholar] [CrossRef]
- Huang, X.; Wang, F.; Zhao, C.; Yang, S.; Cheng, Q.; Tang, Y.; Zhang, F.; Zhang, Y.; Luo, W.; Wang, C.; et al. Dentinogenesis and tooth-alveolar bone complex defects in BMP9/GDF2 knockout mice. Stem Cells Dev. 2019, 28, 683–694. [Google Scholar] [CrossRef]
- Wang, F.; Tao, R.; Zhao, L.; Hao, X.-H.; Zou, Y.; Lin, Q.; Liu, M.M.; Goldman, G.; Luo, D.; Chen, S. Differential lncRNA/mRNA expression profiling and functional network analyses in Bmp2 deletion of mouse dental papilla cells. Front. Genet. 2021, 12, 702540. [Google Scholar] [CrossRef]
- Chen, Z.; Couble, M.-L.; Mouterfi, N.; Magloire, H.; Chen, Z.; Bleicher, F. Spatial and temporal expression of KLF4 and KLF5 during murine tooth development. Arch. Oral Biol. 2009, 54, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-K.; Lee, D.-S.; Park, S.-J.; Cho, K.-H.; Bae, H.-S.; Park, J.-C. Nuclear Factor I-C (NFIC) regulates Dentin Sialophosphoprotein (DSPP) and E-cadherin via control of Krüppel-like Factor 4 (KLF4) during dentinogenesis. J. Biol. Chem. 2014, 289, 28225–28236. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Horrell, J.; Snitow, M.; Cui, J.; Gochnauer, H.; Syrett, C.M.; Kallish, S.; Seykora, J.T.; Liu, F.; Gaillard, D.; et al. WNT10A mutation causes ectodermal dysplasia by impairing progenitor cell proliferation and KLF4-mediated differentiation. Nat. Commun. 2017, 8, 15397. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lin, C.; Zhang, J.; Tao, H.; Liu, H.; Yuan, G.; Chen, Z. Quaking promotes the odontoblastic differentiation of human dental pulp stem cells. J. Cell. Physiol. 2018, 233, 7292–7304. [Google Scholar] [CrossRef]
- Huang, Z.; Yang, R.; Li, R.; Zuo, Y.; Gu, F.; He, M.; Bian, Z. Mesenchymal Mycn participates in odontoblastic lineage commitment by regulating Krüppel-like Factor 4 (Klf4) in mice. Stem. Cell Res. Ther. 2022, 13, 78. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Park, Y.; Seo, Y.; Lee, H.; Park, J. Tubular dentin formation by TGF-β/BMP signaling in dental epithelial cells. Oral Dis. 2022. [Google Scholar] [CrossRef]
- Celil, A.B.; Campbell, P.G. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J. Biol. Chem. 2005, 280, 31353–31359. [Google Scholar] [CrossRef]
- Ge, X.; Li, Z.; Jing, S.; Wang, Y.; Li, N.; Lu, J.; Yu, J. Parathyroid hormone enhances the osteo/odontogenic differentiation of dental pulp stem cells via ERK and P38 MAPK pathways. J. Cell. Physiol. 2019, 235, 1209–1221. [Google Scholar] [CrossRef]
- Peng, L.; Dong, G.; Xu, P.; Ren, L.; Wang, C.; Aragon, M.; Zhou, X.; Ye, L. Expression of Wnt5a in tooth germs and the related signal transduction analysis. Arch. Oral Biol. 2010, 55, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Han, M.; Zheng, B.; Li, Y.-J.; Shu, Y.-N.; Wen, J. Krüppel-like factor 4 interacts with p300 to activate mitofusin 2 gene expression induced by all-trans retinoic acid in VSMCs. Acta Pharmacol. Sin. 2010, 31, 1293–1302. [Google Scholar] [CrossRef][Green Version]
- Greenblatt, M.B.; Kim, J.-M.; Oh, H.; Park, K.H.; Choo, M.-K.; Sano, Y.; Tye, C.; Skobe, Z.; Davis, R.J.; Park, J.M.; et al. p38α MAPK is required for tooth morphogenesis and enamel secretion. J. Biol. Chem. 2015, 290, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Jeong, H.M.; Jin, Y.-H.; Li, H.; Yeo, C.-Y.; Lee, K.-Y. Akt phosphorylates and regulates the osteogenic activity of Osterix. Biochem. Biophys. Res. Commun. 2011, 411, 637–641. [Google Scholar] [CrossRef]
