Human Enzyme PADI4 Binds to the Nuclear Carrier Importin α3
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Expression and Purification
2.3. Prediction and Synthesis of the NLS Regions of PADI4
2.4. Fluorescence
2.4.1. Steady-State Fluorescence
2.4.2. Binding Experiments with PADI4
2.4.3. Binding Experiments with NLS1/2-PADI4
2.5. Circular Dichroism (CD)
2.6. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.6.1. 1D-1H-NMR Spectra
2.6.2. Translational Diffusion NMR (DOSY)
2.6.3. 2D-1H-NMR Spectra
2.7. Isothermal Titration Calorimetry (ITC)
2.8. Biolayer Inteferometry (BLI)
2.8.1. Experimental Design
2.8.2. Fitting of the Sensorgrams
2.9. Molecular Docking
2.10. Western Blot
2.11. Size Exclusion Chromatography (SEC)
3. Results
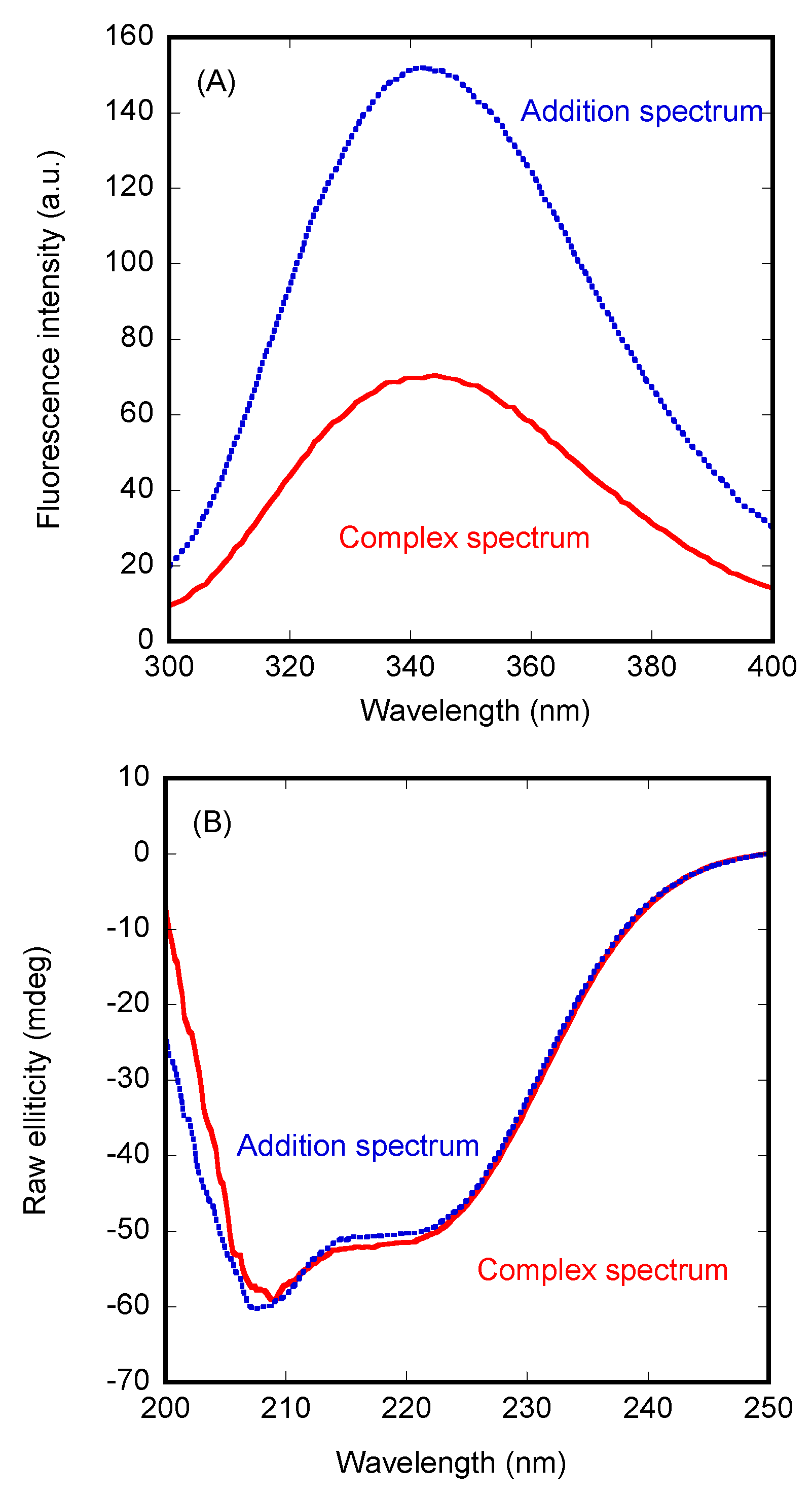
3.1. PADI4 Was Bound to Impα3 and ΔImpα3
3.2. Conformational Features of the Isolated NLSs of PADI4
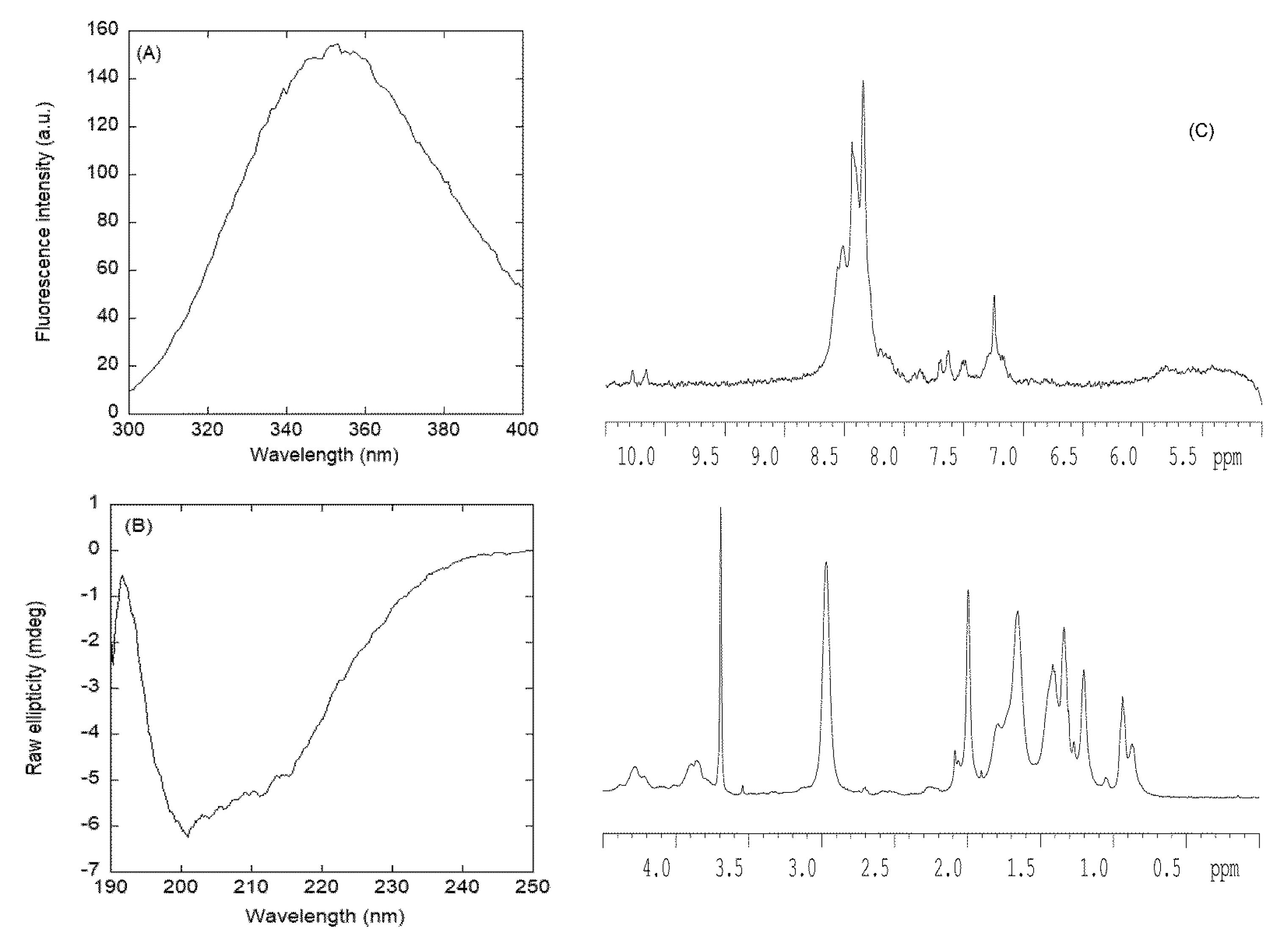
3.2.1. Isolated NLS1-PADI4 Was Oligomeric and Disordered in Solution
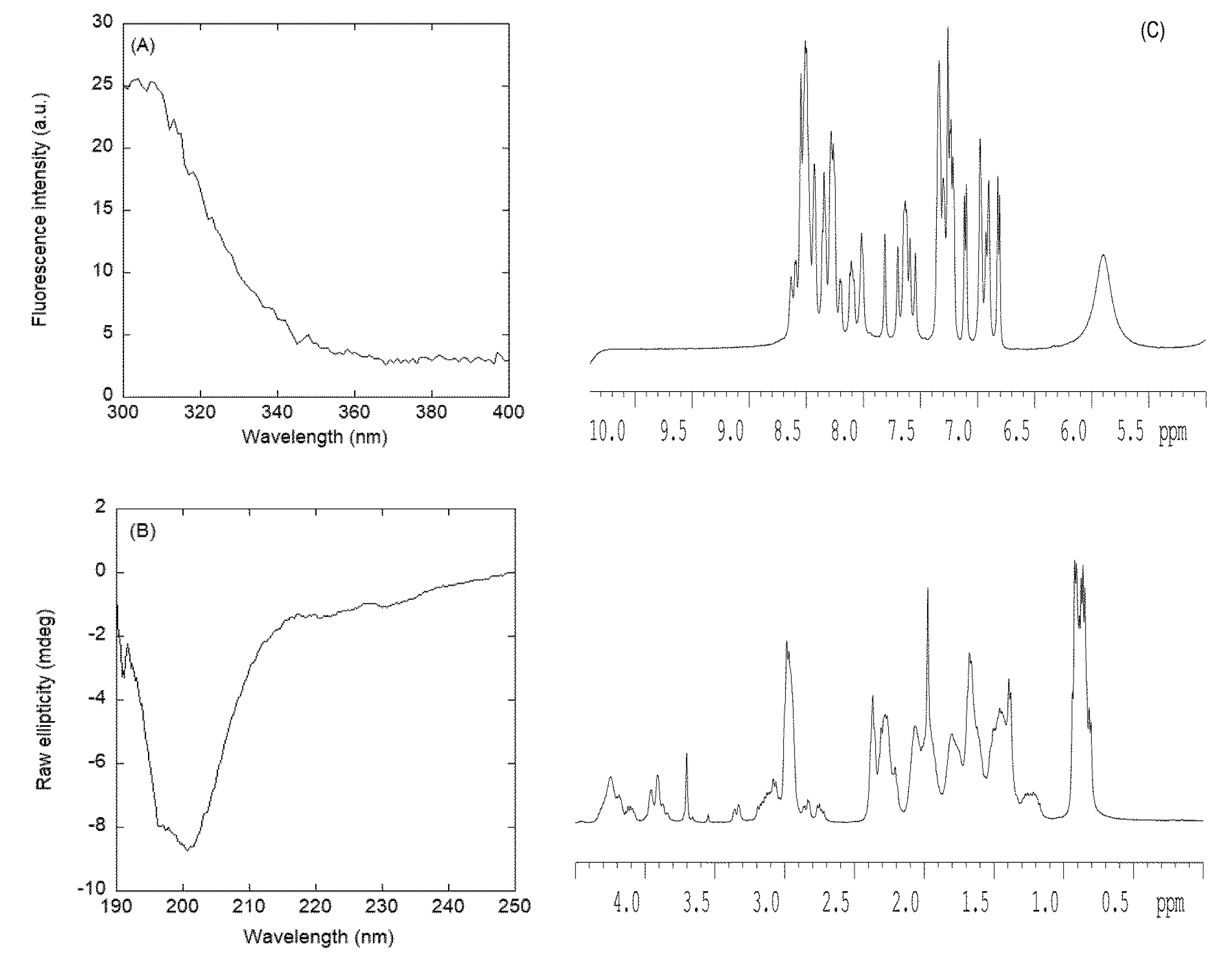
3.2.2. Isolated NLS2-PADI4 Was Monomeric and Disordered in Solution
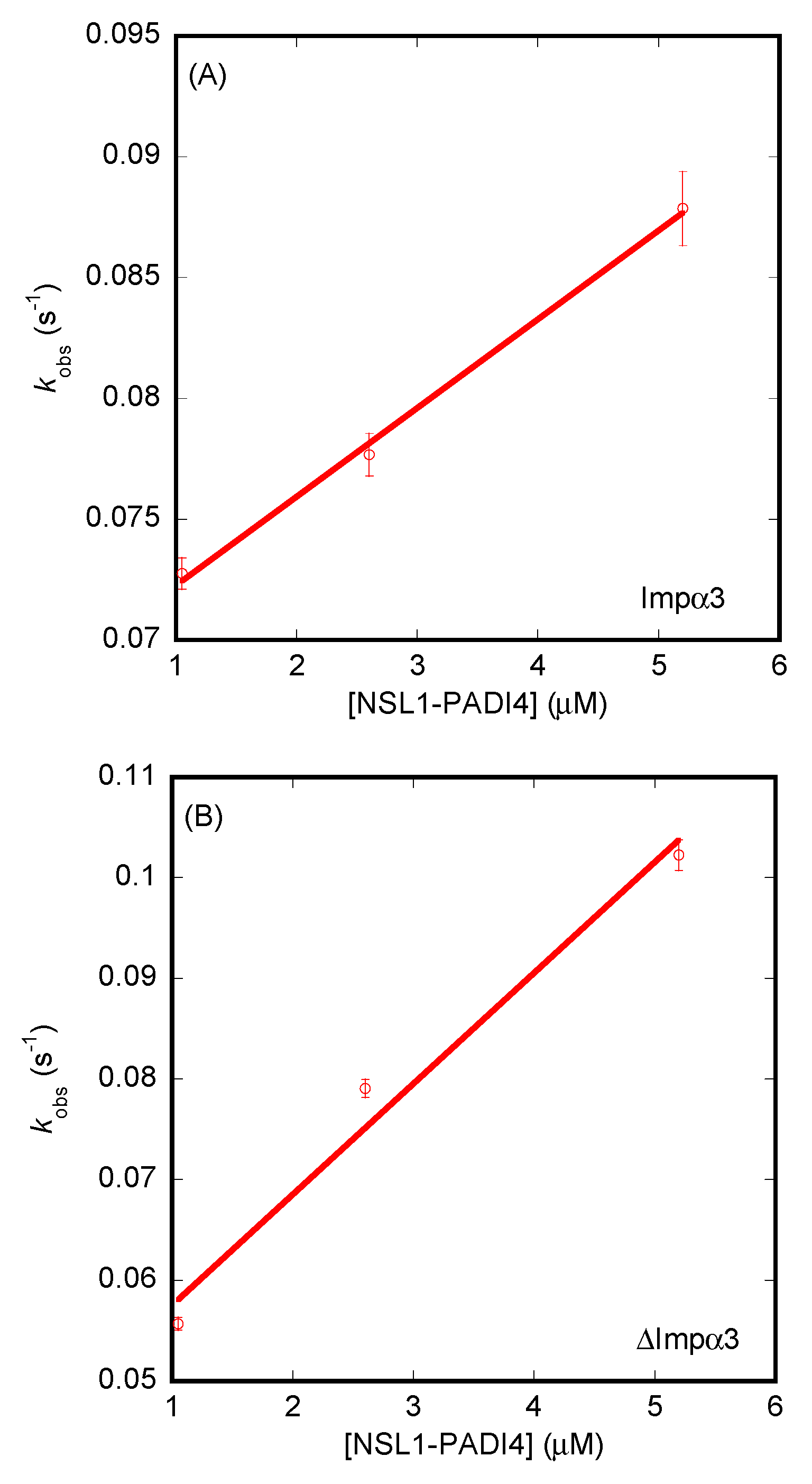
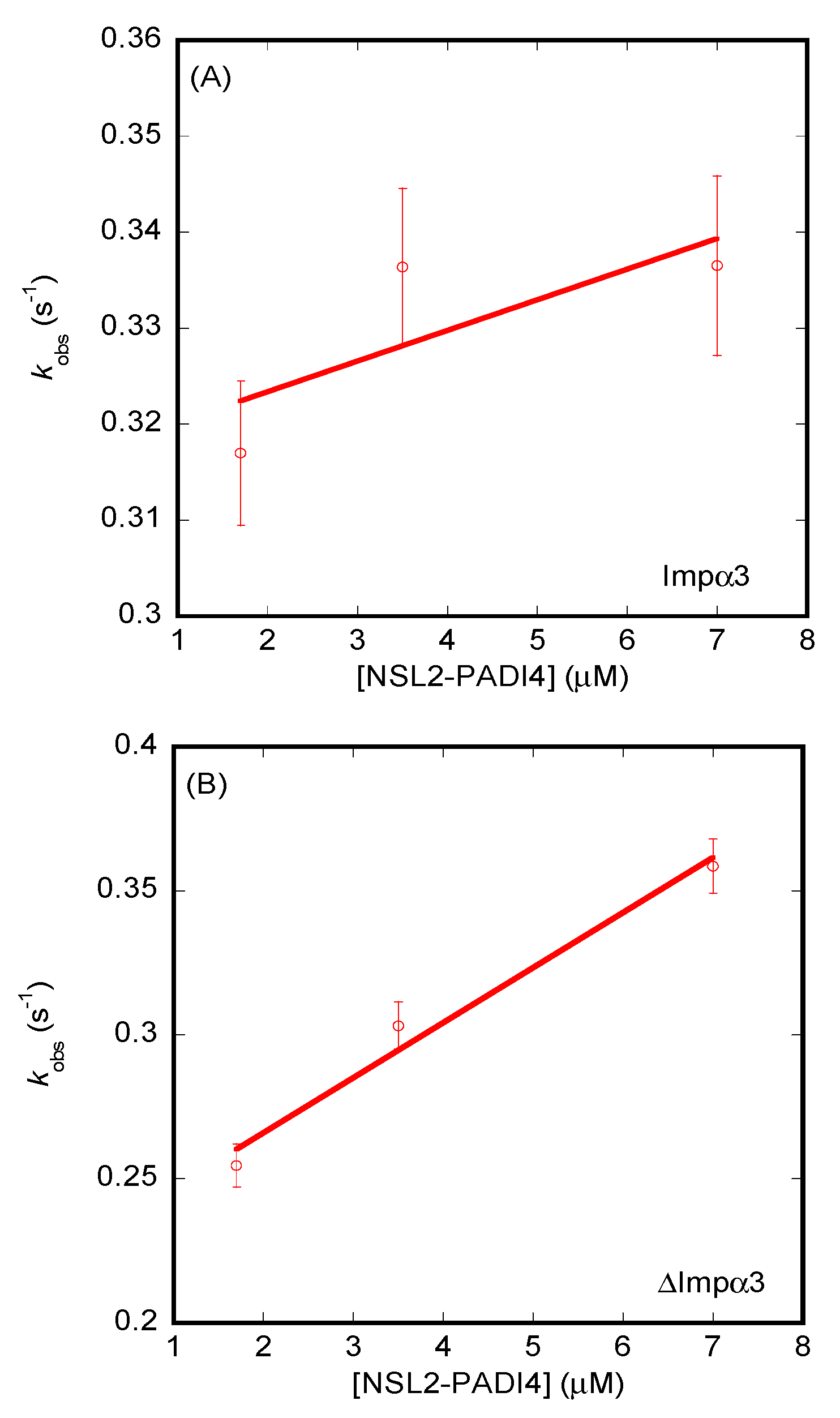
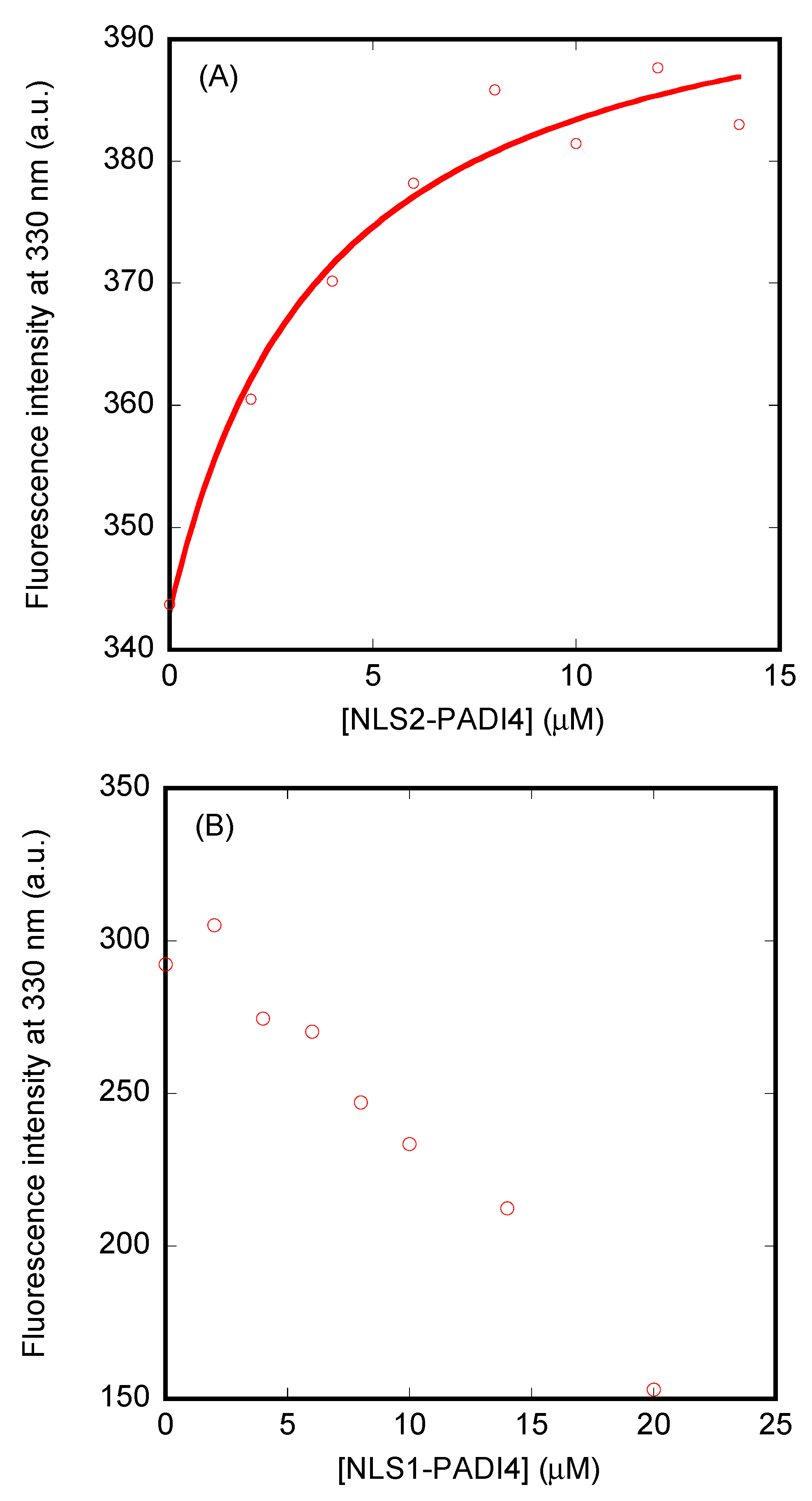
3.3. Isolated NLS1/2-PADI4 Could Bind to Each of the Importin Species
3.4. NLS1/2-PADI4 Could Bind to the Major NLS Binding Site of Impα3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone deimination antagonizes arginine methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Asaga, H.; Yamada, M.; Senshu, T. Selective deimination of vimentin in calcium ionophore-induced apoptosis of mouse peritoneal macrophages. Biochem. Biophys. Res. Commun. 1998, 243, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Assohou-Luty, C.; Raijmakers, R.; Benckhuijsen, W.E.; Stammen-Vogelzangs, J.; De Ru, A.; Van Veelen, P.A.; Franken, K.L.M.C.; Drijfhout, J.W.; Pruijn, G.J.M. The human peptidylarginine deiminases type 2 and type 4 have distinct substrate specificities. Biochim. Biophys. Acta 2014, 1844, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Senshu, T.; Akiyama, K.; Ishigami, A.; Nomura, K. Studies on specificity of peptidylarginine deiminase reactions using an immunochemical probe that recognizes an enzymatically deiminated partial sequence of mouse keratin K1. J. Dermatol. Sci. 1999, 21, 113–126. [Google Scholar] [CrossRef]
- Kizawa, K.; Takahara, H.; Troxler, H.; Kleinert, P.; Mochida, U.; Heizmann, C.W. Specific citrullination causes assembly of a globular S100A3 homotetramer: A putative Ca2+ modulator matures human hair cuticle. J. Biol. Chem. 2008, 283, 5004–5013. [Google Scholar] [CrossRef]
