The Diagnostic Value of Circulating Cell-Free HPV DNA in Plasma from Cervical Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Extraction and HPV Analyses on Cervical Tissue
2.2. Blood Sample Preparation
2.3. ddPCR Analysis for ccfHPV DNA Detection
2.4. Statistics
3. Results
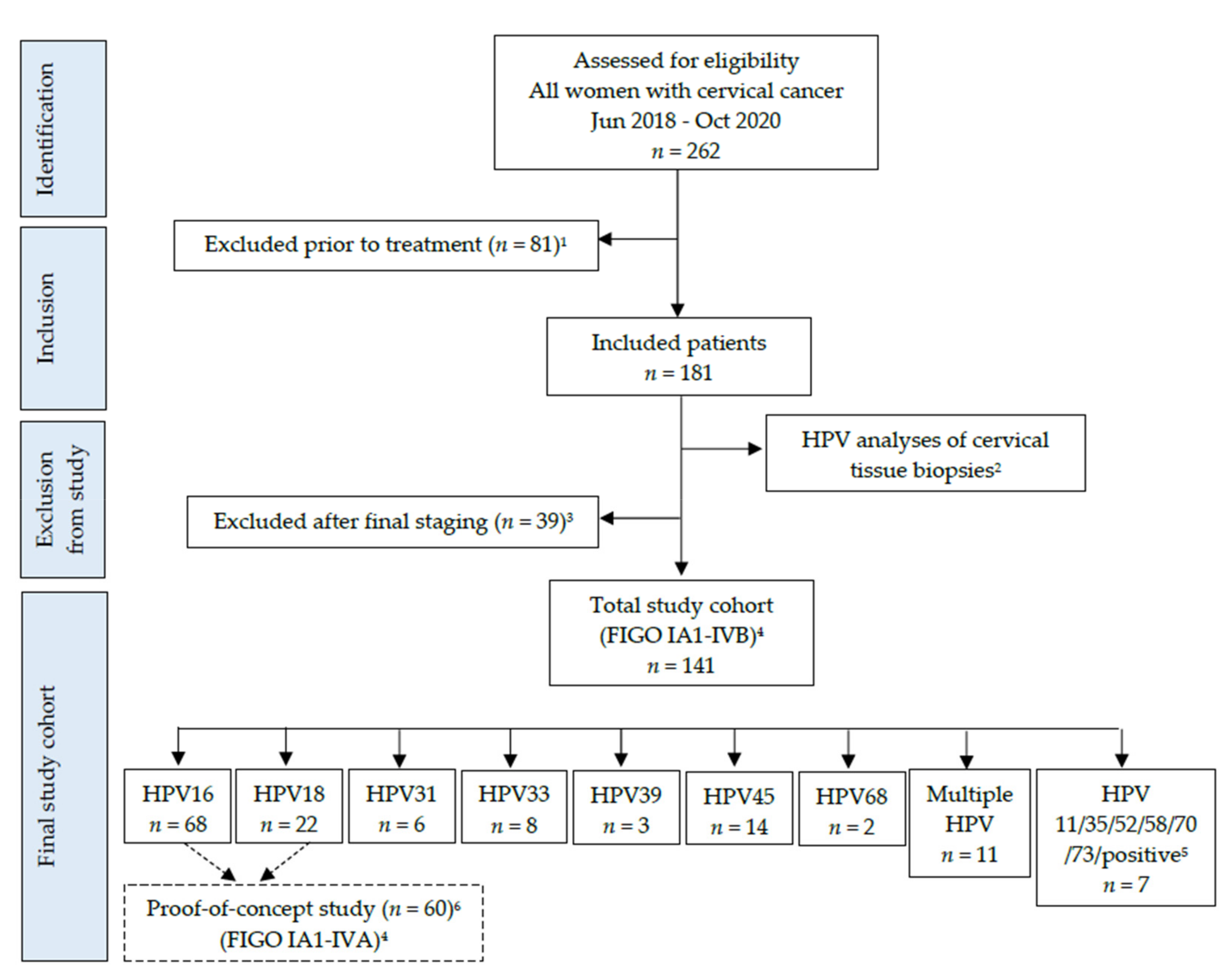
3.1. Patient Characteristics, Histology, and HPV Detection in Cervical Tissue Biopsies
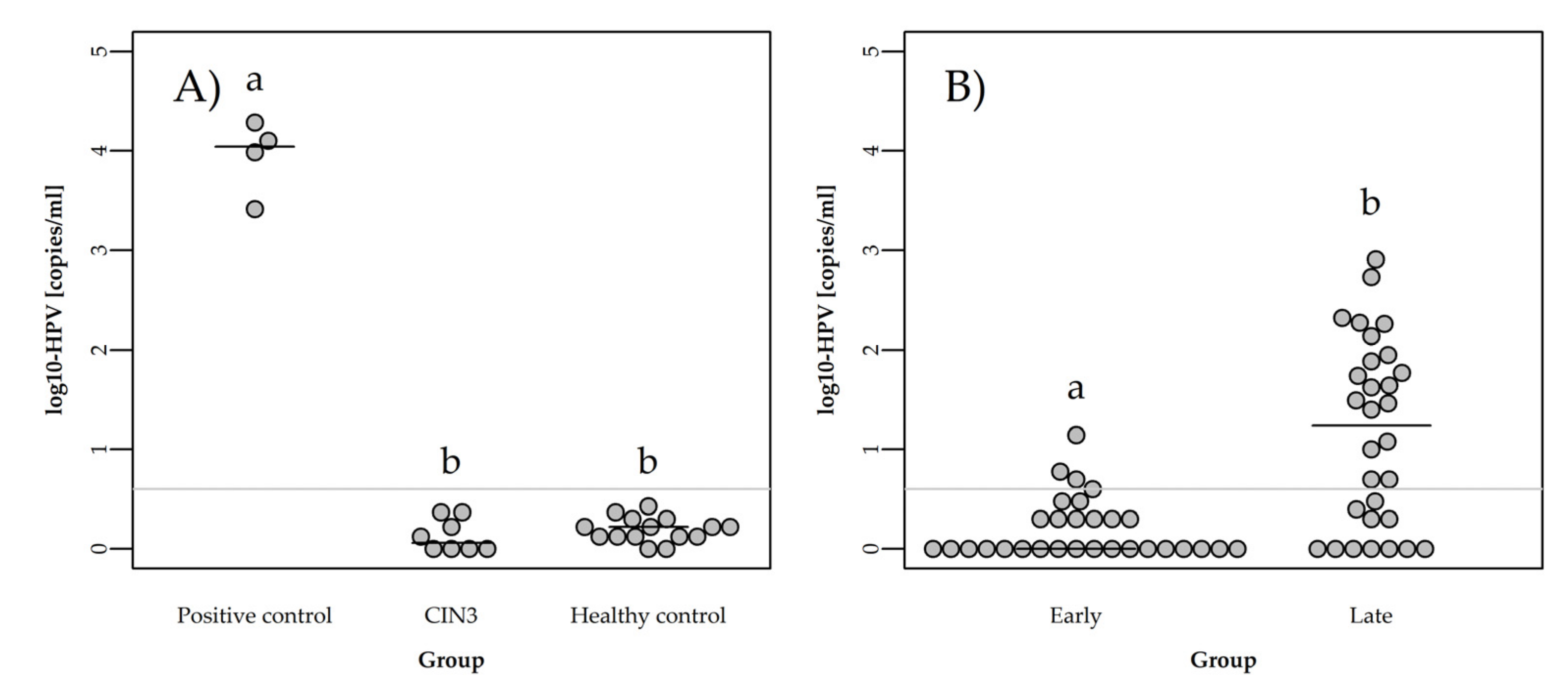
3.2. Detection of ccfHPV16 and ccfHPV18 DNA Using ddPCR
3.3. Quantification of ccfHPV DNA
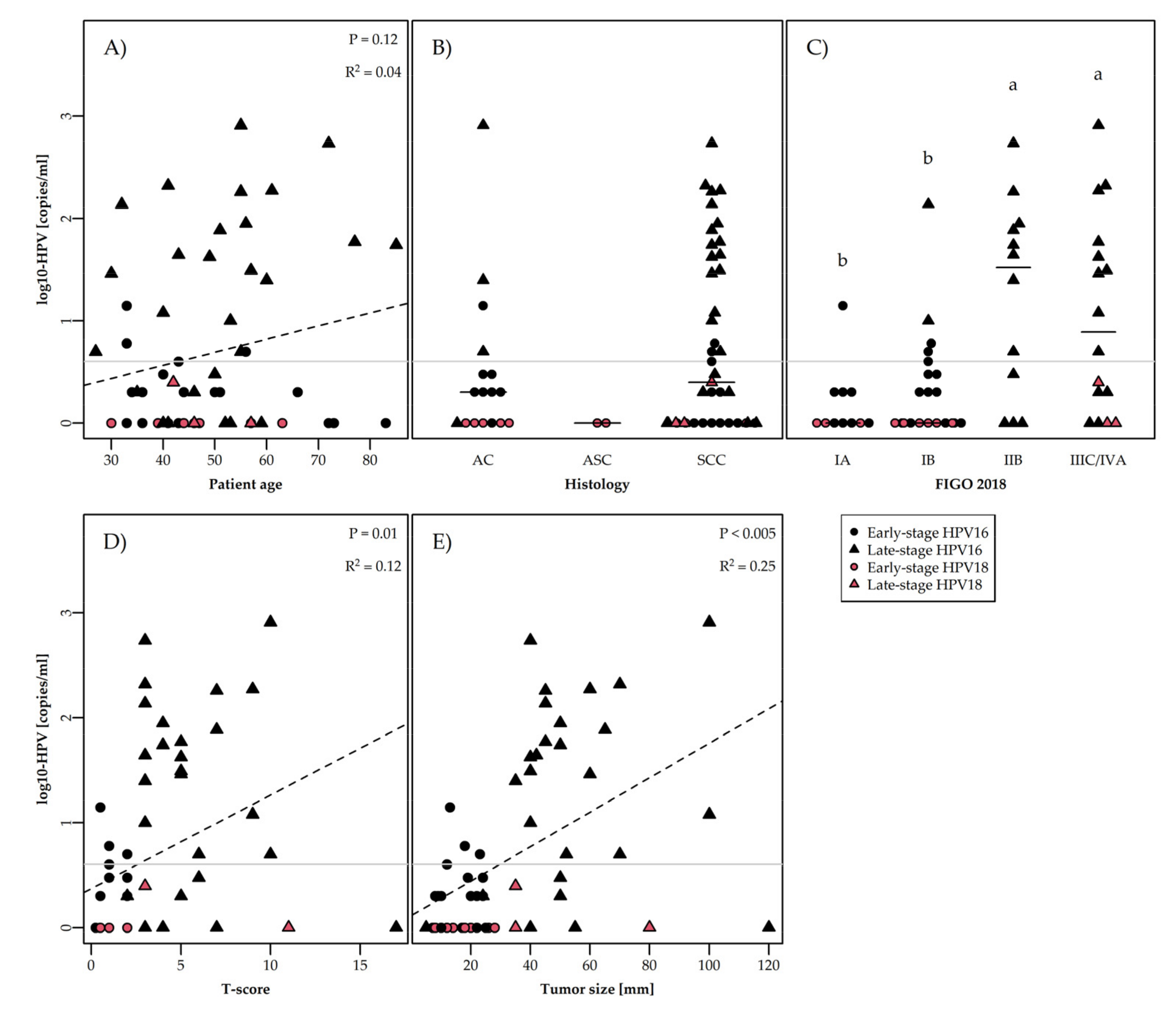
3.4. ccfHPV DNA Level Correlated to Clinical and Biological Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pretorius, R.; Semrad, N.; Watring, W.; Fotheringham, N. Presentation of cervical cancer. Gynecol. Oncol. 1991, 42, 48–53. [Google Scholar] [CrossRef]
- Shapley, M.; Jordan, J.; Croft, P.R. A systematic review of postcoital bleeding and risk of cervical cancer. Br. J. Gen. Pract. 2006, 56, 453–460. [Google Scholar]
- Van Schalkwyk, S.L.; Maree, J.E.; Wright, S.C. Cervical cancer: The route from signs and symptoms to treatment in South Africa. Reprod. Health Matters 2008, 16, 9–17. [Google Scholar] [CrossRef]
- Viikki, M.; Pukkala, E.; Hakama, M. Bleeding symptoms and subsequent risk of gynecological and other cancers. Acta Obstet. Gynecol. Scand. 1998, 77, 564–569. [Google Scholar]
- Yu, C.K.; Chiu, C.; McCormack, M.; Olaitan, A. Delayed diagnosis of cervical cancer in young women. J. Obstet. Gynaecol. 2005, 25, 367–370. [Google Scholar] [CrossRef]
- O’Dowd, T.C.; Parker, S.; Kelly, A. Women’s experiences of general practitioner management of their vaginal symptoms. Br. J. Gen. Pract. 1996, 46, 415–418. [Google Scholar]
- Stapley, S.; Hamilton, W. Gynaecological symptoms reported by young women: Examining the potential for earlier diagnosis of cervical cancer. Fam. Pract. 2011, 28, 592–598. [Google Scholar] [CrossRef] [Green Version]
