Abstract
Ovarian germline stem cells (GSCs) of Drosophila melanogaster provide a valuable in vivo model to investigate how the adult stem cell identity is maintained and the differentiation of the daughter cells is regulated. GSCs are embedded into a specialized cellular microenvironment, the so-called stem cell niche. Besides the complex signaling interactions between the germ cells and the niche cells, the germ cell intrinsic mechanisms, such as chromatin regulation and transcriptional control, are also crucial in the decision about self-renewal and differentiation. The key differentiation regulator gene is the bag of marbles (bam), which is transcriptionally repressed in the GSCs and de-repressed in the differentiating daughter cell. Here, we show that the transcription factor MESR4 functions in the germline to promote GSC daughter differentiation. We find that the loss of MESR4 results in the accumulation of GSC daughter cells which fail to transit from the pre-cystoblast (pre-CB) to the differentiated cystoblast (CB) stage. The forced expression of bam can rescue this differentiation defect. By a series of epistasis experiments and a transcriptional analysis, we demonstrate that MESR4 positively regulates the transcription of bam. Our results suggest that lack of repression alone is not sufficient, but MESR4-mediated transcriptional activation is also required for bam expression.
1. Introduction
Stem cells are undifferentiated cells possessing the unique ability of unlimited cell division capacity. Stem cells can undergo symmetric or asymmetric mitotic divisions to accomplish the dual task of self-renewal and the generation of differentiated cells [1]. The symmetric division of a stem cell can produce only stem cell daughters in some divisions and only differentiated daughters in others. This way, the number of stem cells can expand during development or regeneration while maintaining the balance between stem cells and differentiated cells. Upon asymmetric mitosis of a stem cell, however, one of its daughters remains in the stem cell state to maintain the undifferentiated stem cell pool, while the other daughter cell starts to differentiate to support developmental processes or to ensure tissue homeostasis [2]. The better understanding of the mechanisms behind the decision between self-renewal and differentiation is a key question in stem cell biology. The regulation of the Drosophila female germline stem cells (GSCs) provides an excellent model for studying stem cell maintenance and differentiation processes [3,4,5]. In the Drosophila females, a population of GSCs is located in the niches at the anterior tip of the ovary [6]. Each GSC divides asymmetrically and generates a daughter cell staying anchored at the tip of the niche and remaining in the GSC state, while the other daughter cell becomes a pre-cystoblast (pre-CB) committed to differentiate [7,8]. After mitosis, the GSC and the newly formed pre-CB remain physically connected due to an incomplete cytokinesis. The thin cytoplasmic connection is abscised in the G2 phase of the subsequent cell cycle [9,10,11]. The pre-CB then starts to differentiate and becomes a cystoblast (CB) which undergoes four synchronous, incomplete divisions, and gives rise to a sixteen-cell cyst. One cell of the sixteen-cell cyst matures to an oocyte, and the others differentiate into nurse cells. The differentiation state of the germ cells can be easily followed by visualizing specific cytoplasmic organelles. GSCs, pre-CBs and CBs contain a dot like cytoplasmic organelle, the spectrosome, while the cyst is marked by a branched structure, called the fusome (Figure 1A).
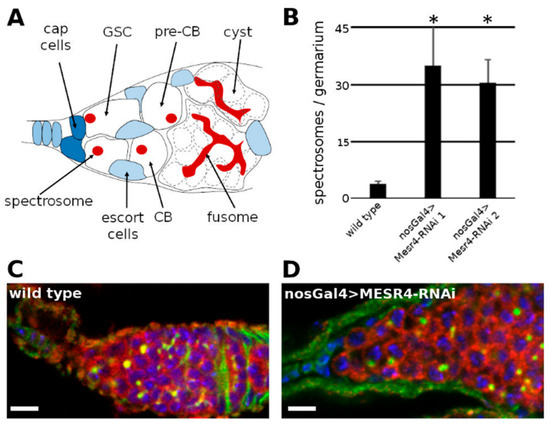
Figure 1.
