WWOX Controls Cell Survival, Immune Response and Disease Progression by pY33 to pS14 Transition to Alternate Signaling Partners
Abstract
1. Introduction
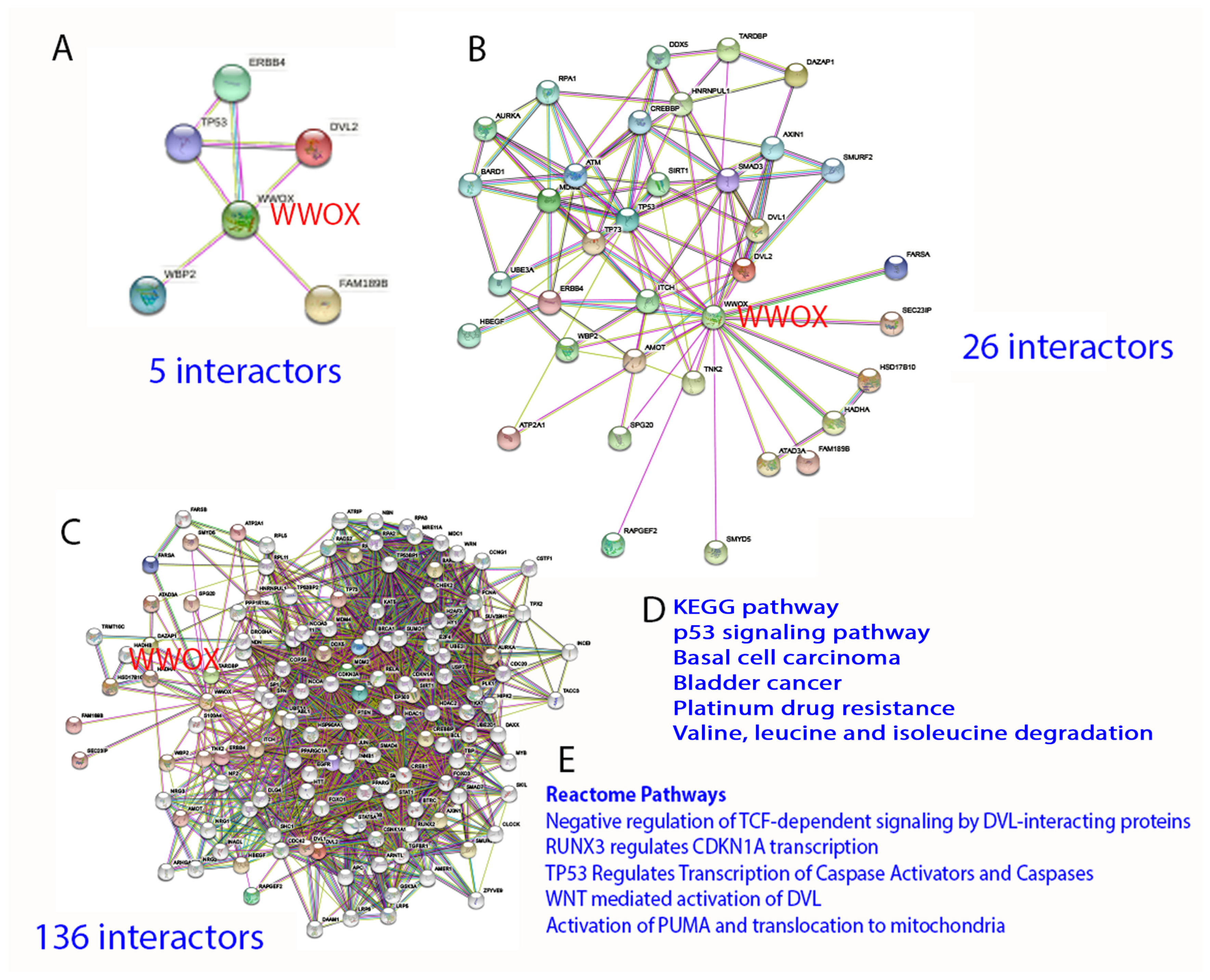
2. Protein Interaction Network in Normal Signaling and Diseases
3. WW Domain-Containing Oxidoreductase (WWOX)
4. WWOX Controls Cell Migration, Cell-Cell Recognition, and Neuronal Heterotopia
5. WWOX Signaling Network
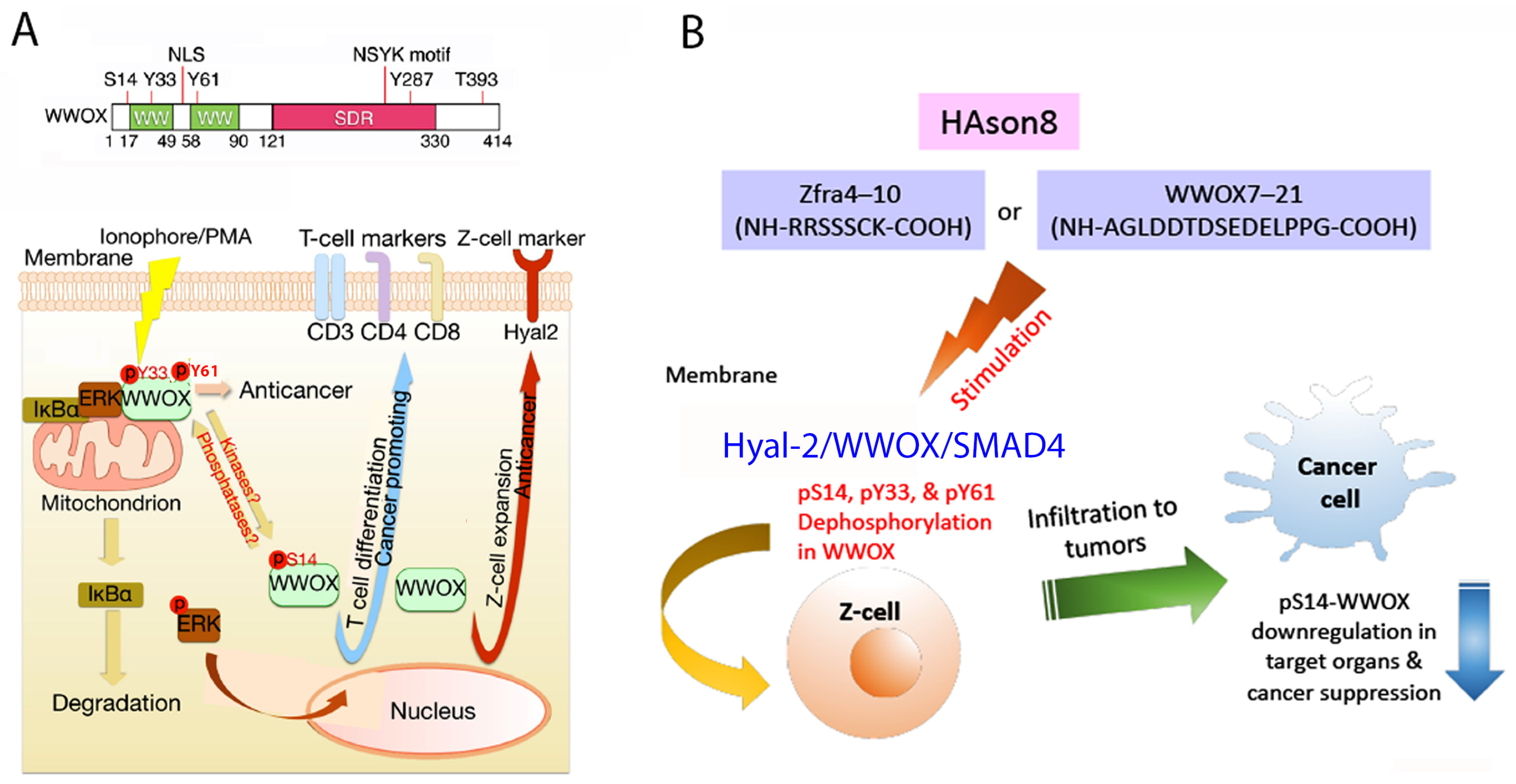
6. pY33 to pS14 Transition in WWOX during Disease Progression
7. WWOX Functional Measurement by Time-Lapse FRET Microscopy
8. TGF-β1 Induction of Initial Driving Force and Then Execution Force for Protein-Protein Binding and Cell Death: TIAF1 Is a Blocker of TGF-β1/SMAD Signaling
9. The Dynamics of WWOX/TIAF1/p53 Triad Formation and Functional Antagonism between p53 and WWOX for Enhancing the Progression of Cancer and Alzheimer’s Disease
10. Identification of HYAL-2/WWOX/SMAD4 Signaling in Regulating Physiological and Pathological Events
11. A WWOX7-21 Epitope Peptide Drives the HYAL-2/WWOX/SMAD4 Signaling
12. Phosphorylation Status of WWOX in the HYAL-2/WWOX/SMAD4 Complex and Disease Progression
13. Zfra4-10 or WWOX7-21 Activates the HYAL-2/WWOX/SMAD4 Signaling for Z Cell Activation and Suppression of Disease Progression In Vivo
14. Zfra-Induced Spleen Z Cell Activation Requires De-Phosphorylation at S14, Y33 and Y61 in WWOX In Vivo
15. Zfra4-10 or WWOX7-21 Increases the Binding of Endogenous WWOX with Intracellular Protein Partners, Which Contributes to Cancer Growth Suppression In Vivo
16. Switching the HYAL-2/WWOX/SMAD4 Signaling from Bubbling Cell Death to Membrane Blebbing by Replacing HYAL-2 with p53
17. Discussion and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gulfidan, G.; Turanli, B.; Beklen, H.; Sinha, R.; Arga, K.Y. Pan-cancer mapping of differential protein-protein interactions. Sci. Rep. 2020, 10, 3272. [Google Scholar] [CrossRef] [PubMed]
- Guda, P.; Chittur, S.V.; Guda, C. Comparative analysis of protein-protein interactions in cancer-associated genes. Genom. Proteom. Bioinform. 2009, 7, 25–36. [Google Scholar] [CrossRef]
- Guda, C.; King, B.R.; Pal, L.R.; Guda, P. A top-down approach to infer and compare domain-domain interactions across eight model organisms. PLoS ONE 2009, 4, e5096. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Goswami, S.; Das, D. Cross β amyloid assemblies as complex catalytic machinery. Chem. Commun. 2021, 57, 7597–7609. [Google Scholar] [CrossRef]
- Capitanio, G.; Papa, F.; Papa, S. The allosteric protein interactions in the proton-motive function of mammalian redox enzymes of the respiratory chain. Biochimie 2021, 189, 1–12. [Google Scholar] [CrossRef]
- Moracci, L.; Crotti, S.; Traldi, P.; Agostini, M.; Cosma, C.; Lapolla, A. Role of mass spectrometry in the study of interactions between amylin and metal ions. Mass Spectrom. Rev. 2021. [Google Scholar] [CrossRef]