- Wang, Y.; Wysocka, J.; Sayegh, J.; Lee, Y.H.; Pertin, J.R.; Leonelli, L.; Sonbuchner, L.S.; McDonald, C.H.; Cook, R.G.; Dou, Y.; et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004, 306, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, A.; Maruyama, N. Importance of research on peptidylarginine deiminase and citrullinated proteins in age-related disease. Geriatr. Gerontol. Int. 2010, 10, S53–S58. [Google Scholar] [CrossRef]
- Klose, R.J.; Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 307–318. [Google Scholar] [CrossRef]
- György, B.; Tóth, E.; Tarcsa, E.; Falus, A.; Buzás, E.I. Citrullination: A posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006, 38, 1662–1677. [Google Scholar] [CrossRef]
- Anzilotti, C.; Pratesi, F.; Tommasi, C.; Migliorini, P. Peptidylarginine deiminase 4 and citrullination in health and disease. Autoimmun. Rev. 2010, 9, 158–160. [Google Scholar] [CrossRef]
- Wang, L.L.; Song, Y.P.; Mi, J.H.; Ding, M.L. Peptidyl arginine deiminase 4 and its potential role in Alzheimer’s disease. Med. Hypotheses 2021, 146, 110466. [Google Scholar] [CrossRef] [PubMed]
- Guerrin, M.; Ishigami, A.; Méchin, M.C.; Nachat, R.; Valmary, S.; Sebbag, M.; Simon, M.; Senshu, T.; Serre, G. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem. J. 2003, 370, 174. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, A.; Ohsawa, T.; Asaga, H.; Akiyama, K.; Kuramoto, M.; Maruyama, N. Human peptidylarginine deiminase type II: Molecular cloning, gene organization, and expression in human skin. Arch. Biochem. Biophys. 2002, 407, 25–31. [Google Scholar] [CrossRef]
- Kanno, T.; Kawada, A.; Yamanouchi, J.; Yosida-Noro, C.; Yoshiki, A.; Shiraiwa, M.; Kusakabe, M.; Manabe, M.; Tezuka, T.; Takahara, H. Human peptidylarginine deiminase type III: Molecular cloning and nucleotide sequence of the cDNA, properties of the recombinant enzyme, and immunohistochemical localization in human skin. J. Investig. Dermatol. 2000, 115, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Chavanas, S.; Méchin, M.C.; Takahara, H.; Kawada, A.; Nachat, R.; Serre, G.; Simon, M. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 2004, 330, 19–27. [Google Scholar] [CrossRef]
- Nakashima, K.; Hagiwara, T.; Ishigami, A.; Nagata, S.; Asaga, H.; Kuramotot, M.; Senshu, T.; Yamada, M. Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1α,25-dihydroxyvitamin D3. J. Biol. Chem. 1999, 274, 27786–27792. [Google Scholar] [CrossRef]
- Dong, S.; Kanno, T.; Yamaki, A.; Kojima, T.; Shiraiwa, M.; Kawada, A.; Méchin, M.C.; Chavanas, S.; Serre, G.; Simon, M.; et al. NF-Y and Sp1/Sp3 are involved in the transcriptional regulation of the peptidylarginine deiminase type III gene (PADI3) in human keratinocytes. Biochem. J. 2006, 397, 449–459. [Google Scholar] [CrossRef]