MESR4 cell-autonomously promotes germ cell differentiation. (A) Schematic representation of a Drosophila niche where the germ cells (white) are surrounded by somatic cells (blue). The germline stem cells (GSCs) reside in the anterior tip of the niche in a specific somatic microenvironment, the stem cell niche, maintained by somatic cells. The GSCs divide continuously and give rise to a self-renewing daughter stem cell and a differentiating daughter cell, called pre-cystoblast (pre-CB). The pre-CB starts to express differentiation factors and becomes cystoblast (CB). The CB undergoes incomplete cell divisions to give rise to 2, 4, 8 and 16-cell cysts. The GSCs, pre-CBs and CBs contain a dot like structure called spectrosome (red) and the more differentiated cysts contain a branched fusome (red). (B) Quantification of single, spectrosome containing cells in wild type and MESR4 silenced niches using two different RNAi lines (35.0 ± 10.2 in MESR4-RNAi line 1, n = 24 and 30.5 ± 6.1 in MESR4-RNAi line 2 n = 24 compared to 3.9 ± 0.8 in wild type control, n = 24). Data are mean ± s.d.; t-test, * p < 0.05. (C,D) Immunostaining of wild type (C) and nosGal4 > MESR4-RNAi (D) niche with germ cell tumor. Spectrosomes and fusomes are labelled with HTS (green), germ cells are labelled for Vasa (red); DAPI is blue. Scale bars are 10 µm.
GSC self-renewal and differentiation are tightly regulated by the germ cell’s intrinsic and extrinsic factors [12]. The extrinsic factors are generated by cap cells and escort cells which surround the GSCs. These somatic cells create the stem cell niche and regulate the GSC differentiation [13,14,15]. The somatic niche cells secret Decapentaplegic (dpp): the Drosophila homolog of the Bone morphogenetic protein [8]. Dpp forms a short-range morphogen gradient and activates the dpp signaling pathway exclusively in the GSCs [16]. The main function of the dpp pathway in the GSCs is to repress the differentiation, and thus maintain the GSCs’ undifferentiated state [17]. Following mitosis of the GSCs, the pre-CBs are displaced from the stem cell niche losing the physical contact with the somatic cap cells, i.e., the source of the inhibitory dpp signal. At this position, the pre-CBs receive less Dpp, which leads to their differentiation into CBs [18].
Besides the extrinsic factors, the germ cell intrinsic programs are crucial to control the differentiation of the GSCs [12]. The transition from self-renewal to differentiation relies on epigenetic mechanisms, mRNA regulation and on protein metabolism. In GSCs, Nanos and Pumilio suppress the translation of mRNAs that promote GSC differentiation [19,20,21]. The miRNA-based (miRNA) silencing machinery also plays an essential role in GSC self-renewal by translational repression of differentiation factors in the GSCs [22,23,24]. In the CBs, the enhanced biogenesis of ribosomes and an increased global protein synthesis have been shown to be required for differentiation [25].
In addition to the regulation of protein turnover and translational repression, the epigenetic control of transcription is also involved in switching from the stem cell fate to the differentiated state [26,27] Epigenetic mechanisms include structural changes of the chromatin or recruitment of transcription regulators, which prevent the ectopic expression of germline differentiation genes in GSCs or activate them in pre-CBs and CBs.
The key regulator gene promoting GSC differentiation is bam. Pre-CBs lacking bam expression fail to differentiate, whereas ectopic bam expression in the GSCs leads to precocious differentiation and GSC loss, indicating that bam is both necessary and sufficient for the differentiation. These observations have led to the simple model in which silenced bam expression in the GSCs maintains self-renewal, and bam de-repression in the pre-CBs promotes differentiation [7,8,17].