- Qyinn, M.A.; Benedet, J.L.; Odicino, F.; Maisonneuve, P.; Beller, U.; Creasman, W.T.; Heintz, A.P.M.; Ngan, H.Y.S.; Pecorelli, S. Carcinoma of the cervix uteri. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int. J. Gynaecol. Obstet. 2006, 95 (Suppl. 1), S43–S103. [Google Scholar]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [Green Version]
- Pornthanakasem, W.; Shotelersuk, K.; Termrungruanglert, W.; Voravud, N.; Niruthisard, S.; Mutirangura, A. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer 2001, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Mazurek, A.M.; Fiszer-Kierzkowska, A.; Rutkowski, T.; Składowski, K.; Pierzyna, M.; Ścieglińska, D.; Woźniak, G.; Głowacki, G.; Kawczyński, R.; Małusecka, E. Optimization of circulating cell-free DNA recovery for KRAS mutation and HPV detection in plasma. Cancer Biomark. 2013, 13, 385–394. [Google Scholar] [CrossRef]
- Gu, Y.; Wan, C.; Qiu, J.; Cui, Y.; Jiang, T.; Zhuang, Z. Circulating HPV cDNA in the blood as a reliable biomarker for cervical cancer: A meta-analysis. PLoS ONE 2020, 15, e0224001. [Google Scholar] [CrossRef]
- Chan, C.K.; Aimagambetova, G.; Ukybassova, T.; Kongrtay, K.; Azizan, A. Human papillomavirus infection and cervical cancer: Epidemiology, screening, and vaccination-review of current perspectives. J. Oncol. 2019, 2019, 3257939. [Google Scholar] [CrossRef]
- Kessis, T.D.; Slebos, R.J.; Nelson, W.G.; Kastan, M.B.; Plunkett, B.S.; Han, S.M.; Lorincz, A.T.; Hedrick, L.; Cho, K.R. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 1993, 90, 3988–3992. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, S.L.; Stremlau, M.; He, X.; Basile, J.R.; Münger, K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 2001, 75, 7583–7591. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.; Allenhoffmann, B.L.; Lambert, P.F. Integration of human papillomavirus type-16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995, 69, 2989–2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Wang, W.; Si, M.; Han, L.; Gao, Q.; Luo, A.; Li, Y.; Lu, Y.; Wang, S.; Ma, D. The physical state of HPV16 infection and its clinical significance in cancer precursor lesion and cervical carcinoma. J. Cancer Res. Clin. Oncol. 2008, 134, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Peitsaro, P.; Johansson, B.; Syrjanen, S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 2002, 40, 886–891. [Google Scholar] [CrossRef] [Green Version]