- Chavanas, S.; Adoue, V.; Méchin, M.C.; Ying, S.; Dong, S.; Duplan, H.; Charveron, M.; Takahara, H.; Serre, G.; Simon, M. Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte. PLoS ONE 2008, 3, e3408. [Google Scholar] [CrossRef]
- Hung, H.C.; Lin, C.Y.; Liao, Y.F.; Hsu, P.C.; Tsay, G.J.; Liu, G.Y. The functional haplotype of peptidylarginine deiminase IV (S55G, A82V and A112G) associated with susceptibility to rheumatoid arthritis dominates apoptosis of acute T leukemia Jurkat cells. Apoptosis 2007, 12, 475–487. [Google Scholar] [CrossRef]
- Li, P.; Yao, H.; Zhang, Z.; Li, M.; Luo, Y.; Thompson, P.R.; Gilmour, D.S.; Wang, Y. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol. Cell. Biol. 2008, 28, 4745–4758. [Google Scholar] [CrossRef]
- Li, P.; Wang, D.; Yao, H.; Doret, P.; Hao, G.; Shen, Q.; Qiu, H.; Zhang, X.; Wang, Y.; Chen, G.; et al. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene 2010, 29, 3153–3162. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Hagiwara, T.; Yamada, M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 2002, 277, 49562–49568. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.E.; Causey, C.P.; Knuckley, B.; Slack-Noyes, J.L.; Thompson, P.R. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr. Opin. Drug Discov. Dev. 2009, 12, 627. [Google Scholar]
- Neira, J.L.; Araujo-Abad, S.; Cámara-Artigas, A.; Rizzuti, B.; Abián, O.; Giudici, A.M.; Velázquez-Campoy, A.; de Juan Romero, C. Biochemical and Biophysical characterization of PADI4 supports its involvement in cancer. Arch. Biochem. Biophys. 2022, 717, 109125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, R.; Gan, Y.; Ying, S. The roles of PAD2- and PAD4-mediated protein citrullination catalysis in cancers. Int. J. Cancer 2021, 148, 267–276. [Google Scholar] [CrossRef]
- Stewart, M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell. Biol. 2007, 8, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Bednenko, J.; Cingolani, G.; Gerace, L. Nucleo-cytoplasmic transport navigating the channel. Traffic 2003, 4, 127–135. [Google Scholar] [CrossRef]
- Christie, M.; Chang, C.-W.; Róna, G.; Smith, K.M.; Stewart, A.G.; Takeda, A.A.S.; Fontes, M.R.M.; Stewart, M.; Vértessy, B.G.; Foorwood, J.K.; et al. Structural biology and regulation of protein import into the nucleus. J. Mol. Biol. 2016, 428, 2060–2090. [Google Scholar] [CrossRef]
- Kobe, B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 1999, 6, 388–397. [Google Scholar] [CrossRef]