- Yang, J.; Perrett, S.; Wu, S. Single Molecule Characterization of Amyloid Oligomers. Molecules 2021, 26, 948. [Google Scholar] [CrossRef]
- Barik, S. The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins. Int. J. Mol. Sci. 2020, 21, 8776. [Google Scholar] [CrossRef]
- Bailly, C.; Vergoten, G. Flurbiprofen as a biphenyl scaffold for the design of small molecules binding to PD-L1 protein dimer. Biochem. Pharmacol. 2020, 178, 114042. [Google Scholar] [CrossRef]
- Meiser, N.; Fuks, C.; Hengesbach, M. Cooperative Analysis of Structural Dynamics in RNA-Protein Complexes by Single-Molecule Förster Resonance Energy Transfer Spectroscopy. Molecules 2020, 25, 2057. [Google Scholar] [CrossRef]
- Chang, N.S.; Pratt, N.; Heath, J.; Schultz, L.; Sleve, D.; Carey, G.B.; Zevotek, N. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J. Biol. Chem. 2001, 276, 3361–3370. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.S.; Hsu, L.J.; Lin, Y.S.; Lai, F.J.; Sheu, H.M. WW domain-containing oxidoreductase: A candidate tumor suppressor. Trends Mol. Med. 2007, 13, 12–22. [Google Scholar] [CrossRef]
- Abu-Remaileh, M.; Joy-Dodson, E.; Schueler-Furman, O.; Aqeilan, R.I. Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells. J. Biol. Chem. 2015, 290, 30728–30735. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Lee, J.; Abba, M.C.; Chen, J.; Aldaz, C.M. Delineating WWOX Protein Interactome by Tandem Affinity Purification-Mass Spectrometry: Identification of Top Interactors and Key Metabolic Pathways Involved. Front. Oncol. 2018, 8, 591. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.T.; Liu, C.C.; Chen, S.T.; Yap, Y.V.; Chang, N.S.; Sze, C.I. WW domain-containing oxidoreductase in neuronal injury and neurological diseases. Oncotarget 2014, 5, 11792–11799. [Google Scholar] [CrossRef]
- Chang, N.S. Bubbling cell death: A hot air balloon released from the nucleus in the cold. Exp. Biol. Med. 2016, 241, 1306–1315. [Google Scholar] [CrossRef]
- Chen, S.J.; Lin, P.W.; Lin, H.P.; Huang, S.S.; Lai, F.J.; Sheu, H.M.; Hsu, L.J.; Chang, N.S. UV irradiation/cold shock-mediated apoptosis is switched to bubbling cell death at low temperatures. Oncotarget 2015, 6, 8007–8018. [Google Scholar] [CrossRef]
- Hsu, L.J.; Chiang, M.F.; Sze, C.I.; Su, W.P.; Yap, Y.V.; Lee, I.T.; Kuo, H.L.; Chang, N.S. HYAL-2-WWOX-SMAD4 Signaling in Cell Death and Anticancer Response. Front. Cell Dev. Biol. 2016, 4, 141. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Lee, K.T.; Sun, T.Y.; Sze, C.I.; Huang, S.S.; Hsu, L.J.; Chang, N.S. WWOX and Its Binding Proteins in Neurodegeneration. Cells 2021, 10, 1781. [Google Scholar] [CrossRef]
- Aldaz, C.M.; Hussain, T. WWOX Loss of Function in Neurodevelopmental and Neurodegenerative Disorders. Int. J. Mol. Sci. 2020, 21, 8922. [Google Scholar] [CrossRef]