- Pett, M.R.; Herdman, M.T.; Palmer, R.D.; Yeo, G.S.; Shivji, M.K.; Stanley, M.A.; Coleman, N. Selection of cervical keratinocytes containing integrated HPV16 associates with episome loss and an endogenous antiviral response. Proc. Natl. Acad. Sci. USA 2006, 103, 3822–3827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vernon, S.D.; Unger, E.R.; Miller, D.L.; Lee, D.R.; Reeves, W.C. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int. J. Cancer 1997, 74, 50–56. [Google Scholar] [CrossRef]
- Cullen, A.P.; Reid, R.; Campion, M.; Lörincz, A.T. Analysis of the physical state of different human papillomavirus dnas in intraepithelial and invasive cervical neoplasm. J. Virol. 1991, 65, 606–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, K.; Herrington, C.S.; Stickland, J.E.; Evans, M.F.; McGee, J.O. Episomal and integrated human papillomavirus in cervical neoplasia shown by nonisotopic insitu hybridization. J. Clin. Pathol. 1991, 44, 990–996. [Google Scholar] [CrossRef] [Green Version]
- Kristiansen, E.; Jenkins, A.; Holm, R. Coexistence of Episomal and Integrated Hpv16 DNA in Squamous-Cell Carcinoma of the Cervix. J. Clin. Pathol. 1994, 47, 253–256. [Google Scholar] [CrossRef] [Green Version]
- Matsukura, T.; Koi, S.; Sugase, M. Both episomal and integrated forms of human papillomavirus type-16 are involved in invasive cervical cancers. Virology 1989, 172, 63–72. [Google Scholar] [CrossRef]
- Unger, E.R.; Vernon, S.D.; Thoms, W.W.; Nisenbaum, R.; Spann, C.O.; Horowitz, I.R.; Icenogle, J.P.; Reeves, W.C. Human papillomavirus and disease-free survival in figo stage Ib cervical-cancer. J. Infect. Dis. 1995, 172, 1184–1190. [Google Scholar] [CrossRef]