- Choi, Y.H.; Choi, H.-J.; Lee, K.-Y.; Oh, J.-W. Akt1 regulates phosphorylation and osteogenic activity of Dlx3. Biochem. Biophys. Res. Commun. 2012, 425, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Pande, S.; Browne, G.; Padmanabhan, S.; Zaidi, S.K.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J. Cell. Physiol. 2013, 228, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zheng, B.; Zhang, X.-H.; He, M.; Guo, Z.-W.; Wen, J.-K. PPAR-γ agonist stabilizes KLF4 protein via activating Akt signaling and reducing KLF4 ubiquitination. Biochem. Biophys. Res. Commun. 2013, 443, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-G.; Kim, H.-J.; Park, H.-J.; Kim, Y.-J.; Yoon, W.-J.; Lee, S.-J.; Ryoo, H.-M.; Cho, J.-Y. Runx2 phosphorylation induced by fibroblast growth factor-2/protein kinase C pathways. Proteomics 2006, 6, 1166–1174. [Google Scholar] [CrossRef]
- Chew, Y.C.; Adhikary, G.; Wilson, G.M.; Reece, E.A.; Eckert, R.L. Protein Kinase C (PKC) δ suppresses keratinocyte proliferation by increasing p21Cip1 level by a KLF4 transcription factor-dependent mechanism. J. Biol. Chem. 2011, 286, 28772–28782. [Google Scholar] [CrossRef]
- Jeong, H.M.; Jin, Y.-H.; Choi, Y.H.; Yum, J.; Choi, J.-K.; Yeo, C.-Y.; Lee, K.-Y. PKC signaling inhibits osteogenic differentiation through the regulation of Msx2 function. Biochim. Biophys. Acta 2012, 1823, 1225–1232. [Google Scholar] [CrossRef]
- He, S.; Choi, Y.H.; Choi, J.-K.; Yeo, C.-Y.; Chun, C.; Lee, K.Y. Protein kinase a regulates the osteogenic activity of Osterix. J. Cell. Biochem. 2014, 115, 1808–1815. [Google Scholar] [CrossRef]
- Li, H.; Jeong, H.M.; Choi, Y.H.; Kim, J.H.; Choi, J.-K.; Yeo, C.-Y.; Jeong, H.G.; Jeong, T.C.; Chun, C.; Lee, K.Y. Protein kinase A phosphorylates Dlx3 and regulates the function of Dlx3 during osteoblast differentiation. J. Cell. Biochem. 2014, 115, 2004–2011. [Google Scholar] [CrossRef]
- Palazzo, E.; Kellett, M.D.; Cataisson, C.; Bible, P.W.; Bhattacharya, S.; Sun, H.-W.; Gormley, A.C.; Yuspa, S.H.; Morasso, M.I. A novel DLX3–PKC integrated signaling network drives keratinocyte differentiation. Cell Death Differ. 2017, 24, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, M.; Jia, X.; Hou, W.; Yang, J.; Zhao, H.; Wang, G.; Wang, J. The homeoprotein Msx1 cooperates with Pkn1 to prevent terminal differentiation in myogenic precursor cells. Biochimie 2019, 162, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Unterbrink, A.; Kadapakkam, S.; Dong, J.; Gu, T.T.; Dickson, J.; Chuang, H.-H.; MacDougall, M. Regulation of the cell type-specific dentin sialophosphoprotein gene expression in mouse odontoblasts by a novel transcription repressor and an activator CCAAT-binding factor. J. Biol. Chem. 2004, 279, 42182–42191. [Google Scholar] [CrossRef]
- He, W.-X.; Niu, Z.-Y.; Zhao, S.-L.; Jin, W.-L.; Gao, J.; Smith, A.J. TGF-β activated Smad signalling leads to a Smad3-mediated down-regulation of DSPP in an odontoblast cell line. Arch. Oral Biol. 2004, 49, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Ramachandran, A.; Peterson, M.C.; Hao, J.; Kolstø, A.-B.; Friedman, A.D.; George, A. The CCAAT enhancer-binding protein (C/EBP)β and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. J. Biol. Chem. 2004, 279, 45423–45432. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, H.; Jeong, B.; Oh, S.; Hur, S.; Lee, B.; Kim, S.; Nör, J.; Koh, J.; Hwang, Y. Transcriptional factor ATF6 is involved in odontoblastic differentiation. J. Dent. Res. 2014, 93, 483–489. [Google Scholar] [CrossRef]