- Liu, C.C.; Ho, P.C.; Lee, I.T.; Chen, Y.A.; Chu, C.H.; Teng, C.C.; Wu, S.N.; Sze, C.I.; Chiang, M.F.; Chang, N.S. WWOX Phosphorylation, Signaling, and Role in Neurodegeneration. Front. Neurosci. 2018, 12, 563. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Abu-Odeh, M.; Bar-Mag, T.; Huang, H.; Kim, T.; Salah, Z.; Abdeen, S.K.; Sudol, M.; Reichmann, D.; Sidhu, S.; Kim, P.M.; et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J. Biol. Chem. 2014, 289, 8865–8880. [Google Scholar] [CrossRef] [PubMed]
- Bouteille, N.; Driouch, K.; Hage, P.E.; Sin, S.; Formstecher, E.; Camonis, J.; Lidereau, R.; Lallemand, F. Inhibition of the Wnt/beta-catenin pathway by the WWOX tumor suppressor protein. Oncogene 2009, 28, 2569–2580. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.S.; Doherty, J.; Ensign, A. JNK1 physically interacts with WW domain-containing oxidoreductase (WOX1) and inhibits WOX1-mediated apoptosis. J. Biol. Chem. 2003, 278, 9195–9202. [Google Scholar] [CrossRef]
- Hsu, L.J.; Schultz, L.; Hong, Q.; Van Moer, K.; Heath, J.; Li, M.Y.; Lai, F.J.; Lin, S.R.; Lee, M.H.; Lo, C.P.; et al. Transforming growth factor beta1 signaling via interaction with cell surface Hyal-2 and recruitment of WWOX/WOX1. J. Biol. Chem. 2009, 284, 16049–16059. [Google Scholar] [CrossRef]
- Aldaz, C.M.; Ferguson, B.W.; Abba, M.C. WWOX at the crossroads of cancer, metabolic syndrome related traits and CNS pathologies. Biochim. Biophys. Acta 2014, 1846, 188–200. [Google Scholar] [CrossRef]
- Chen, Y.A.; Lu, C.Y.; Cheng, T.Y.; Pan, S.H.; Chen, H.F.; Chang, N.S. WW Domain-Containing Proteins YAP and TAZ in the Hippo Pathway as Key Regulators in Stemness Maintenance, Tissue Homeostasis, and Tumorigenesis. Front. Oncol. 2019, 9, 60. [Google Scholar] [CrossRef]
- Bendinelli, P.; Maroni, P.; Matteucci, E.; Desiderio, M.A. HGF and TGFβ1 differently influenced Wwox regulatory function on Twist program for mesenchymal-epithelial transition in bone metastatic versus parental breast carcinoma cells. Mol. Cancer 2015, 14, 112. [Google Scholar] [CrossRef]
- Chang, R.; Song, L.; Xu, Y.; Wu, Y.; Dai, C.; Wang, X.; Sun, X.; Hou, Y.; Li, W.; Zhan, X.; et al. Loss of Wwox drives metastasis in triple-negative breast cancer by JAK2/STAT3 axis. Nat. Commun. 2018, 9, 3486. [Google Scholar] [CrossRef]
- Taouis, K.; Driouch, K.; Lidereau, R.; Lallemand, F. Molecular Functions of WWOX Potentially Involved in Cancer Development. Cells 2021, 10, 1051. [Google Scholar] [CrossRef] [PubMed]
- Abu-Odeh, M.; Salah, Z.; Herbel, C.; Hofmann, T.G.; Aqeilan, R.I. WWOX, the common fragile site FRA16D gene product, regulates ATM activation and the DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 4716–4725. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Pekarsky, Y.; Herrero, J.J.; Palamarchuk, A.; Letofsky, J.; Druck, T.; Trapasso, F.; Han, S.Y.; Melino, G.; Huebner, K.; et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc. Natl. Acad. Sci. USA 2004, 101, 4401–4406. [Google Scholar] [CrossRef] [PubMed]