- Jeannot, E.; Becette, V.; Campitelli, M.; Calméjane, M.A.; Lappartient, E.; Ruff, E.; Saada, S.; Holmes, A.; Bellet, D.; Sastre-Garau, X. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J. Pathol. Clin. Res. 2016, 2, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.M.; Pai, S.I.; Rha, S.H.; Hildesheim, A.; Kurman, R.J.; Schwartz, P.E.; Mortel, R.; McGowan, L.; Greenberg, M.D.; Barnes, W.A.; et al. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol. Biomark. Prev. 2002, 11, 3–6. [Google Scholar]
- Liu, V.W.; Tsang, P.; Yip, A.; Ng, T.Y.; Wong, L.C.; Ngan, H.Y. Low incidence of HPV DNA in sera of pretreatment cervical cancer patients. Gynecol. Oncol. 2001, 82, 269–272. [Google Scholar] [CrossRef]
- Shimada, T.; Yamaguchi, N.; Nishida, N.; Yamasaki, K.; Miura, K.; Katamine, S.; Masuzaki, H. Human Papillomavirus DNA in Plasma of Patients with HPV16 DNA-positive Uterine Cervical Cancer. Jpn. J. Clin. Oncol. 2010, 40, 420–424. [Google Scholar] [CrossRef]
- Cheung, T.H.; Yim, S.F.; Yu, M.Y.; Worley, M.J., Jr.; Fiascone, S.J.; Chiu, R.W.; Lo, K.W.; Siu, N.S.; Wong, M.C.; Yeung, A.C.; et al. Liquid biopsy of HPV DNA in cervical cancer. J. Clin. Virol. 2019, 114, 32–36. [Google Scholar] [CrossRef]
- Rungkamoltip, P.; Temisak, S.; Piboonprai, K.; Japrung, D.; Thangsunan, P.; Chanpanitkitchot, S.; Chaowawanit, W.; Chandeying, N.; Tangjitgamol, S.; Iempridee, T. Rapid and ultrasensitive detection of circulating human papillomavirus E7 cell-free DNA as a cervical cancer biomarker. Exp. Biol. Med. 2021, 246, 654–666. [Google Scholar] [CrossRef]
- Kang, Z.; Stevanović, S.; Hinrichs, C.S.; Cao, L. Circulating cell-free DNA for metastatic cervical cancer detection, genotyping, and monitoring. Clin. Cancer Res. 2017, 23, 6856–6862. [Google Scholar] [CrossRef] [Green Version]
- Clementi, M.; Bagnarelli, P. Are three generations of quantitative molecular methods sufficient in medical virology? Brief review. New Microbiol. 2015, 38, 437–441. [Google Scholar]
- Sathish, N.; Abraham, P.; Peedicayil, A.; Sridharan, G.; John, S.; Shaji, R.V.; Chandy, G. HPV DNA in plasma of patients with cervical carcinoma. J. Clin. Virol. 2004, 31, 204–209. [Google Scholar] [CrossRef]
- Kay, P.; Allan, B.; Denny, L.; Hoffman, M.; Williamson, A.L. Detection of HPV 16 and HPV 18 DNA in the blood of patients with cervical cancer. J. Med. Virol. 2005, 75, 435–439. [Google Scholar] [CrossRef]