- Zhou, C.; Yang, G.; Chen, M.; Wang, C.; He, L.; Xiang, L.; Chen, D.; Ling, J.; Mao, J.J. Lhx8 mediated Wnt and TGFβ pathways in tooth development and regeneration. Biomaterials 2015, 63, 35–46. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Q.; Wang, H.; Li, W.; Wang, F.; Wan, C.; Deng, S.; Chen, H.; Yin, Y.; Li, X.; et al. Klf5 mediates odontoblastic differentiation through regulating dentin-specific extracellular matrix gene expression during mouse tooth development. Sci. Rep. 2017, 7, 46746. [Google Scholar] [CrossRef]
- Martín-González, J.; Pérez-Pérez, A.; Cabanillas-Balsera, D.; Vilariño-García, T.; Sánchez-Margalet, V.; Segura-Egea, J.J. Leptin stimulates DMP-1 and DSPP expression in human dental pulp via MAPK 1/3 and PI3K signaling pathways. Arch. Oral Biol. 2018, 98, 126–131. [Google Scholar] [CrossRef]
- Deng, Z.; Yan, W.; Dai, X.; Chen, M.; Qu, Q.; Wu, B.; Zhao, W. N-cadherin regulates the odontogenic differentiation of dental pulp stem cells via β-catenin activity. Front. Cell Dev. Biol. 2021, 9, 661116. [Google Scholar] [CrossRef]
- Harland, R.M. A protein scaffold plays matchmaker for chordin. Cell 2008, 134, 718–719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gajos-Michniewicz, A.; Piastowska, A.W.; Russell, J.A.; Ochedalski, T. Follistatin as a potent regulator of bone metabolism. Biomarkers 2010, 15, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Khokha, M.K.; Hsu, D.; Brunet, L.J.; Dionne, M.; Harland, R.M. Gremlin is the BMP antagonist required for maintenance of SHH and FGF signals during limb patterning. Nat. Genet. 2003, 34, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Guha, U.; Gomes, W.A.; Kobayashic, T.; Pestell, R.G.; Kessler, J.A. In vivo evidence that BMP signaling is necessary for apoptosis in the mouse limb. Dev. Biol. 2002, 249, 108–120. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Zheng, Y.; Yuan, G.; Yang, G.; He, F.; Chen, Y. BMP activity is required for tooth development from the lamina to bud stage. J. Dent. Res. 2012, 91, 690–695. [Google Scholar] [CrossRef]
- Gong, Y.; Krakow, D.; Marcelino, J.M.; Wilkin, D.J.; Chitayat, D.; Babul-Hirji, R.; Hudgins, L.; Cremers, C.W.; Cremers, F.P.; Brunner, H.G.; et al. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat. Genet. 1999, 21, 302–304. [Google Scholar] [CrossRef]
- Brown, D.J.; Kim, T.B.; Petty, E.M.; Downs, C.A.; Martin, D.M.; Strouse, P.J.; Moroi, S.E.; Milunsky, J.M.; Lesperance, M.M. Autosomal dominant stapes ankylosis with broad thumbs and toes, hyperopia, and skeletal anomalies is caused by heterozygous nonsense and frameshift mutations in NOG, the gene encoding noggin. Am. J. Hum. Genet. 2002, 71, 618–624. [Google Scholar] [CrossRef]
- Moffett, S.P.; Dillon, K.A.; Yerges, L.M.; Goodrich, L.J.; Nestlerode, C.; Bunker, C.H.; Wheeler, V.W.; Patrick, A.L.; Zmuda, J.M. Identification and association analysis of single nucleotide polymorphisms in the human noggin (NOG) gene and osteoporosis phenotypes. Bone 2009, 44, 999–1002. [Google Scholar] [CrossRef]
- Gutiérrez-Prieto, S.J.; Torres-López, D.M.; García-Robayo, D.A.; Rey-Cubillos, J.A.; Gómez-Rodríguez, M. Clinical and molecular study of the NOG gene in families with mandibular micrognathism. Eur. J. Dent. 2021, 15, 746–754. [Google Scholar] [CrossRef]
- Warren, S.M.; Brunet, L.J.; Harland, R.M.; Economides, A.; Longaker, M.T. The BMP antagonist noggin regulates cranial suture fusion. Nature 2003, 422, 625–629. [Google Scholar] [CrossRef]