- Schuchardt, B.J.; Mikles, D.C.; Bhat, V.; McDonald, C.B.; Sudol, M.; Farooq, A. Allostery mediates ligand binding to WWOX tumor suppressor via a conformational switch. J. Mol. Recognit. 2015, 28, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Chou, P.Y.; Lin, S.R.; Lee, M.H.; Schultz, L.; Sze, C.I.; Chang, N.S. A p53/TIAF1/WWOX triad exerts cancer suppression but may cause brain protein aggregation due to p53/WWOX functional antagonism. Cell Commun. Signal. 2019, 17, 76. [Google Scholar] [CrossRef]
- Chen, Y.A.; Sie, Y.D.; Liu, T.Y.; Kuo, H.L.; Chou, P.Y.; Chen, Y.J.; Lee, K.T.; Chen, P.J.; Chen, S.T.; Chang, N.S. Normal cells repel WWOX-negative or -dysfunctional cancer cells via WWOX cell surface epitope 286–299. Commun. Biol. 2021, 4, 753. [Google Scholar] [CrossRef]
- Chou, P.Y.; Lai, F.J.; Chen, Y.A.; Sie, Y.D.; Kuo, H.L.; Su, W.P.; Wu, C.Y.; Liu, T.Y.; Wen, K.Y.; Hsu, L.J.; et al. Strategies by which WWOX-deficient metastatic cancer cells utilize to survive via dodging, compromising, and causing damage to WWOX-positive normal microenvironment. Cell Death Discov. 2019, 5, 97. [Google Scholar] [CrossRef]
- Sze, C.I.; Su, M.; Pugazhenthi, S.; Jambal, P.; Hsu, L.J.; Heath, J.; Schultz, L.; Chang, N.S. Down-regulation of WW domain-containing oxidoreductase induces Tau phosphorylation in vitro. A potential role in Alzheimer’s disease. J. Biol. Chem. 2004, 279, 30498–30506. [Google Scholar] [CrossRef]
- Wang, H.Y.; Juo, L.I.; Lin, Y.T.; Hsiao, M.; Lin, J.T.; Tsai, C.H.; Tzeng, Y.H.; Chuang, Y.C.; Chang, N.S.; Yang, C.N.; et al. WW domain-containing oxidoreductase promotes neuronal differentiation via negative regulation of glycogen synthase kinase 3β. Cell Death Differ. 2012, 19, 1049–1059. [Google Scholar] [CrossRef]
- Cheng, Y.Y.; Chou, Y.T.; Lai, F.J.; Jan, M.S.; Chang, T.H.; Jou, I.M.; Chen, P.S.; Lo, J.Y.; Huang, S.S.; Chang, N.S.; et al. Wwox deficiency leads to neurodevelopmental and degenerative neuropathies and glycogen synthase kinase 3β-mediated epileptic seizure activity in mice. Acta Neuropathol. Commun. 2020, 8, 6. [Google Scholar] [CrossRef]
- Steinberg, D.J.; Aqeilan, R.I. WWOX-Related Neurodevelopmental Disorders: Models and Future Perspectives. Cells 2021, 10, 3082. [Google Scholar] [CrossRef] [PubMed]
- Repudi, S.; Steinberg, D.J.; Elazar, N.; Breton, V.L.; Aquilino, M.S.; Saleem, A.; Abu-Swai, S.; Vainshtein, A.; Eshed-Eisenbach, Y.; Vijayaragavan, B.; et al. Neuronal deletion of Wwox, associated with WOREE syndrome, causes epilepsy and myelin defects. Brain 2021, 144, 3061–3077. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Lai, F.J.; Hsu, L.J.; Lo, C.P.; Cheng, C.L.; Lin, S.R.; Lee, M.H.; Chang, J.Y.; Subhan, D.; Tsai, M.S.; et al. Dramatic co-activation of WWOX/WOX1 with CREB and NF-kappaB in delayed loss of small dorsal root ganglion neurons upon sciatic nerve transection in rats. PLoS ONE 2009, 4, e7820. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.J.; Hong, Q.; Chen, S.T.; Kuo, H.L.; Schultz, L.; Heath, J.; Lin, S.R.; Lee, M.H.; Li, D.Z.; Li, Z.L.; et al. Hyaluronan activates Hyal-2/WWOX/Smad4 signaling and causes bubbling cell death when the signaling complex is overexpressed. Oncotarget 2017, 8, 19137–19155. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Donati, V.; Palamarchuk, A.; Trapasso, F.; Kaou, M.; Pekarsky, Y.; Sudol, M.; Croce, C.M. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005, 65, 6764–6772. [Google Scholar] [CrossRef]
- Yang, T.; Xu, R.; Huo, J.; Wang, B.; Du, X.; Dai, B.; Zhu, M.; Zhan, Y.; Zhang, D.; Zhang, Y. WWOX activation by toosendanin suppresses hepatocellular carcinoma metastasis through JAK2/Stat3 and Wnt/β-catenin signaling. Cancer Lett. 2021, 513, 50–62. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, C.J.; Tian, Y.; Wang, Y.P.; Han, Z.G.; Li, X.C. Ectopic overexpression of COTE1 promotes cellular invasion of hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2012, 13, 5799–5804. [Google Scholar] [CrossRef]
- Chang, N.S.; Doherty, J.; Ensign, A.; Schultz, L.; Hsu, L.J.; Hong, Q. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J. Biol. Chem. 2005, 280, 43100–43108. [Google Scholar] [CrossRef]
- Jin, C.; Ge, L.; Ding, X.; Chen, Y.; Zhu, H.; Ward, T.; Wu, F.; Cao, X.; Wang, Q.; Yao, X. PKA-mediated protein phosphorylation regulates ezrin-WWOX interaction. Biochem. Biophys. Res. Commun. 2006, 341, 784–791. [Google Scholar] [CrossRef]
- Lee, M.H.; Shih, Y.H.; Lin, S.R.; Chang, J.Y.; Lin, Y.H.; Sze, C.I.; Kuo, Y.M.; Chang, N.S. Zfra restores memory deficits in Alzheimer’s disease triple-transgenic mice by blocking aggregation of TRAPPC6AΔ, SH3GLB2, tau, and amyloid β, and inflammatory NF-κB activation. Alzheimers Dement. 2017, 3, 189–204. [Google Scholar] [CrossRef]
- Lee, M.H.; Su, W.P.; Wang, W.J.; Lin, S.R.; Lu, C.Y.; Chen, Y.A.; Chang, J.Y.; Huang, S.S.; Chou, P.Y.; Ye, S.R.; et al. Zfra activates memory Hyal-2+ CD3- CD19- spleen cells to block cancer growth, stemness, and metastasis in vivo. Oncotarget 2015, 6, 3737–3751. [Google Scholar] [CrossRef] [PubMed]
- Su, W.P.; Wang, W.J.; Sze, C.I.; Chang, N.S. Zfra induction of memory anticancer response via a novel immune cell. Oncoimmunology 2016, 5, e1213935. [Google Scholar] [CrossRef] [PubMed]
- Su, W.P.; Wang, W.J.; Chang, J.Y.; Ho, P.C.; Liu, T.Y.; Wen, K.Y.; Kuo, H.L.; Chen, Y.J.; Huang, S.S.; Subhan, D.; et al. Therapeutic Zfra4-10 or WWOX7-21 Peptide Induces Complex Formation of WWOX with Selective Protein Targets in Organs that Leads to Cancer Suppression and Spleen Cytotoxic Memory Z Cell Activation In Vivo. Cancers 2020, 12, 2189. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Ho, P.C.; Nagarajan, G.; Chen, Y.A.; Kuo, H.L.; Subhan, D.; Su, W.P.; Chang, J.Y.; Lu, C.Y.; Chang, K.T.; et al. WWOX Possesses N-Terminal Cell Surface-Exposed Epitopes WWOX(7-21) and WWOX(7-11) for Signaling Cancer Growth Suppression and Prevention In Vivo. Cancers 2019, 11, 1818. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.C. The Contrast Formation in Optical Microscopy. In Handbook of Biological Confocal Microscopy; Pawley, J.B., Ed.; Springer: Boston, MA, USA, 2006; pp. 162–206. [Google Scholar]
- Huang, S.S.; Su, W.P.; Lin, H.P.; Kuo, H.L.; Wei, H.L.; Chang, N.S. Role of WW Domain-containing Oxidoreductase WWOX in Driving T Cell Acute Lymphoblastic Leukemia Maturation. J. Biol. Chem. 2016, 291, 17319–17331. [Google Scholar] [CrossRef]
- Che, Y.; Khavari, P.A. Research Techniques Made Simple: Emerging Methods to Elucidate Protein Interactions through Spatial Proximity. J. Investig. Dermatol. 2017, 137, e197–e203. [Google Scholar] [CrossRef]
- Kuo, H.L.; Ho, P.C.; Huang, S.S.; Chang, N.S. Chasing the signaling run by tri-molecular time-lapse FRET microscopy. Cell Death Discov. 2018, 4, 45. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, J.; Sun, W.; Huo, Y.; Zhang, L.; Hao, P.; Wang, H.; Zhuang, M. A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 2018, 15, 715–722. [Google Scholar] [CrossRef]
- Imani, M.; Mohajeri, N.; Rastegar, M.; Zarghami, N. Recent advances in FRET-Based biosensors for biomedical applications. Anal. Biochem. 2021, 630, 114323. [Google Scholar] [CrossRef]
- Feng, X.A.; Poyton, M.F.; Ha, T. Multicolor single-molecule FRET for DNA and RNA processes. Curr. Opin. Struct. Biol. 2021, 70, 26–33. [Google Scholar] [CrossRef]
- Nakamura, A.; Goto, Y.; Kondo, Y.; Aoki, K. Shedding light on developmental ERK signaling with genetically encoded biosensors. Development 2021, 148, dev199767. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Lin, S.R.; Chang, J.Y.; Schultz, L.; Heath, J.; Hsu, L.J.; Kuo, Y.M.; Hong, Q.; Chiang, M.F.; Gong, C.X.; et al. TGF-β induces TIAF1 self-aggregation via type II receptor-independent signaling that leads to generation of amyloid β plaques in Alzheimer’s disease. Cell Death Dis. 2010, 1, e110. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Chiang, M.F.; Lin, S.R.; Lee, M.H.; He, H.; Chou, P.Y.; Chen, S.J.; Chen, Y.A.; Yang, L.Y.; Lai, F.J.; et al. TIAF1 self-aggregation in peritumor capsule formation, spontaneous activation of SMAD-responsive promoter in p53-deficient environment, and cell death. Cell Death Dis. 2012, 3, e302. [Google Scholar] [CrossRef]
- Hong, Q.; Hsu, L.J.; Chou, P.Y.; Chou, Y.T.; Lu, C.Y.; Chen, Y.A.; Chang, N.S. Self-aggregating TIAF1 in lung cancer progression. Transl. Respir. Med. 2013, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Chang, N.S. WWOX dysfunction induces sequential aggregation of TRAPPC6AΔ, TIAF1, tau and amyloid β, and causes apoptosis. Cell Death Discov. 2015, 1, 15003. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.S. Introduction to a thematic issue for WWOX. Exp. Biol. Med. 2015, 240, 281–284. [Google Scholar] [CrossRef]