- Zaidi, S.K.; Young, D.W.; Javed, A.; Pratap, J.; Mortecino, M.; van Wijnen, A.; Lian, J.B.; Stein, J.L.; Stein, G.S. Nuclear microenvironments in biological control and cancer. Nat. Rev. Cancer 2007, 7, 454–463. [Google Scholar] [CrossRef]
- Fukasawa, K. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer 2007, 7, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Chahine, M.N.; Pierce, G.N. Therapeutic targeting of nuclear protein import in pathological cell conditions. Pharmacol. Rev. 2009, 61, 358–372. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, K.; Tanaka, S.; Mogushi, K.; Aihara, A.; Murakata, A.; Matsumura, S.; Mitsunori, Y.; Yasen, M.; Ban, D.; Noguchi, N.; et al. Importin a1 as a novel prognostic target for hepatocellular carcinoma. Ann. Surg. Oncol. 2011, 18, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tao, T.; Yan, W.; Feng, Y.; Wang, Y.; Cai, J.; You, Y.; Jiang, T.; Jiang, C. Upregulation of miR-181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci. Rep. 2015, 5, 13072. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lu, C.; Wei, J.; Guo, Y.; Liu, W.; Luo, L.; Fish, G.; Li, L. Inhibition of KPNA4 attenuates prostate cancer metastasis. Oncogene 2017, 36, 2868–2878. [Google Scholar] [CrossRef]
- Xu, M.; Liang, H.; Li, K.; Zhu, S.; Yao, Z.; Xu, R.; Lin, N. Value of KPNA4 as a diagnostic and prognostic biomarker for hepatocellular carcinoma. Aging 2021, 13, 5263–5283. [Google Scholar] [CrossRef]
- Li, X.; Yu, M.; Yang, C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J. Cell. Biochem. 2020, 121, 2258–2267. [Google Scholar] [CrossRef]
- Wang, D.; Cui, Y.; Xu, A.; Zhao, L.; Li, P. MiR-596 activated by EP300 controls the tumorigenesis in epithelial ovarian cancer by declining BRD4 and KPNA4. Cancer Cell. Int. 2020, 20, 447. [Google Scholar] [CrossRef]
- Thiele, S.; Stanelle-Bertram, S.; Beck, S.; Kouassi, N.M.; Zickler, M.; Müller, M.; Tuku, B.; Resa-Infante, P.; van Riel, D.; Alawi, M.; et al. Cellular Importin-α3 expression dynamics in the lung regulate antiviral response pathways against influenza A virus infection. Cell Rep. 2020, 31, 107549. [Google Scholar] [CrossRef]
- Smith, K.M.; Tsimbalyuk, S.; Edwards, M.G.; Cross, E.M.; Batra, J.; Soares da Costa, T.P.; Aragao, D.; Basler, C.F.; Forwood, J.K. Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun. 2018, 9, 3703. [Google Scholar] [CrossRef]
- Neira, J.L.; Rizzuti, B.; Jiménez-Alesanco, A.; Abián, O.; Velázquez-Campoy, A.; Iovanna, J.L. The paralogue of the intrinsically disordered Nuclear Protein 1 has a nuclear localization sequence that binds to human importin α3. Int. J. Mol. Sci. 2020, 21, 7428. [Google Scholar] [CrossRef] [PubMed]