- Wijgerde, M.; Karp, S.; McMahon, J.; McMahon, A.P. Noggin antagonism of BMP4 signaling controls development of the axial skeleton in the mouse. Dev. Biol. 2005, 286, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.D.; Du, Z.; Pereira, R.C.; Kimble, R.B.; Economides, A.N.; Jorgetti, V.; Canalis, E. Skeletal overexpression of noggin results in osteopenia and reduced bone formation. Endocrinology 2003, 144, 1972–1978. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-B.; Li, Y.; Schneider, A.; Yu, W.; Rajendren, G.; Iqbal, J.; Yamamoto, M.; Alam, M.; Brunet, L.J.; Blair, H.C.; et al. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin-overexpressing mice. J. Clin. Investig. 2003, 112, 924–934. [Google Scholar] [CrossRef]
- Yuan, G.; Yang, G.; Zheng, Y.; Zhu, X.; Chen, Z.; Zhang, Z.; Chen, Y. The non-canonical BMP and Wnt/β-catenin signaling pathways orchestrate early tooth development. Development 2015, 142, 128–139. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Auh, Q.-S.; Kang, S.-K.; Kim, H.-J.; Lee, J.-W.; Noh, K.; Jang, J.-H.; Kim, E.-C. Combined effects of dentin sialoprotein and bone morphogenetic protein-2 on differentiation in human cementoblasts. Cell Tissue Res. 2014, 357, 119–132. [Google Scholar] [CrossRef]
- Gazzerro, E.; Gangji, V.; Canalis, E. Bone morphogenetic proteins induce the expression of noggin, which limits their activity in cultured rat osteoblasts. J. Clin. Investig. 1998, 102, 2106–2114. [Google Scholar] [CrossRef] [PubMed]
- Kantaputra, P.; Kaewgahya, M.; Hatsadaloi, A.; Vogel, P.; Kawasaki, K.; Ohazama, A.; Cairns, J.K. GREMLIN 2 mutations and dental anomalies. J. Dent. Res. 2015, 94, 1646–1652. [Google Scholar] [CrossRef]
- Vogel, P.; Liu, J.; Platt, K.A.; Read, R.W.; Thiel, M.; Vance, R.B.; Brommage, R. Malformation of incisor teeth in Grem2−/− mice. Veter. Pathol. 2014, 52, 224–229. [Google Scholar] [CrossRef]
- Nagatomo, K.J.; Tompkins, K.A.; Fong, H.; Zhang, H.; Foster, B.L.; Chu, E.Y.; Murakami, A.; Stadmeyer, L.; Canalis, E.; Somerman, M.J. Transgenic overexpression of gremlin results in developmental defects in enamel and dentin in mice. Connect. Tissue Res. 2008, 49, 391–400. [Google Scholar] [CrossRef]
- Wang, C.-L.; Xiao, F.; Wang, C.-D.; Zhu, J.-F.; Shen, C.; Zuo, B.; Wang, H.; Li, D.; Wang, X.-Y.; Feng, W.-J.; et al. Gremlin2 suppression increases the BMP-2-induced osteogenesis of human bone marrow-derived mesenchymal stem cells via the BMP-2/Smad/Runx2 signaling pathway. J. Cell. Biochem. 2016, 118, 286–297. [Google Scholar] [CrossRef]
- Liu, H.; Han, X.; Yang, H.; Cao, Y.; Zhang, C.; Du, J.; Diao, S.; Fan, Z. GREM1 inhibits osteogenic differentiation, senescence and BMP transcription of adipose-derived stem cells. Connect. Tissue Res. 2020, 62, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; He, Y.; Li, Y.; Shi, C.; Wei, Z.; Zhao, R.; Han, Y.; Pan, L.; Yang, J.; Hou, T.Z. Gremlin aggravates periodontitis via activating the NF-κB signaling pathway. J. Periodontol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Abreu, J.G.; Coffinier, C.; Larraín, J.; Oelgeschläger, M.; De Robertis, E. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene 2002, 287, 39–47. [Google Scholar] [CrossRef]
- Itoh, N.; Ohta, H. Secreted bone morphogenetic protein antagonists of the chordin family. Biomol. Concepts 2010, 1, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, S.; Sasai, Y.; Lu, B.; De Robertis, E.M. Dorsoventral patterning in xenopus: Inhibition of ventral signals by direct binding of chordin to BMP-4. Cell 1996, 86, 589–598. [Google Scholar] [CrossRef]