In the GSCs, the expression of bam is suppressed by the transcription factor Mad, which is activated by phosphorylation in response to the dpp signal [17]. Phosphorylated Mad (pMad) translocates into the nucleus to directly suppress bam transcription. In the pre-CBs, however, turnover of pMad is shifted towards its degradation, which leads to the de-repression of bam transcription and to differentiation [28]. Recently, the transcription factor Krüppel (Kr) has been demonstrated to be involved in the temporal regulation of bam transcription independently of the dpp signaling. [29]. Krüppel suppresses bam transcription in the germ cells at the larval stages preventing their precocious differentiation into CBs.
Besides trans acting factors, cis regulatory elements are essential for the proper temporal control of bam transcription. A suppressor element, located upstream of the transcription start site, associates with Kr to maintain the repressed state of bam in the larval germ cells [29]. Another 18 bp long silencer element (SE) in the 5′UTR of the bam gene mediates the dpp-dependent suppression of bam transcription in the GSCs. The bam transcriptional regulator contains an enhancer element responsible for germ cell specific bam expression. In the GSCs, the Dpp signal maintains the silencer in an active state and bam transcription is suppressed. At the pre-CB stage, the SE becomes inactive and the bam transcription commences under the control of the active enhancer elements, which switches the state of the pre-CB to the CB fate [17,30].
While the negative regulation of bam expression is well known, the positive regulation is poorly studied. Here, we demonstrate that MESR4 is a trans acting factor positively regulating bam transcription to promote differentiation of the GSCs. GSCs lacking MESR4 initiate their differentiation into pre-CBs, but fail to adopt the differentiated CB fate, which leads to the accumulation of pre-CBs in the niche. We propose a model in which MESR4 promotes bam expression in the pre-CBs to support the transition of the committed pre-CB to the differentiated CB state.
2. Materials and Methods
2.1. Drosophila Stocks and Genetics
Flies were raised at 25 °C unless specified otherwise. The strains used in this study include: bamP-Bam:GFP and bamPΔSE-Bam:GFP [30], MESR479 [31], bam-Gal4 and bamP-GFP [30], hs-bam [7], Pgc:GFP [32] nos-Gal4Vp16 [33], c587-Gal4 (Bl#67747), osk-Gal4VP16 (BL#44242), Pmatalpha4-Gal4VP16 (Bl#706), P{TRiP.HMC04881}attP40 (MESR4-RNAi-1 Bl#57564)), P{TRiP.GL00462}attP2 (MESR4-RNAi-2 Bl#35618), P{TRiP.HMS00029}attP2 (Bam-RNAi Bl#33631), bgcnEY00974 (Bl#20106), vasa-Cas9 (Bl#51323), MESR4-GFP.FPTB (Bl#67731), bgcn1 (Bl#6054), Df(2R)BSC136 (Bl#9424).
The MESR4ΔPHD allele was generated by CRISPR/Cas9 mediated genome engineering [34]. To generate the MESR4ΔPHD allele, target sites of two sgRNAs were inserted into the pCFD4 vector (Addgene, 49411) and introduced into the attP2 docking site on the Drosophila genome by standard transformation methods [34] (Table 1). MESR4+; Vasa-cas9/attP2-pCFD4-MESR4-gRNA males were generated and the mutagenized second chromosomes were isolated. The MESR4ΔPHD deletion allele was confirmed by sequencing.

Table 1.
List of sgRNAs used for MESR4 mutant generation.
2.2. Heat Shock
Two-day-old nosGal4 > MESR4-RNAi; hs-bam females were heat-shocked at 37 °C twice for 1 h, separated by a 2 h recovery period at 25 °C. For the control, nosGa4 > MESR4-RNAi; hs-bam flies without heat shock were used. The phenotypes were examined one day after heat shocks.
2.3. Immunohistochemistry
Immunostainings were performed as described earlier [35,36]. The following primary antibodies were used: anti-vasa (1:300, DSHB), anti-HTS (1:20, DSHB, 1B1), anti-GFP (1:500, Invitrogen, Waltham, MA, USA, A-11122), anti-Smad3 (1:100, abcam, ab52903), anti-cycB (1:30 DSHB, F2F4). The tissues were observed with a Leica SP5 or with Zeiss LSM800 confocal microscope.