- Neira, J.L.; Rizzuti, B.; Jiménez-Alesanco, A.; Palomino-Schätzlein, M.; Abián, O.; Velázquez-Campoy, A.; Iovanna, J.L. A phosphorylation-induced switch in the nuclear localization sequence of the intrinsically disordered NUPR1 hampers binding to importin. Biomolecules 2020, 10, 1313. [Google Scholar] [CrossRef] [PubMed]
- Neira, J.L.; Jiménez-Alesanco, A.; Rizzuti, B.; Velázquez-Campoy, A. The nuclear localization sequence of the epigenetic factor RYBP binds to human importin α3. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140670. [Google Scholar] [CrossRef]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Arita, K.; Hashimoto, H.; Shimizu, T.; Nakashima, K.; Yamada, M.; Sato, M. Structural basis for Ca2+-induced activation of human PAD4. Nat. Struct. Mol. Biol. 2004, 11, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Neira, J.L.; Hornos, F.; Bacarizo, J.; Cámara-Artigas, A.; Gómez, J. The monomeric species of the regulatory domain of tyrosine hydroxylase has a low conformational stability. Biochemistry 2017, 55, 3418–3431. [Google Scholar] [CrossRef]
- Birdsall, B.; King, R.W.; Wheeler, M.R.; Lewis, C.A., Jr.; Goode, S.; Dunlap, R.B.; Roberts, G.C. Correction for light absorption in fluorescence studies of protein-ligand interactions. Anal. Biochem. 1983, 132, 353–361. [Google Scholar] [CrossRef]
- Beckett, D. Measurement and analysis of equilibrium binding titrations: A beginner’s guide. Methods Enzymol. 2011, 488, 1–16. [Google Scholar] [CrossRef]
- Royer, C.A.; Scarlatta, S.F. Fluorescence approaches to quantifying biomolecular interactions. Methods Enzymol. 2008, 450, 79–106. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, J.; Fairbrother, W.J.; Palmer, A.G.; Skelton, N.J. Protein NMR Spectroscopy: Principles and Practice; Academic Press: New York, NY, USA, 1996. [Google Scholar]
- Piotto, M.; Saudek, V.; Sklenar, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 1992, 2, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D.K.; Grimshaw, S.B.; Receveur, V.; Dobson, C.M.; Jones, J.A.; Smith, L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR technique. Biochemistry 1999, 38, 16424–16431. [Google Scholar] [CrossRef]
- Marion, M.; Wüthrich, K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983, 11, 967–975. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Kumar, A.; Ernst, R.R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Cavanagh, J.; Rance, M. Suppression of cross-relaxation effects in TOCSY spectra via a modified DIPSI-2 mixing sequence. J. Magn. Reson. 1992, 96, 670–678. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; John Wiley and Sons: New York, NY, USA, 1986. [Google Scholar]