- Larrain, J.; Bachiller, D.; Lu, B.; Agius, E.; Piccolo, S.; De Robertis, E. BMP-binding modules in chordin: A model for signalling regulation in the extracellular space. Development 2000, 127, 821–830. [Google Scholar] [CrossRef]
- Nakayama, N.; Han, C.-Y.E.; Scully, S.; Nishinakamura, R.; He, C.; Zeni, L.; Yamane, H.; Chang, D.; Yu, D.; Yokota, T.; et al. A novel chordin-like protein inhibitor for bone morphogenetic proteins expressed preferentially in mesenchymal cell lineages. Dev. Biol. 2001, 232, 372–387. [Google Scholar] [CrossRef]
- Nakayama, N.; Han, C.-Y.E.; Cam, L.; Lee, J.I.; Pretorius, J.; Fisher, S.; Rosenfeld, R.; Scully, S.; Nishinakamura, R.; Duryea, D.; et al. A novel chordin-like BMP inhibitor, CHL2, expressed preferentially in chondrocytes of developing cartilage and osteoarthritic joint cartilage. Development 2004, 131, 229–240. [Google Scholar] [CrossRef]
- Stottmann, R.W.; Anderson, R.M.; Klingensmith, J. The BMP antagonists chordin and noggin have essential but redundant roles in mouse mandibular outgrowth. Dev. Biol. 2001, 240, 457–473. [Google Scholar] [CrossRef]
- Zhang, N.; Ferguson, C.M.; O’Keefe, R.J.; Puzas, J.E.; Rosier, R.N.; Reynolds, P.R. A role for the BMP antagonist chordin in endochondral ossification. J. Bone Miner. Res. 2002, 17, 293–300. [Google Scholar] [CrossRef]
- Ryan, A.K.; Goodship, J.A.; Wilson, D.I.; Philip, N.; Levy, A.; Seidel, H.; Schuffenhauer, S.; Oechsler, H.; Belohradsky, B.; Prieur, M.; et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: A European collaborative study. J. Med. Genet. 1997, 34, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Bachiller, D.; Klingensmith, J.; Shneyder, N.; Tran, U.; Anderson, R.; Rossant, J.; De Robertis, E.M. The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development 2003, 130, 3567–3578. [Google Scholar] [CrossRef] [PubMed]
- Webb, T.R.; Matarin, M.; Gardner, J.C.; Kelberman, D.; Hassan, H.; Ang, W.; Michaelides, M.; Ruddle, J.B.; Pennell, C.E.; Yazar, S.; et al. X-linked megalocornea caused by mutations in CHRDL1 identifies an essential role for ventroptin in anterior segment development. Am. J. Hum. Genet. 2012, 90, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Liu, Y.; Shu, G.; Chen, C.; Sullivan, D.A.; Kam, W.R.; Hann, S.; Fowler, M.; Warman, M.L. Ocular manifestations of chordin-like 1 knockout mice. Cornea 2020, 39, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Petryk, A.; Shimmi, O.; Jia, X.; Carlson, A.E.; Tervonen, L.; Jarcho, M.P.; O’Connor, M.B.; Gopalakrishnan, R. Twisted gastrulation and chordin inhibit differentiation and mineralization in MC3T3-E1 osteoblast-like cells. Bone 2005, 36, 617–626. [Google Scholar] [CrossRef]
- Liu, T.; Li, B.; Zheng, X.-F.; Jiang, S.-D.; Zhou, Z.-Z.; Xu, W.-N.; Zheng, H.-L.; Wang, C.-D.; Zhang, X.-L.; Jiang, L.-S. Chordin-like 1 improves osteogenesis of bone marrow mesenchymal stem cells through enhancing BMP4-SMAD pathway. Front. Endocrinol. 2019, 10, 00360. [Google Scholar] [CrossRef]
- World Health Organization. Dental Diseases and Oral Health. 2003. Available online: https://www.who.int/oral_health/publications/en/orh_fact_sheet.pdf (accessed on 19 November 2020).
- World Health Organization. Oral Health. 2017. Available online: https://www.who.int/publications/i/item/WHO-NMH-NHD-17.12 (accessed on 19 November 2020).
- Medina-Fernandez, I.; Celiz, A.D. Acellular biomaterial strategies for endodontic regeneration. Biomater. Sci. 2018, 7, 506–519. [Google Scholar] [CrossRef]