- Anbarasan, T.; Bourdon, J.C. The Emerging Landscape of p53 Isoforms in Physiology, Cancer and Degenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6257. [Google Scholar] [CrossRef]
- Joruiz, S.M.; Beck, J.A.; Horikawa, I.; Harris, C.C. The Δ133p53 Isoforms, Tuners of the p53 Pathway. Cancers 2020, 12, 3422. [Google Scholar] [CrossRef]
- Free, R.B.; Hazelwood, L.A.; Sibley, D.R. Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr. Protoc. Neurosci. 2009, 46, 5–28. [Google Scholar] [CrossRef]
- Vojtek, A.B.; Hollenberg, S.M. Ras-Raf interaction: Two-hybrid analysis. Methods Enzymol. 1995, 255, 331–342. [Google Scholar]
- Chang, N.S. The non-ankyrin C terminus of Ikappa Balpha physically interacts with p53 in vivo and dissociates in response to apoptotic stress, hypoxia, DNA damage, and transforming growth factor-beta 1-mediated growth suppression. J. Biol. Chem. 2002, 277, 10323–10331. [Google Scholar] [CrossRef] [PubMed]
- Deeks, E.D. Ceritinib: A Review in ALK-Positive Advanced NSCLC. Target. Oncol. 2016, 11, 693–700. [Google Scholar] [CrossRef]
- Hsu, L.J.; Schultz, L.; Mattison, J.; Lin, Y.S.; Chang, N.S. Cloning and characterization of a small-size peptide Zfra that regulates the cytotoxic function of tumor necrosis factor by interacting with JNK1. Biochem. Biophys. Res. Commun. 2005, 327, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.J.; Hong, Q.; Schultz, L.; Kuo, E.; Lin, S.R.; Lee, M.H.; Lin, Y.S.; Chang, N.S. Zfra is an inhibitor of Bcl-2 expression and cytochrome c release from the mitochondria. Cell. Signal. 2008, 20, 1303–1312. [Google Scholar] [CrossRef]
- Huppert, J.; Closhen, D.; Croxford, A.; White, R.; Kulig, P.; Pietrowski, E.; Bechmann, I.; Becher, B.; Luhmann, H.J.; Waisman, A.; et al. Cellular mechanisms of IL-17-induced blood-brain barrier disruption. FASEB J. 2010, 24, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.T.; Chuang, J.I.; Wang, J.P.; Tsai, M.S.; Li, H.; Chang, N.S. Expression of WW domain-containing oxidoreductase WOX1 in the developing murine nervous system. Neuroscience 2004, 124, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.T.; Chuang, J.I.; Cheng, C.L.; Hsu, L.J.; Chang, N.S. Light-induced retinal damage involves tyrosine 33 phosphorylation, mitochondrial and nuclear translocation of WW domain-containing oxidoreductase in vivo. Neuroscience 2005, 130, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.F.; Chen, S.T.; Lo, C.P.; Sze, C.I.; Chang, N.S.; Chen, Y.J. Expression of WW domain-containing oxidoreductase WOX1 in human nervous system tumors. Anal. Cell. Pathol. 2013, 36, 133–147. [Google Scholar] [CrossRef]