- Kjaergaard, M.; Brander, S.; Poulsen, F.M. Random coil chemical shifts for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 2011, 49, 139–149. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Poulsen, F.M. Sequence correction of random coil chemical shifts: Correlation between neighbour correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 2011, 50, 157–165. [Google Scholar] [CrossRef]
- Frenzel, D.; Willbolds, D. Kinetic titration series with biolayer interferometry. PLoS ONE 2014, 9, e106882. [Google Scholar] [CrossRef]
- Pantoja-Uceda, D.; Neira, J.L.; Saelices, L.; Robles-Rengel, R.; Florencio, F.J.; Muro-Pastor, M.I.; Santoro, J. Dissecting the binding between glutamine synthetase and its two natively unfolded protein inhibitors. Biochemistry 2016, 55, 3370–3382. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Koyama, M.; Matsuura, Y. Crystal structure of importin-alpha 3 bound to the nuclear localization signal of Ran-binding protein 3. Biochem. Biophys. Res Commun. 2017, 491, 609–613. [Google Scholar] [CrossRef]
- Nakada, R.; Matsuura, Y. Crystal structure of importin-α bound to the nuclear localization signal of Epstein-Barr virus EBNA-LP protein. Protein Sci. 2017, 26, 1231–1235. [Google Scholar] [CrossRef][Green Version]
- Grande, F.; Rizzuti, B.; Occhiuzzi, M.A.; Ioele, G.; Casacchia, T.; Gelmini, F.; Guzzi, R.; Garofalo, A.; Statti, B. Identification by molecular docking of homoisoflavones from Leopoldia comosa as ligands of estrogen receptors. Molecules 2018, 23, 894. [Google Scholar] [CrossRef]
- Paketurytė, V.; Petrauskas, V.; Zubrienė, A.; Abian, O.; Bastos, M.; Chen, W.Y.; Moreno, M.J.; Krainer, G.; Linkuvienė, V.; Sedivy, A.; et al. Uncertainty in protein-ligand binding constants: Asymmetric confidence intervals versus standard errors. Eur. Biophys. J. 2021, 50, 661–670. [Google Scholar] [CrossRef]
- Lobley, A.; Whitmore, L.; Wallace, B.A. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 2002, 18, 211–212. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. DICHROWEB an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Shi, Z.; Woody, R.W.; Kallenbach, N.R. Is poly-proline II a major backbone conformation in unfolded proteins? Adv. Protein Chem. 2002, 62, 163–240. [Google Scholar] [CrossRef]
- Danielsson, J.; Jarvet, J.; Damberg, P.; Gräslund, A. Translational diffusion measured by PFG-NMR on full length and fragments of the Alzheimer Aβ(1–40) peptide. Determination of hydrodynamic radii of random coil peptides of varying length. Magn. Reson. Chem. 2002, 40, S89–S97. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta Proteins Proteom 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Price, N.C. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef] [PubMed]