- GBD 2017 Oral Disorders Collaborators; Bernabe, E.; Marcenes, W.; Hernandez, C.R.; Bailey, J.; Abreu, L.G.; Alipour, V.; Amini, S.; Arabloo, J.; Arefi, Z.; et al. Global, regional, and national levels and trends in burden of oral conditions from 1990 to 2017: A systematic analysis for the global burden of disease 2017 study. J. Dent. Res. 2020, 99, 362–373. [Google Scholar] [CrossRef]
- Kazeminia, M.; Abdi, A.; Shohaimi, S.; Jalali, R.; Vaisi-Raygani, A.; Salari, N.; Mohammadi, M. Dental caries in primary and permanent teeth in children’s worldwide, 1995 to 2019: A systematic review and meta-analysis. Head Face Med. 2020, 16, 22. [Google Scholar] [CrossRef]
- Soares, D.G.; Bordini, E.A.F.; Swanson, W.B.; Costa, C.A.D.S.; Bottino, M.C. Platform technologies for regenerative endodontics from multifunctional biomaterials to tooth-on-a-chip strategies. Clin. Oral Investig. 2021, 25, 4749–4779. [Google Scholar] [CrossRef]
- Burke, F.J.T.; Lucarotti, P.S.K. How long do direct restorations placed within the general dental services in England and Wales survive? Br. Dent. J. 2008, 206, E2. [Google Scholar] [CrossRef] [PubMed]
- Tziafas, D.; Smith, A.; Lesot, H. Designing new treatment strategies in vital pulp therapy. J. Dent. 1999, 28, 77–92. [Google Scholar] [CrossRef]
- Goldberg, M.; Smith, A.J. Cells and extracellular matrices of dentin and pulp: A biological basis for repair and tissue engineering. Crit. Rev. Oral Biol. Med. 2004, 15, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.E.; Garcia-Godoy, F.; Hargreaves, K.M. Regenerative endodontics: A review of current status and a call for action. J. Endod. 2007, 33, 377–390. [Google Scholar] [CrossRef]
- Langer, R.; Vacanti, J.P. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. SHED: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef]
- Sonoyama, W.; Liu, Y.; Yamaza, T.; Tuan, R.S.; Wang, S.; Shi, S.; Huang, G.T. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: A pilot study. J. Endod. 2008, 34, 166–171. [Google Scholar] [CrossRef]
- Nakashima, M.; Iohara, K.; Murakami, M.; Nakamura, H.; Sato, Y.; Ariji, Y.; Matsushita, K. Pulp regeneration by transplantation of dental pulp stem cells in pulpitis: A pilot clinical study. Stem Cell Res. Ther. 2017, 8, 61. [Google Scholar] [CrossRef]
- Xuan, K.; Li, B.; Guo, H.; Sun, W.; Kou, X.; He, X.; Zhang, Y.; Sun, J.; Liu, A.; Liao, L.; et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci. Transl. Med. 2018, 10, eaaf3227. [Google Scholar] [CrossRef]
- Nakashima, M. Induction of dentin formation on canine amputated pulp by recombinant human bone morphogenetic proteins (BMP)-2 and -4. J. Dent. Res. 1994, 73, 1515–1522. [Google Scholar] [CrossRef]
- Kirker-Head, C. Potential applications and delivery strategies for bone morphogenetic proteins. Adv. Drug Deliv. Rev. 2000, 43, 65–92. [Google Scholar] [CrossRef]
- Goldberg, M.; Six, N.; Decup, F.; Buch, D.; Majd, E.S.; Lasfargues, J.-J.; Salih, E.; Stanislawski, L. Application of bioactive molecules in pulp-capping situations. Adv. Dent. Res. 2001, 15, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Iohara, K.; Nakashima, M.; Ito, M.; Ishikawa, M.; Nakasima, A.; Akamine, A. Dentin regeneration by dental pulp stem cell therapy with recombinant human bone morphogenetic protein 2. J. Dent. Res. 2004, 83, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Ogawa, M.; Hata, Y.; Bessho, K. Acceleration effect of human recombinant bone morphogenetic protein-2 on differentiation of human pulp cells into odontoblasts. J. Endod. 2004, 30, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Chakka, L.R.J.; Vislisel, J.; Vidal, C.D.M.P.; Biz, M.T.; Salem, A.K.; Cavalcanti, B.N. Application of BMP-2/FGF-2 gene–activated scaffolds for dental pulp capping. Clin. Oral Investig. 2020, 24, 4427–4437. [Google Scholar] [CrossRef]