- Widschwendter, A.; Blassnig, A.; Wiedemair, A.; Müller-Holzner, E.; Müller, H.M.; Marth, C. Human papillomavirus DNA in sera of cervical cancer patients as tumor marker. Cancer Lett. 2003, 202, 231–239. [Google Scholar] [CrossRef]
- Yang, H.J.; Liu, V.W.S.; Tsang, P.C.K.; Yip, A.M.W.; Tam, K.F.; Wong, L.C.; Ng, T.Y.; Ngan, H.Y.S. Quantification of human papillomavirus DNA in the plasma of patients with cervical cancer. Int. J. Gynecol. Cancer 2004, 14, 903–910. [Google Scholar] [CrossRef]
- Gnanamony, M.; Peedicayil, A.; Subhashini, J.; Ram, T.S.; Rajasekar, A.; Gravitt, P.; Abraham, P. Detection and quantitation of HPV 16 and 18 in plasma of Indian women with cervical cancer. Gynecol. Oncol. 2010, 116, 447–451. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Lindegaard, J.C.; Petric, P.; Lindegaard, A.M.; Tanderup, K.; Fokdal, L.U. Evaluation of a New Prognostic Tumor Score in Locally Advanced Cervical Cancer Integrating Clinical Examination and Magnetic Resonance Imaging. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 754–763. [Google Scholar] [CrossRef]
- Lindegaard, J.C.; Petric, P.; Schmid, M.P.; Nesvacil, N.; Haie-Meder, C.; Fokdal, L.U.; Sturdza, A.E.; Hoskin, P.; Mahantshetty, U.; Segedin, B.; et al. Prognostic implications of uterine cervical cancer regression during chemoradiation evaluated by the T-score in the multicenter XXX study. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 379–389. [Google Scholar] [CrossRef]
- Cibula, D.; Pötter, R.; Planchamp, F.; Avall-Lundqvist, E.; Fischerova, D.; Haie-Meder, C.; Köhler, C.; Landoni, F.; Lax, S.; Lindegaard, J.C.; et al. The European Society of Gynaecological Oncology/European Society for Radiotherapy and Oncology/European Society of Pathology guidelines for the management of patients with cervical cancer. Radiother. Oncol. 2018, 127, 404–416. [Google Scholar] [CrossRef]
- Bhatla, N.; Berek, J.S.; Cuello Fredes, M.; Denny, L.A.; Grenman, S.; Karunaratne, K.; Kehoe, S.T.; Konishi, I.; Olawaiye, A.B.; Prat, J.; et al. Revised FIGO staging for carcinoma of the cervix uteri. Int. J. Gynecol. Obstet. 2019, 145, 129–135. [Google Scholar] [CrossRef] [PubMed]