- Lo, C.P.; Hsu, L.J.; Li, M.Y.; Hsu, S.Y.; Chuang, J.I.; Tsai, M.S.; Lin, S.R.; Chang, N.S.; Chen, S.T. MPP+-induced neuronal death in rats involves tyrosine 33 phosphorylation of WW domain-containing oxidoreductase WOX1. Eur. J. Neurosci. 2008, 27, 1634–1646. [Google Scholar] [CrossRef]
- Ludes-Meyers, J.H.; Kil, H.; Bednarek, A.K.; Drake, J.; Bedford, M.T.; Aldaz, C.M. WWOX binds the specific proline-rich ligand PPXY: Identification of candidate interacting proteins. Oncogene 2004, 23, 5049–5055. [Google Scholar] [CrossRef]
- McDonald, C.B.; Buffa, L.; Bar-Mag, T.; Salah, Z.; Bhat, V.; Mikles, D.C.; Deegan, B.J.; Seldeen, K.L.; Malhotra, A.; Sudol, M.; et al. Biophysical basis of the binding of WWOX tumor suppressor to WBP1 and WBP2 adaptors. J. Mol. Biol. 2012, 422, 58–74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reuven, N.; Shanzer, M.; Shaul, Y. Tyrosine phosphorylation of WW proteins. Exp. Biol. Med. 2015, 240, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, H.; Huang, Y.F.; Li, M.L.; Cheng, J.H.; Hu, P.; Lu, C.H.; Zhang, Y.; Liu, N.; Tzeng, C.M.; et al. WW domain-binding protein 2: An adaptor protein closely linked to the development of breast cancer. Mol. Cancer 2017, 16, 128. [Google Scholar] [CrossRef] [PubMed]
- Saigo, C.; Kito, Y.; Takeuchi, T. Cancerous Protein Network That Inhibits the Tumor Suppressor Function of WW Domain-Containing Oxidoreductase (WWOX) by Aberrantly Expressed Molecules. Front. Oncol. 2018, 8, 350. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Ruthel, G.; Freedman, B.D.; Harty, R.N. WWOX-Mediated Degradation of AMOTp130 Negatively Affects Egress of Filovirus VP40 Virus-Like Particles. J. Virol. 2022, 96, e0202621. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.-Y.; Nagarajan, G.; Chiang, M.-F.; Huang, S.-S.; Lin, T.-C.; Chen, Y.-A.; Sze, C.-I.; Chang, N.-S. WWOX Controls Cell Survival, Immune Response and Disease Progression by pY33 to pS14 Transition to Alternate Signaling Partners. Cells 2022, 11, 2137. https://doi.org/10.3390/cells11142137
Liu T-Y, Nagarajan G, Chiang M-F, Huang S-S, Lin T-C, Chen Y-A, Sze C-I, Chang N-S. WWOX Controls Cell Survival, Immune Response and Disease Progression by pY33 to pS14 Transition to Alternate Signaling Partners. Cells. 2022; 11(14):2137. https://doi.org/10.3390/cells11142137
Chicago/Turabian StyleLiu, Tsung-Yun, Ganesan Nagarajan, Ming-Fu Chiang, Shenq-Shyang Huang, Tzu-Chia Lin, Yu-An Chen, Chun-I Sze, and Nan-Shan Chang. 2022. "WWOX Controls Cell Survival, Immune Response and Disease Progression by pY33 to pS14 Transition to Alternate Signaling Partners" Cells 11, no. 14: 2137. https://doi.org/10.3390/cells11142137
APA StyleLiu, T.-Y., Nagarajan, G., Chiang, M.-F., Huang, S.-S., Lin, T.-C., Chen, Y.-A., Sze, C.-I., & Chang, N.-S. (2022). WWOX Controls Cell Survival, Immune Response and Disease Progression by pY33 to pS14 Transition to Alternate Signaling Partners. Cells, 11(14), 2137. https://doi.org/10.3390/cells11142137