- Um, I.-W.; Ku, J.-K.; Kim, Y.-K.; Lee, B.-K.; Leem, D.H. Histological review of demineralized dentin matrix as a carrier of rhBMP-2. Tissue Eng. Part B Rev. 2020, 26, 284–293. [Google Scholar] [CrossRef]
- Wang, W.; Dang, M.; Zhang, Z.; Hu, J.; Eyster, T.W.; Ni, L.; Ma, P.X. Dentin regeneration by stem cells of apical papilla on injectable nanofibrous microspheres and stimulated by controlled BMP-2 release. Acta Biomater. 2016, 36, 63–72. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Summary of Safety and Effectiveness Data; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2007. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf5/P050053b.pdf (accessed on 30 December 2018).
- King, G.N.; Hughes, F.J. Bone morphogenetic protein-2 stimulates cell recruitment and cementogenesis during early wound healing. J. Clin. Periodontol. 2001, 28, 465–475. [Google Scholar] [CrossRef]
- King, G.N.; Cochrant, D.L. Factors that modulate the effects of bone morphogenetic protein-induced periodontal regeneration: A critical review. J. Periodontol. 2002, 73, 925–936. [Google Scholar] [CrossRef]
- Jones, A.A.; Buser, D.; Schenk, R.K.; Wozney, J.; Cochran, D.L. The effect of rhBMP-2 around endosseous implants with and without membranes in the canine model. J. Periodontol. 2006, 77, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Jung, R.E.; Weber, F.E.; Thoma, D.S.; Ehrbar, M.; Cochran, D.L.; Hämmerle, C.H.F. Bone morphogenetic protein-2 enhances bone formation when delivered by a synthetic matrix containing hydroxyapatite/tricalciumphosphate. Clin. Oral Implant. Res. 2008, 19, 188–195. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, R.M.; Spin-Neto, R.; Junior, E.M.; Pereira, L.A.V.D.; Wikesjo, U.M.; Susin, C. Alveolar ridge and maxillary sinus augmentation using rhBMP-2: A systematic review. Clin. Implant Dent. Relat. Res. 2013, 17, e192–e201. [Google Scholar] [CrossRef] [PubMed]
- Talley, A.D.; Kalpakci, K.N.; Shimko, D.A.; Zienkiewicz, K.J.; Cochran, D.L.; Guelcher, S.A. Effects of recombinant human bone morphogenetic protein-2 dose and ceramic composition on new bone formation and space maintenance in a canine mandibular ridge saddle defect model. Tissue Eng. Part A 2016, 22, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Batool, F.; Strub, M.; Petit, C.; Bugueno, I.M.; Bornert, F.; Clauss, F.; Huck, O.; Kuchler-Bopp, S.; Benkirane-Jessel, N. Periodontal tissues, maxillary jaw bone, and tooth regeneration approaches: From animal models analyses to clinical applications. Nanomaterials 2018, 8, 337. [Google Scholar] [CrossRef]
- Boda, S.K.; Almoshari, Y.; Wang, H.; Wang, X.; Reinhardt, R.A.; Duan, B.; Wang, D.; Xie, J. Mineralized nanofiber segments coupled with calcium-binding BMP-2 peptides for alveolar bone regeneration. Acta Biomater. 2018, 85, 282–293. [Google Scholar] [CrossRef]
- Le, B.; Too, J.; Tan, T.; Smith, R.; Nurcombe, V.; Cool, S.; Yu, N. Application of a BMP2-binding heparan sulphate to promote periodontal regeneration. Eur. Cells Mater. 2021, 42, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, M.; Reddi, A.H. The application of bone morphogenetic proteins to dental tissue engineering. Nat. Biotechnol. 2003, 21, 1025–1032. [Google Scholar] [CrossRef]