- De Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milbury, C.A.; Zhong, Q.; Lin, J.; Williams, M.; Olson, J.; Link, D.R.; Hutchison, B. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol. Detect. Quantif. 2014, 1, 8–22. [Google Scholar] [CrossRef] [Green Version]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef]
- Dahlstrom, K.R.; Li, G.; Hussey, C.S.; Vo, J.T.; Wei, Q.; Zhao, C.; Sturgis, E.M. Circulating Human Papillomavirus DNA as a Marker for Disease Extent and Recurrence Among Patients With Oropharyngeal Cancer. Cancer 2015, 121, 3455–3464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallay, C.; Miranda, E.; Schaefer, S.; Catarino, R.; Jacot-Guillarmod, M.; Menoud, P.A.; Guerry, F.; Achtari, C.; Sahli, R.; Vassilakos, P.; et al. Human papillomavirus (HPV) contamination of gynaecological equipment. Sex. Transm. Infect. 2016, 92, 19–23. [Google Scholar] [CrossRef]
- Roden, R.B.S.; Lowy, D.R.; Schiller, J.T. Papillomavirus is resistant to desiccation. J. Infect. Dis. 1997, 176, 1076–1079. [Google Scholar] [CrossRef] [Green Version]
- Strauss, S.; Sastry, P.; Sonnex, C.; Edwards, S.; Gray, J. Contamination of environmental surfaces by genital human papillomaviruses. Sex. Transm. Infect. 2002, 78, 135–138. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.M.; Chien, T.Y.; Huang, S.H.; Lee, B.H.; Chang, S.F. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol. Oncol. 2006, 102, 54–60. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, Z.; Xu, R.; Ke, Y. Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology. Oncotarget 2016, 7, 5852. [Google Scholar] [CrossRef] [Green Version]
- Anayannis, N.V.; Schlecht, N.F.; Ben-Dayan, M.; Smith, R.V.; Belbin, T.J.; Ow, T.J.; Blakaj, D.M.; Burk, R.D.; Leonard, S.M.; Woodman, C.B.; et al. Association of an intact E2 gene with higher HPV viral load, higher viral oncogene expression, and improved clinical outcome in HPV16 positive head and neck squamous cell carcinoma. PLoS ONE 2018, 13, e0191581. [Google Scholar] [CrossRef]