- Woody, R.W. Circular dichroism. Methods Enzymol. 1995, 246, 34–71. [Google Scholar] [CrossRef] [PubMed]
- De Barros, A.C.; Takeda, A.A.S.; Dreyer, T.R.; Velázquez-Campoy, A.; Kobe, B.; Fontes, M.R.M. DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway. Biochimie 2018, 146, 87–96. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Rizzuti, B.; Pey, A.L.; Soubeyran, P.; Vidal, M.; Urrutia, R.; Iovanna, J.L.; Neira, J.L. Intrinsically disordered chromatin protein NUPR1 binds to the C-terminal region of Polycomb RING1B. Proc. Natl. Acad. Sci. USA 2017, 114, E6332–E6341. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Rizzuti, B.; Abián, O.; Velázquez-Campoy, A.; Iovanna, J.L.; Neira, J.L. Amphipathic helical peptides hamper protein-protein interactions of the intrinsically disordered chromatin nuclear protein 1 (NUPR1). Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1283–1295. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Xia, Y.; Lan, W.; Zhou, Z.; Huang, C.; Peng, L.; Soubeyran, P.; Velázquez-Campoy, A.; Abián, O.; Rizzuti, B.; et al. Ligand-based design identifies a potent NUPR1 inhibitor exerting anticancer activity via necroptosis. J. Clin. Investig. 2019, 129, 2500–2513. [Google Scholar] [CrossRef]
- Rizzuti, B.; Lan, W.; Santofimia-Castaño, P.; Zhou, Z.; Velázquez-Campoy, A.; Abián, O.; Peng, L.; Neira, J.L.; Xia, Y.; Iovanna, J.L. Design of inhibitors of the intrinsically disordered protein NUPR1: Balance between drug affinity and target function. Biomolecules 2021, 11, 1453. [Google Scholar] [CrossRef]
- Lan, W.; Santofimia-Castaño, P.; Swayden, M.; Xia, Y.; Zhou, Z.; Audebert, S.; Camoin, L.; Huang, C.; Peng, L.; Jiménez-Alesanco, A.; et al. ZZW-115-dependent inhibition of NUPR1 nuclear translocation sensitizes cancer cells to genotoxic agents. JCI Insight 2020, 5, 138117. [Google Scholar] [CrossRef]
- Miyatake, H.; Sanjoh, A.; Unzai, S.; Matsuda, G.; Tatsumi, Y.; Miyamoto, Y.; Dohmae, N.; Aida, Y. Crystal structure of human Importin α1 (Rch1) revealing a potential autoinhibition mode involving homodimerization. PLoS ONE 2015, 10, e0115995. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Counago, R.M.; Williams, S.J.; Boden, M.; Kobe, B. Distinctive conformation of minor site-specific nuclear localization signals bound to importin-alpha. Traffic 2013, 14, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Loveland, K.L.; Yoneda, Y. Nuclear importin α and its physiological importance. Commun. Integr. Biol. 2012, 5, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, D.S.; Corbett, A.H.; Mason, D.A.; Harreman, M.T.; Adam, S.A. Importin α: A multipurpose nuclear-transport receptor. Trends Cell Biol. 2004, 14, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-L.; Chiang, Y.-H.; Liu, G.-Y.; Hung, H.-C. Functional role of dimerization of human peptidylarginine deiminase 4 (PAD4). PLoS ONE 2011, 6, e21314. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Lee, C.-Y.; Huang, Y.-N.; Chen, H.-Y.; Liu, G.-Y.; Hung, H.-C. Probing the roles of calcium binding sites during the folding of human peptidylarginine deiminase 4. Sci Rep. 2017, 7, 2429. [Google Scholar] [CrossRef]
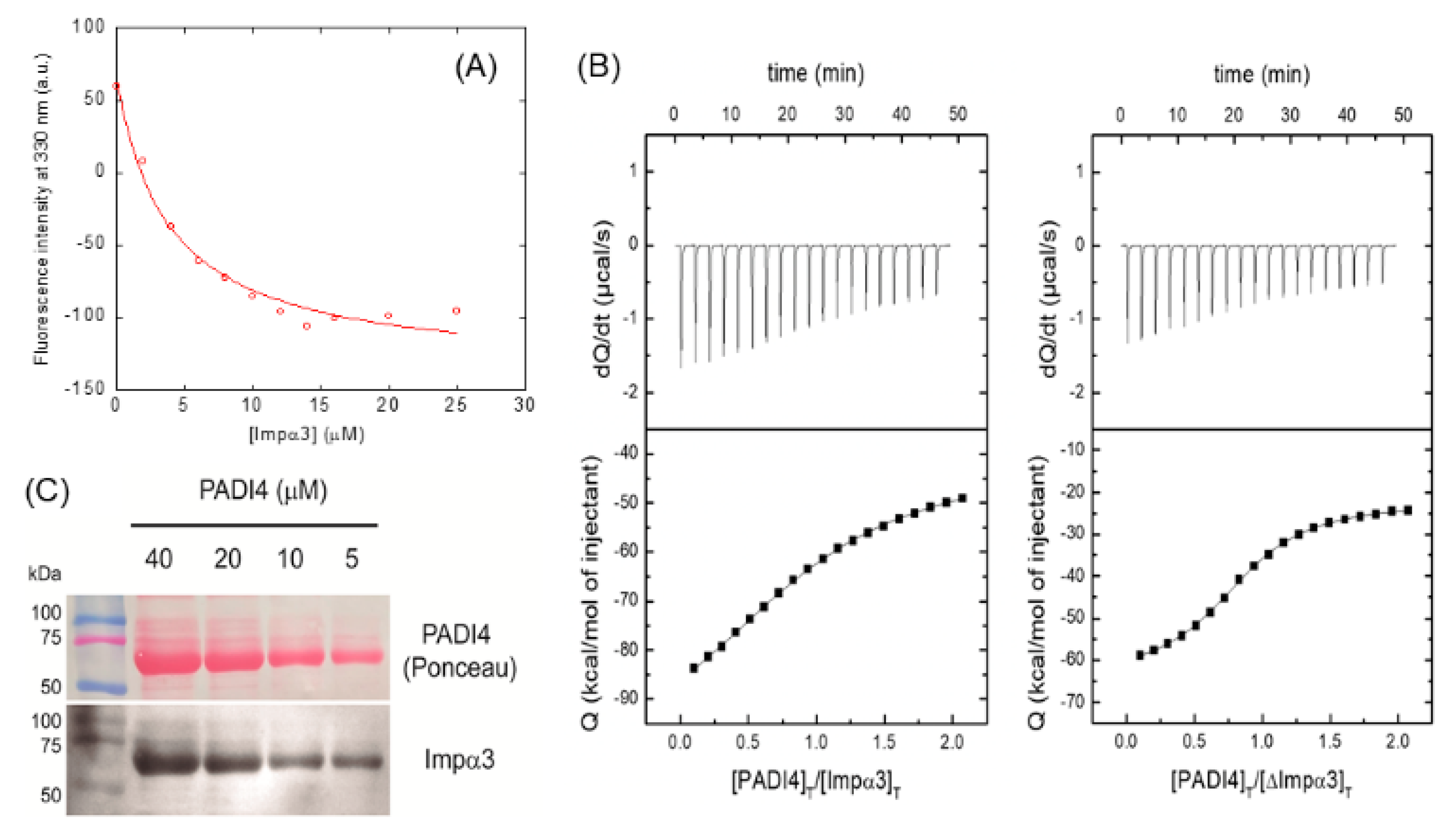

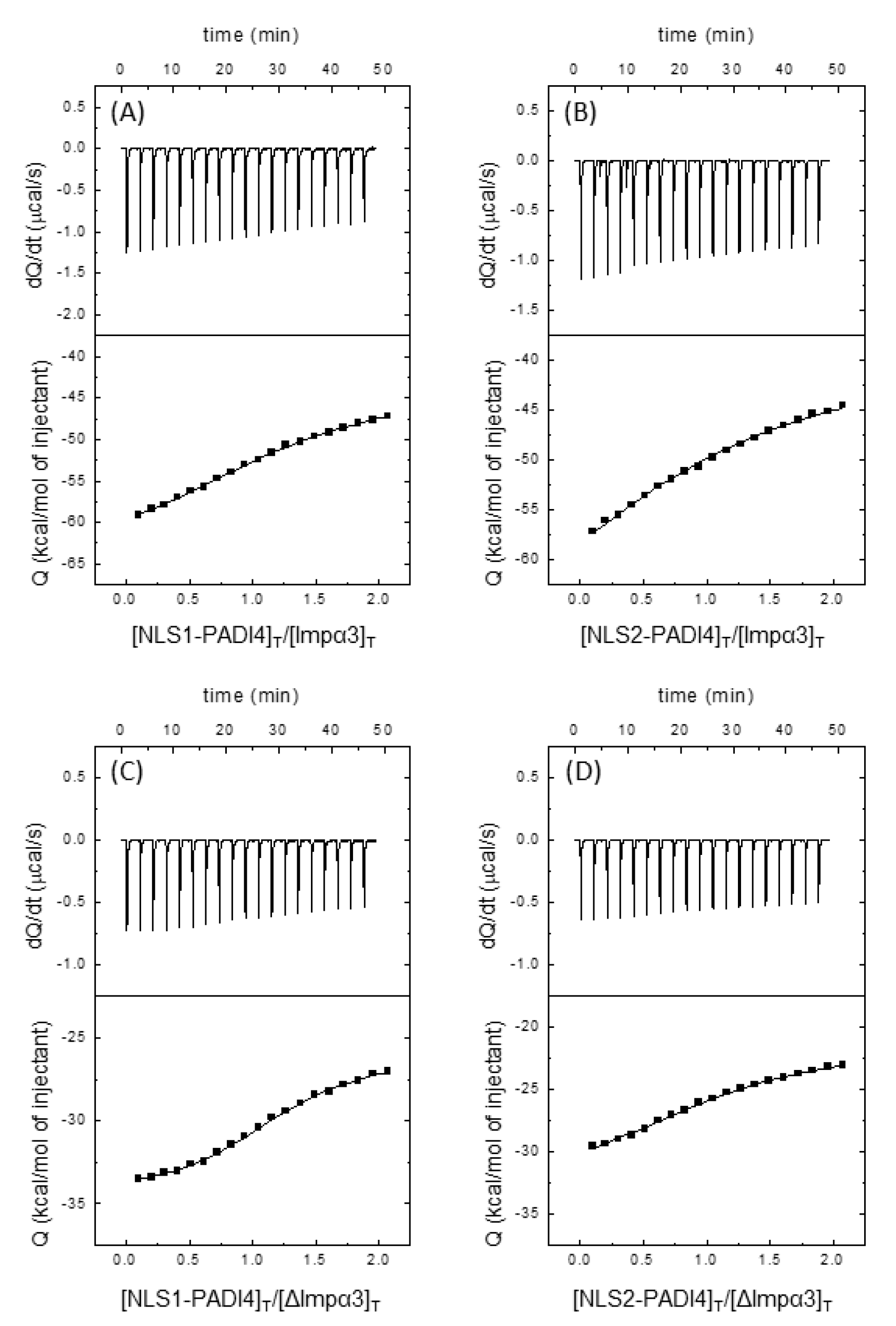
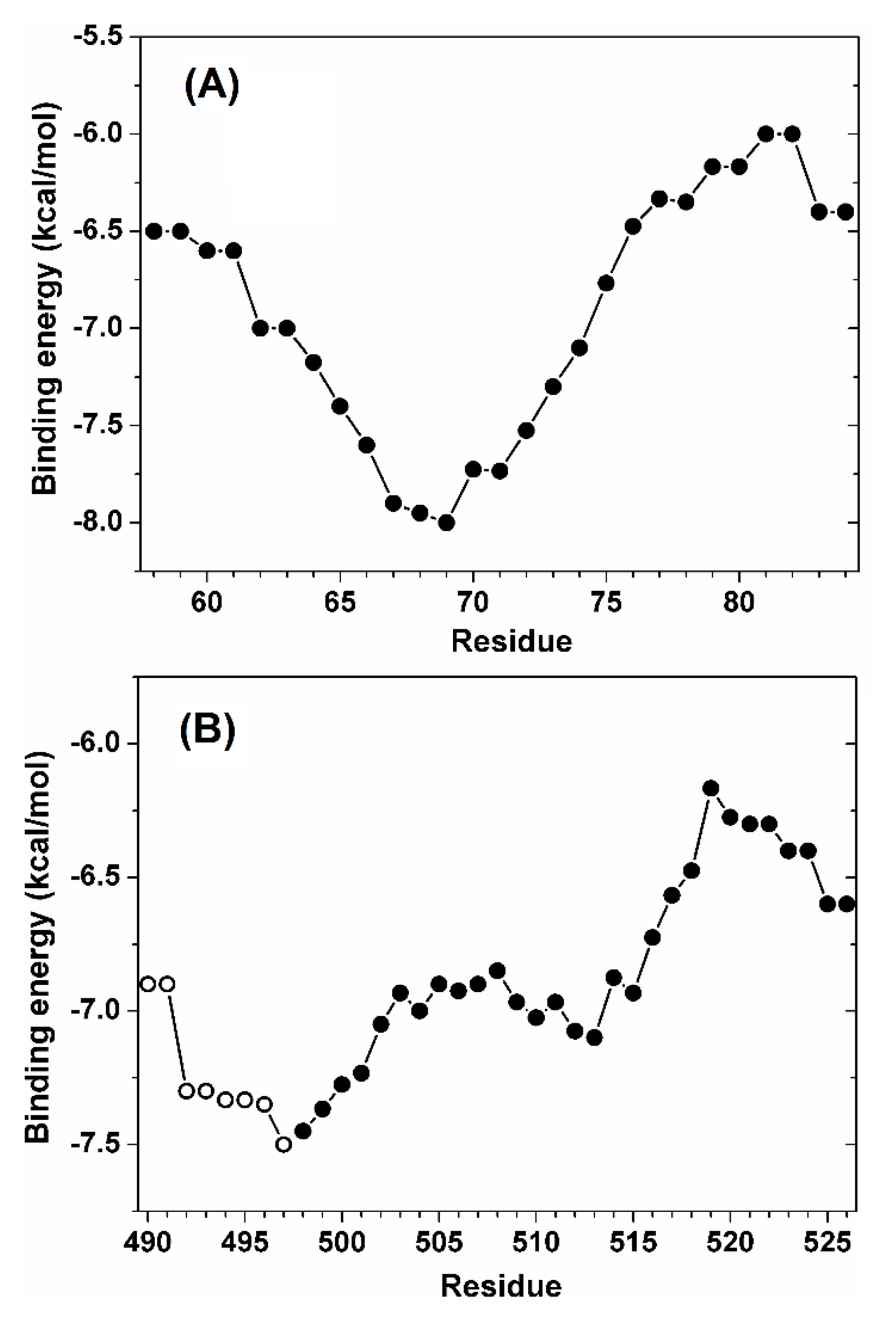
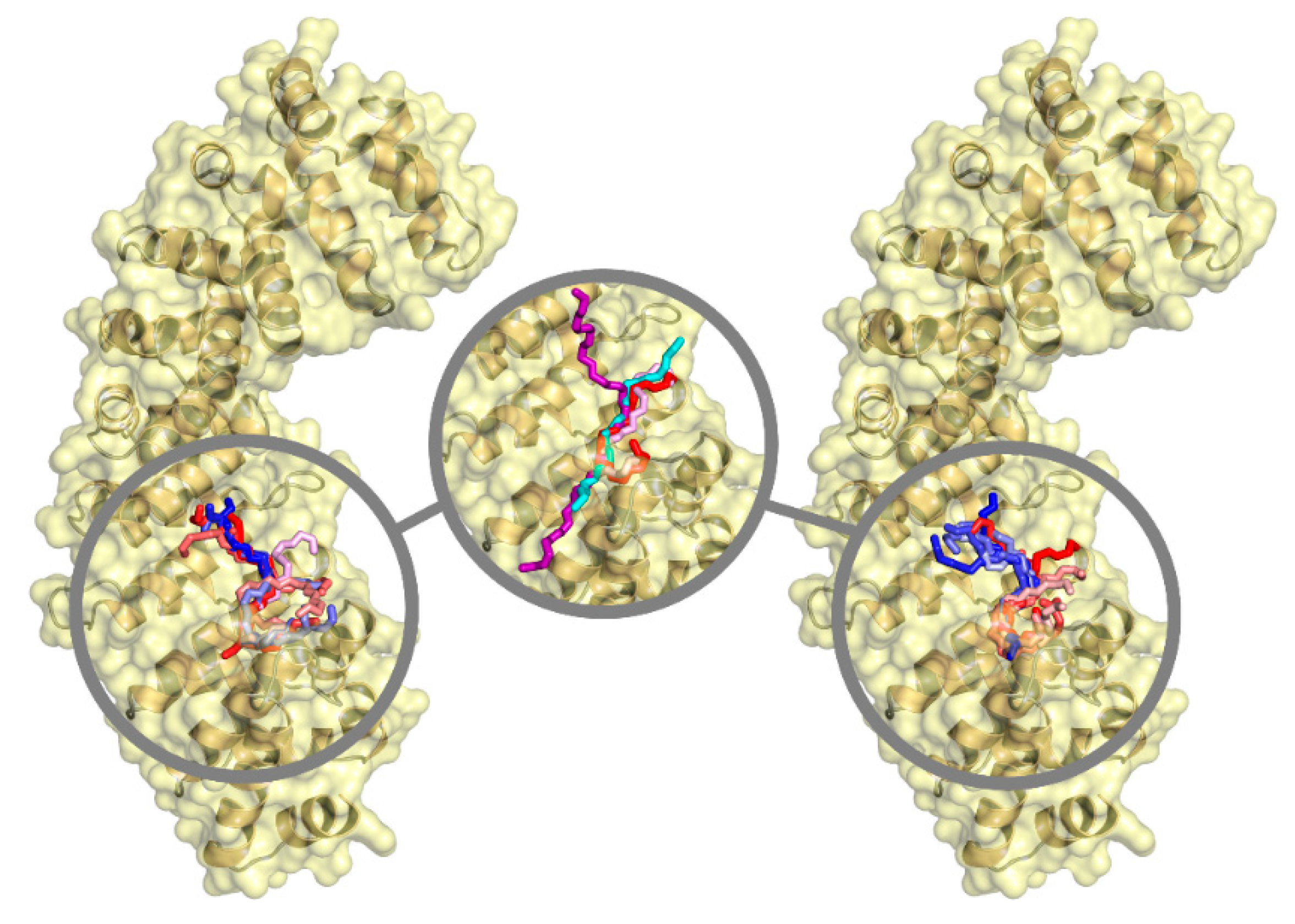
| Ka (105 M−1) | Kd (μM) | ΔH (kcal/mol) | ΔG (kcal/mol) | -TΔS (kcal/mol) | n | ||
|---|---|---|---|---|---|---|---|
| PADI4 | Impα3 | 2.1 (1.7, 2.5) | 4.8 (4.0, 5.9) | −65.5 (−72.0, −60.5) | −7.3 | −58.2 | 0.97 (0.94, 1.00) |
| ΔImpα3 | 7.8 (7.3, 8.5) | 1.3 (1.2, 1.4) | −38.4 (−39.5, 37.2) | −8.0 | −30.4 | 0.84 (0.83, 0.85) | |
| NLS1 | Impα3 | 2.3 (1.6, 3.0) | 4.3 (3.3, 6.3) | −21.3 (−25.3, −18.8) | −7.3 | −14.0 | 1.24 (1.15, 1.37) |
| ΔImpα3 | 6.5 (4.9, 8.3) | 1.5 (1.2, 2.0) | −8.7 (−9.6, −8.0) | −7.9 | −0.8 | 1.22 (1.16, 1.29) | |
| NLS2 | Impα3 | 0.43 (0.31, 0.57) | 23 (18, 32) | −35.7 (−39.8, −31.1) | −6.3 | −29.4 | 1.08 (0.98, 1.20) |
| ΔImpα3 | 2.3 (1.5, 3.2) | 4.3 (3.2, 6.7) | −11.2 (−13.7, −9.8) | −7.3 | −3.9 | 1.10 (1.02, 1.23) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neira, J.L.; Rizzuti, B.; Abián, O.; Araujo-Abad, S.; Velázquez-Campoy, A.; de Juan Romero, C. Human Enzyme PADI4 Binds to the Nuclear Carrier Importin α3. Cells 2022, 11, 2166. https://doi.org/10.3390/cells11142166
Neira JL, Rizzuti B, Abián O, Araujo-Abad S, Velázquez-Campoy A, de Juan Romero C. Human Enzyme PADI4 Binds to the Nuclear Carrier Importin α3. Cells. 2022; 11(14):2166. https://doi.org/10.3390/cells11142166
Chicago/Turabian StyleNeira, José L., Bruno Rizzuti, Olga Abián, Salomé Araujo-Abad, Adrián Velázquez-Campoy, and Camino de Juan Romero. 2022. "Human Enzyme PADI4 Binds to the Nuclear Carrier Importin α3" Cells 11, no. 14: 2166. https://doi.org/10.3390/cells11142166
APA StyleNeira, J. L., Rizzuti, B., Abián, O., Araujo-Abad, S., Velázquez-Campoy, A., & de Juan Romero, C. (2022). Human Enzyme PADI4 Binds to the Nuclear Carrier Importin α3. Cells, 11(14), 2166. https://doi.org/10.3390/cells11142166