- Nakashima, M. Bone morphogenetic proteins in dentin regeneration for potential use in endodontic therapy. Cytokine Growth Factor Rev. 2005, 16, 369–376. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Kim, S.-Y.; Park, S.-H.; Kim, J.-J.; Jang, J.-H.; Kim, E.-C. Effects of recombinant dentin sialoprotein in dental pulp cells. J. Dent. Res. 2012, 91, 407–412. [Google Scholar] [CrossRef]
- Ozer, A.; Yuan, G.H.; Yang, G.B.; Wang, F.; Li, W.; Yang, Y.; Guo, F.; Gao, Q.P.; Shoff, L.; Chen, Z.; et al. Domain of dentine sialoprotein mediates proliferation and differentiation of human periodontal ligament stem cells. PLoS ONE 2013, 8, e81655. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Yuan, G.; Luo, D.; Zhang, L.; Lin, H.; Liu, H.; Chen, L.; Yang, G.; Chen, S.; Chen, Z. The Dentin Sialoprotein (DSP) domain regulates dental mesenchymal cell differentiation through a novel surface receptor. Sci. Rep. 2016, 6, 29666. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, L.; Chen, Z.; Wu, L.; Feng, J.; Wang, F.; Shoff, L.; Li, X.; Donly, K.J.; MacDougall, M.; et al. Dentin sialoprotein facilitates dental mesenchymal cell differentiation and dentin formation. Sci. Rep. 2017, 7, 300. [Google Scholar] [CrossRef] [PubMed]
- James, A.W.; Lachaud, G.; Shen, J.; Asatrian, G.; Nguyen, V.; Zhang, X.; Ting, K.; Soo, C. A review of the clinical side effects of bone morphogenetic protein-2. Tissue Eng. Part B Rev. 2016, 22, 284–297. [Google Scholar] [CrossRef]
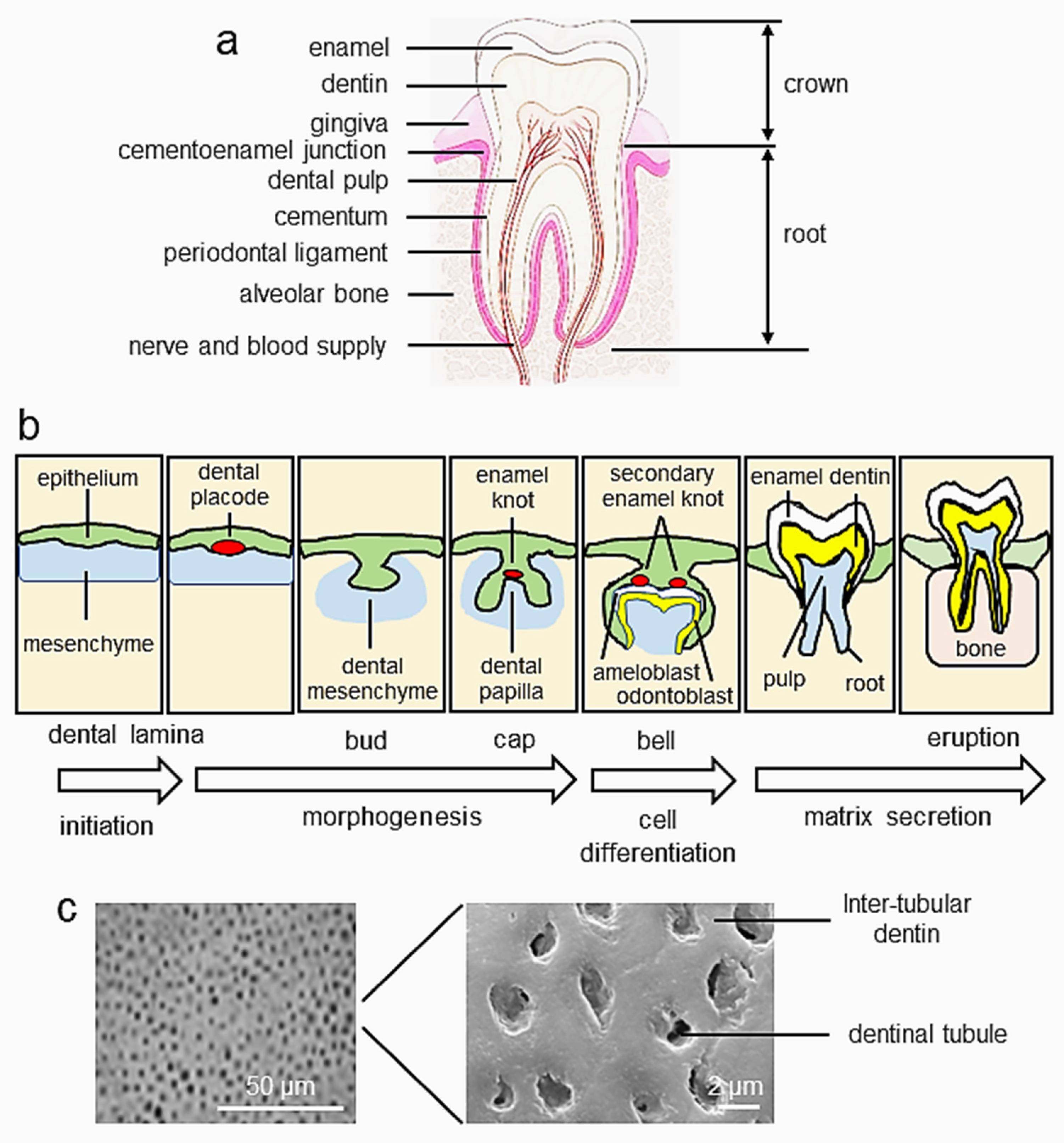

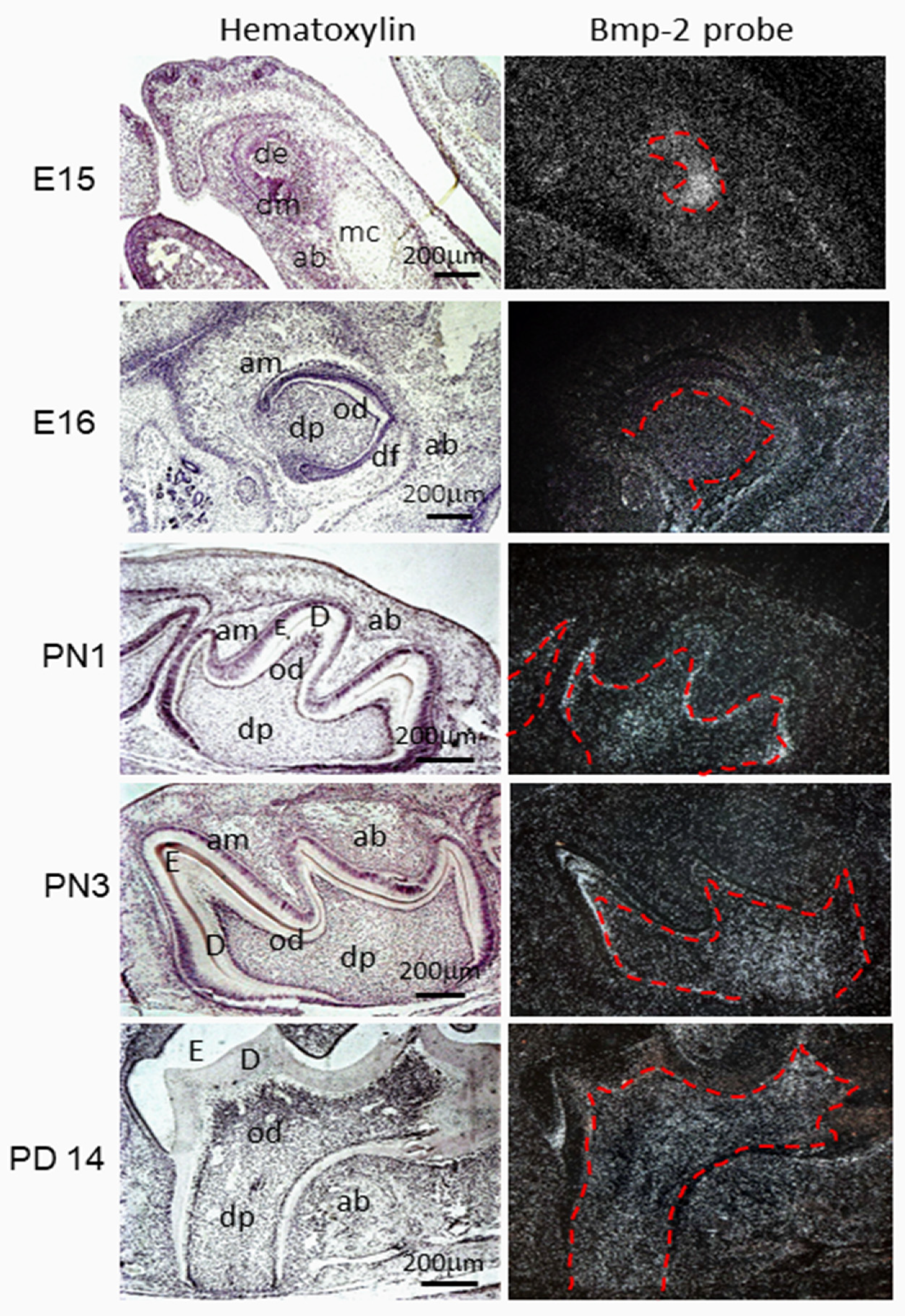
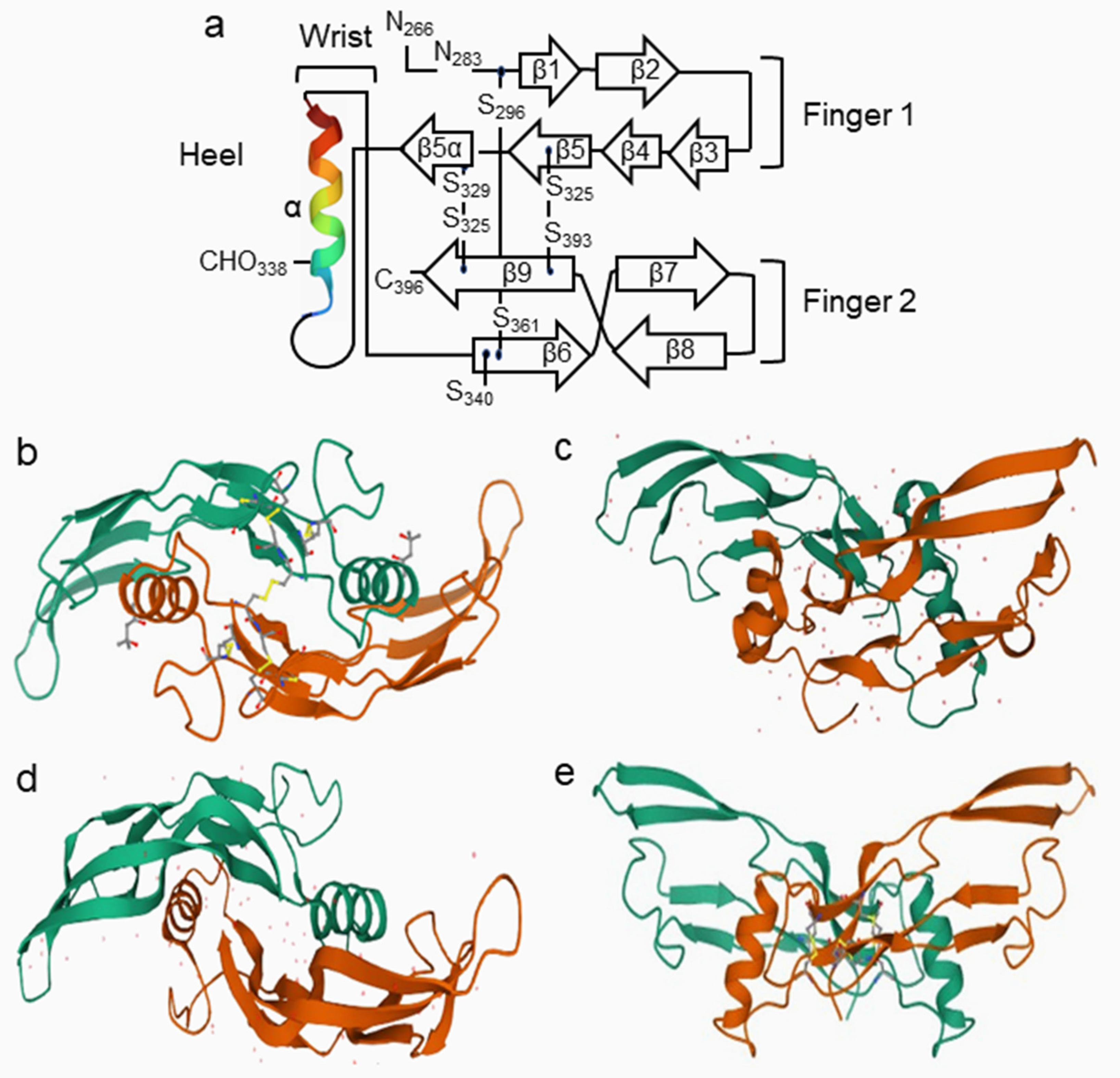
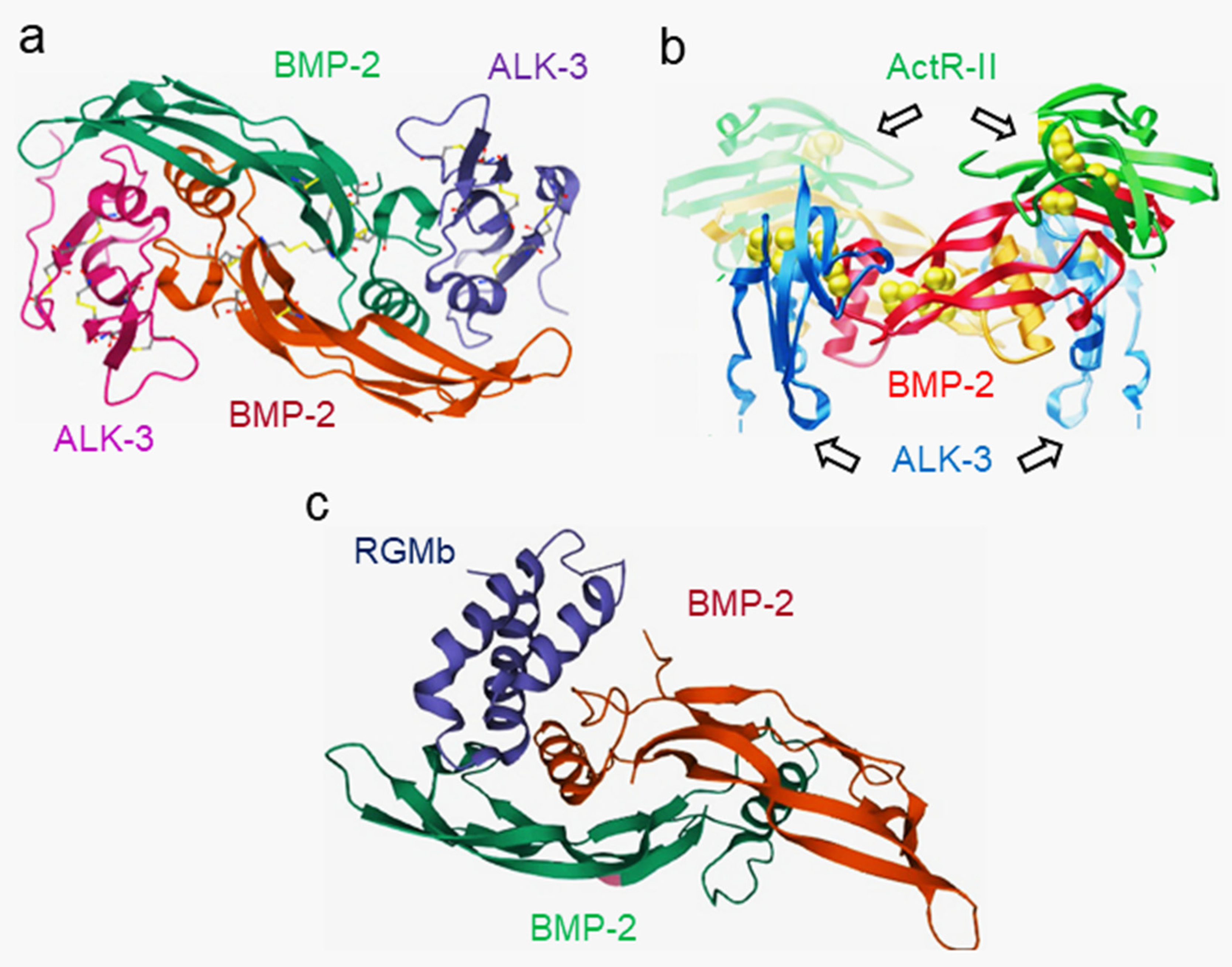

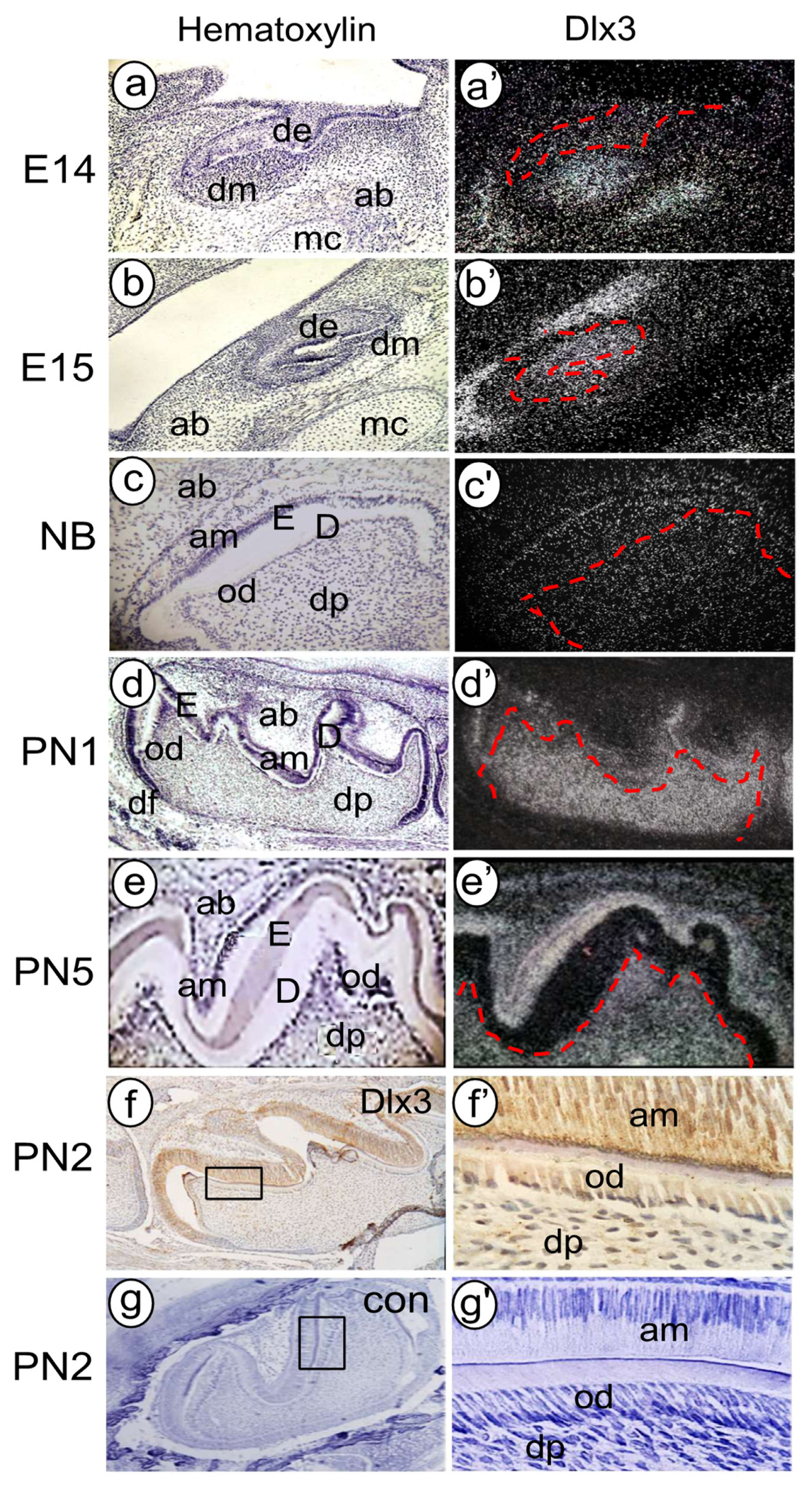
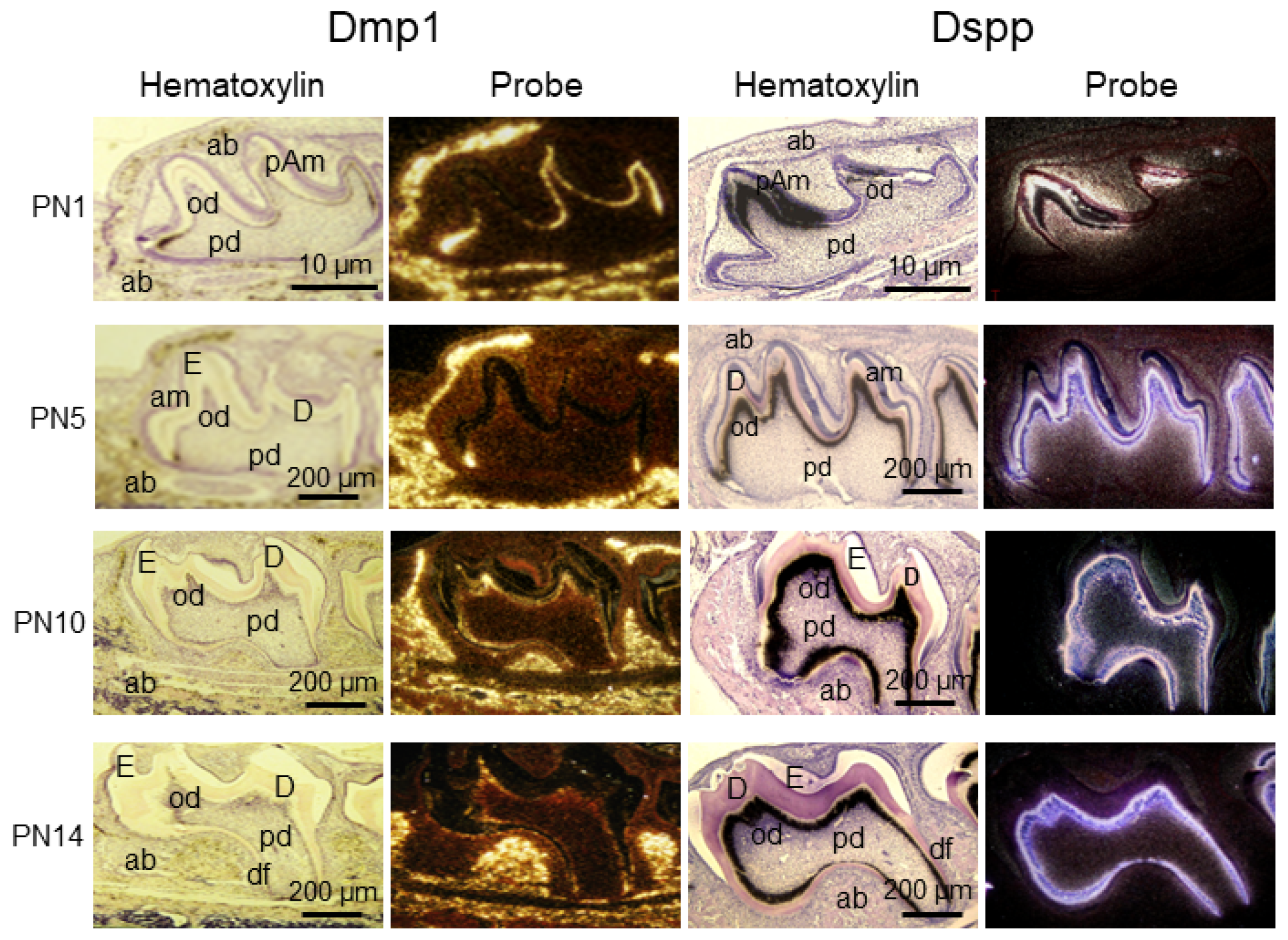
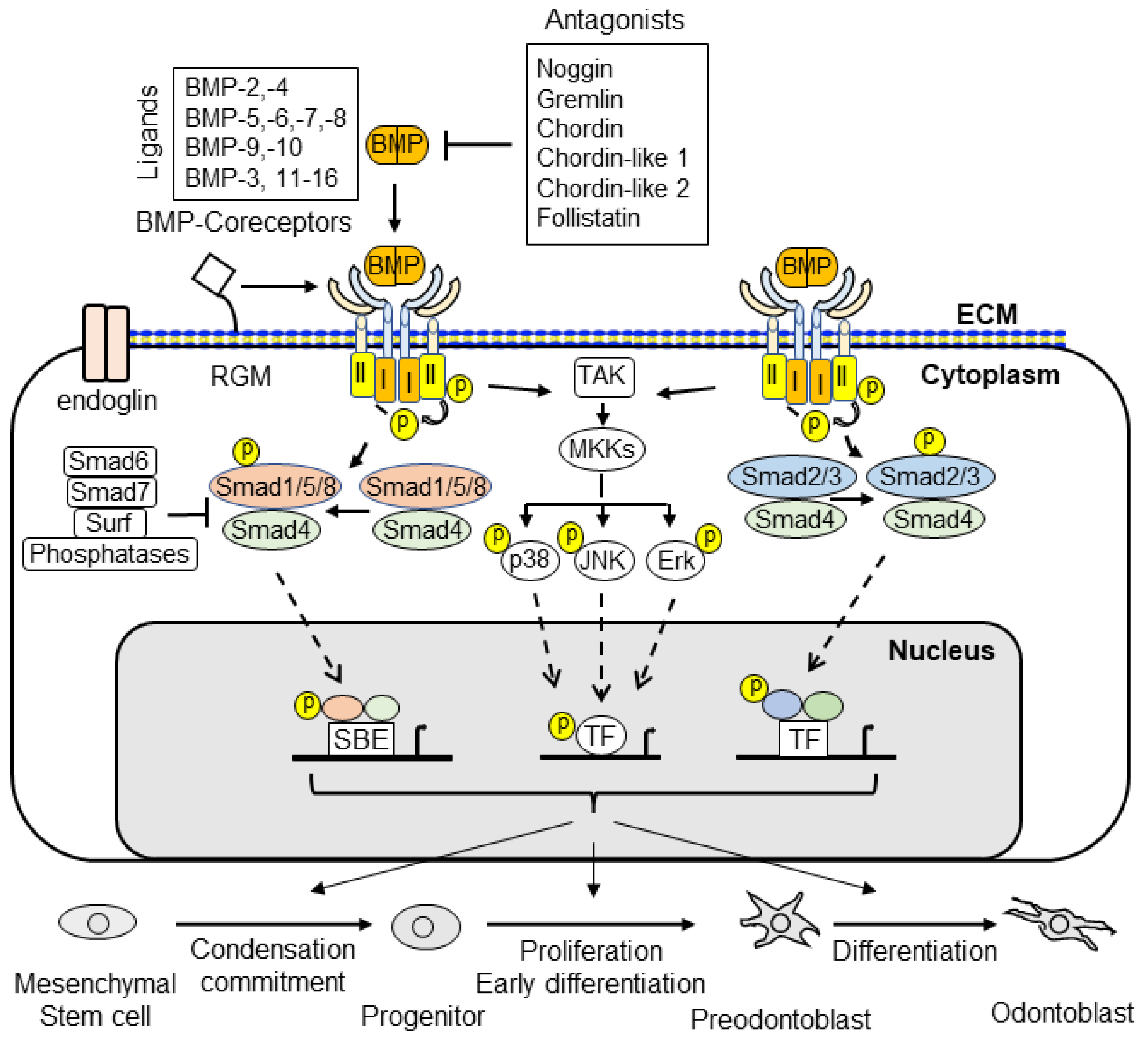

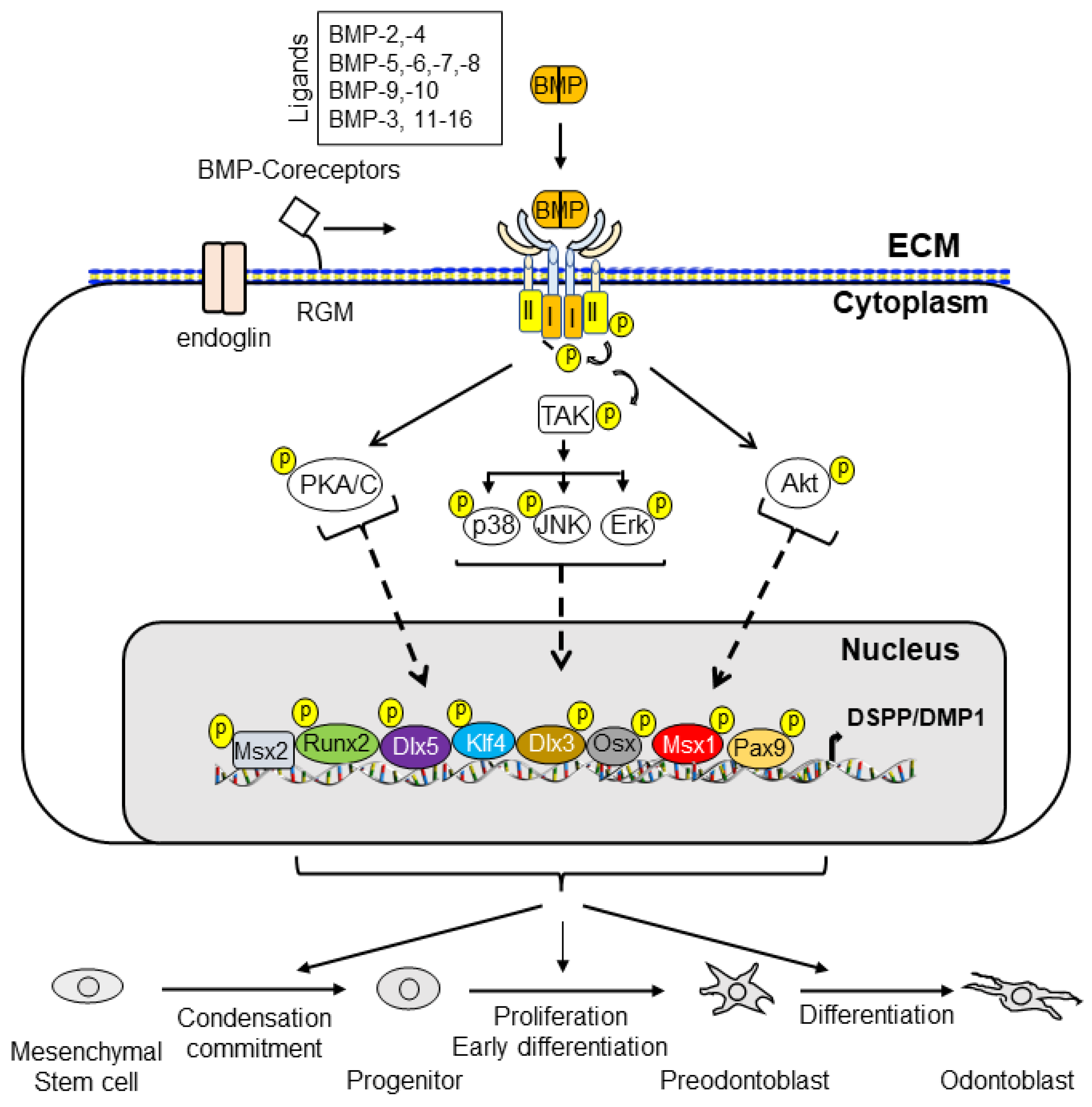

| Genes | Initiate | Bud | Cap | Bell | Dif. & Sec. | Root | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DE | DM | DE | DM | DE | DM | DE | DM | DE | DM | DE | DM | |
| BMP−2 | +/− | − | + | + | + | +/− | + | + | + | + | +/− | + |
| BMP−3 | n/a | n/a | +/− | n/a | +/− | + | n/a | + | n/a | + | n/a | + |
| BMP−4 | +/− | + | +/− | + | + | + | +/− | + | +/− | + | +/− | + |
| BMP−5 | − | − | − | − | − | − | +/− | − | + | +/− | + | − |
| BMP−6 | − | − | + | + | + | + | − | + | − | + | + | + |
| BMP−7 | + | +/− | + | − | + | − | + | + | + | + | + | + |
| Alk−1 | n/a | n/a | + | + | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Alk−2 | n/a | − | + | + | n/a | n/a | n/a | n/a | + | + | + | + |
| Alk−3 | n/a | n/a | + | n/a | + | + | + | + | + | + | + | + |
| Alk−5 | n/a | n/a | + | + | + | + | + | + | + | + | n/a | n/a |
| Alk−6 | n/a | n/a | + | + | + | − | + | + | + | + | + | + |
| Bmpr−II | n/a | n/a | + | + | + | + | + | + | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Goldman, G.; MacDougall, M.; Chen, S. BMP Signaling Pathway in Dentin Development and Diseases. Cells 2022, 11, 2216. https://doi.org/10.3390/cells11142216
Liu M, Goldman G, MacDougall M, Chen S. BMP Signaling Pathway in Dentin Development and Diseases. Cells. 2022; 11(14):2216. https://doi.org/10.3390/cells11142216
Chicago/Turabian StyleLiu, Mengmeng, Graham Goldman, Mary MacDougall, and Shuo Chen. 2022. "BMP Signaling Pathway in Dentin Development and Diseases" Cells 11, no. 14: 2216. https://doi.org/10.3390/cells11142216
APA StyleLiu, M., Goldman, G., MacDougall, M., & Chen, S. (2022). BMP Signaling Pathway in Dentin Development and Diseases. Cells, 11(14), 2216. https://doi.org/10.3390/cells11142216