| Patients | Early Stage Subgroup, N = 30 | Late-Stage Subgroup, N = 30 |
|---|---|---|
| Age (mean ± SD) | 47 ± 13 | 51 ± 13 |
| Histology 1 | n (%) | n (%) |
| SCC | 15 (50.0) | 26 (86.7) |
| ASC | 2 (6.7) | |
| AC | 13 (43.3) | 4 (13.3) |
| FIGO 2018 2 | n (%) | n (%) |
| IA1 | 1 (3.3) | |
| IA2 | 10 (33.3) | |
| IB1 | 7 (23.3) | |
| IB2 | 12 (40.0) | |
| IB3 | 2 (6.7) | |
| IIB | 12 (40.0) | |
| IIIC1 | 13 (43.3) | |
| IIIC2 | 2 (6.7) | |
| IVA | 1 (3.3) | |
| T score (mean ± SD) 3 | 1.2 ± 0.7 | 5.6 ± 3.3 |
| Tumor size (mm) (mean ± SD) 4 | 15.4 ± 6.6 | 49.83 ± 19.7 |
| HPV genotype tissue 5 | n (%) | n (%) |
| 16 | 21 (70.0) | 27 (90.0) |
| 18 | 9 (30.0) | 3 (10.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bønløkke, S.; Stougaard, M.; Sorensen, B.S.; Booth, B.B.; Høgdall, E.; Nyvang, G.-B.; Lindegaard, J.C.; Blaakær, J.; Bertelsen, J.; Fuglsang, K.; et al. The Diagnostic Value of Circulating Cell-Free HPV DNA in Plasma from Cervical Cancer Patients. Cells 2022, 11, 2170. https://doi.org/10.3390/cells11142170
Bønløkke S, Stougaard M, Sorensen BS, Booth BB, Høgdall E, Nyvang G-B, Lindegaard JC, Blaakær J, Bertelsen J, Fuglsang K, et al. The Diagnostic Value of Circulating Cell-Free HPV DNA in Plasma from Cervical Cancer Patients. Cells. 2022; 11(14):2170. https://doi.org/10.3390/cells11142170
Chicago/Turabian StyleBønløkke, Sara, Magnus Stougaard, Boe Sandahl Sorensen, Berit Bargum Booth, Estrid Høgdall, Gitte-Bettina Nyvang, Jacob Christian Lindegaard, Jan Blaakær, Jesper Bertelsen, Katrine Fuglsang, and et al. 2022. "The Diagnostic Value of Circulating Cell-Free HPV DNA in Plasma from Cervical Cancer Patients" Cells 11, no. 14: 2170. https://doi.org/10.3390/cells11142170
APA StyleBønløkke, S., Stougaard, M., Sorensen, B. S., Booth, B. B., Høgdall, E., Nyvang, G.-B., Lindegaard, J. C., Blaakær, J., Bertelsen, J., Fuglsang, K., Strube, M. L., Lenz, S., & Steiniche, T. (2022). The Diagnostic Value of Circulating Cell-Free HPV DNA in Plasma from Cervical Cancer Patients. Cells, 11(14), 2170. https://doi.org/10.3390/cells11142170