2.4. Quantitative PCR
For the quantitative PCR, total RNA was prepared using the Reliaprep RNA tissue Miniprep System (Promega, Medison, WI, USA, Z6111). To ensure comparability of the ovaries, bgcn1/Df(2R)BSC136 mutant ovaries were used as a control and MESR4 and bam silencing was performed on bgcn1/Df(2R)BSC136 mutant background. Ovaries were collected from 2-days-old bgcn1/Df(2R)BSC136 and nosGal4>MESR4-RNAi and nosGal4 > bam-RNAi ovaries [37]. To achieve a comparable reduction of bam expression in MESR4-RNAi and bam-RNAi females, nosGal4 > bam-RNAi females were raised at 18 °C.
OligodT primers were used to synthetize cDNAs (First Strand cDNA Synthesis kit, ThermoScientific, Waltham, MA, USA, K1612). qPCR reactions were performed using SensiFAST SYBR Hi-ROX Kit (Bioline, London, UK, BIO-92005). The thermal cycling condition consisted of 95 °C for 30 min, 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, 35 cycles. Melt: 60–95 °C, 1 °C each step. For each reaction, there were three technical replicates and three biological replicates. An Rp49 transcript was used for internal control (Table 2). Rotor-Gene Q Series software was used for data analysis.

Table 2.
List of primers used for RT-qPCR.
3. Results
3.1. MESR4 Promotes GSC Differentiation in a Cell Autonomous Manner
MESR4 has been shown to be required for the proper germ cell development [31]. To determine the spatial and temporal requirement of MESR4, we silenced it in the ovarian somatic and germ cells at distinct stages of the germ cell differentiation by expressing MESR4-shRNAs with tissue specific Gal4 drivers with various expression patterns in the ovary.
First, we investigated how a somatic MESR4 function affects germline behavior. Therefore, the c587Gal4 line was used to induce MESR4 silencing in all somatic cell types of the ovary and the differentiation state of the germ cells was analyzed. By immunolabelling the spectrosomes and the fusomes, no GSC differentiation defects were detectable in c587Gal4 > MESR4-shRNA niches (Figure S1B). The adult c587Gal4 > MESR4-shRNA females were fertile, indicating that the MESR4 function in the ovarian soma is dispensable for germ cell development.
To examine the MESR4 function in the germline, we expressed two independent MESR4-shRNAs (GL00462 and HMS00029) by Gal4 drivers specifically active in the germ cells at various stages of the oogenesis. The oskGal4 and matTubGal4 drivers were used to silence MESR4 outside of the GSC niche, at the later stages of the germ cell development. No defects were detected in oskGal4 > MESR4-shRNA and matTubGal4 > MESR4-shRNA ovaries, indicating that MESR4 is required for the early germ cell development exclusively in the niche (Figure S1C,D).
Next, we silenced MESR4 by the nosGal4 driver which started to express in the GSCs, and its expression persisted throughout the oogenesis. While wild type control niches carried less than four spectrosome-containing cells (3.9 ± 0.8, n = 24) which could be GSCs, pre-CBs or CBs, the niches of the nosGal4 > MESR4-shRNA females contained many spectrosome-containing single germ cells (35.0 ± 10.2 in MESR4-RNAi line 1, n = 24 and 30.5 ± 6.1 in MESR4-RNAi line 2, n = 24) which formed germ cell tumors (Figure 1C,D). The accumulation of spectrosome-containing cells in the niches indicates that germ cells failed to differentiate. The silencing of MESR4 with both MESR4-shRNA lines resulted in identical differentiation defects confirming the specificity of the tumorous MESR4-RNAi phenotype (Figure 1B). In summary, these results have revealed that MESR4 is required for germ cell development and functions exclusively in the germline where it promotes the differentiation of the germ cells in a cell autonomous manner.
3.2. MESR4 Regulates Germ Cell Differentiation at the Pre-CB Stage
The accumulated undifferentiated germ cells observed in the nosGal4 > MESR4-shRNA niches could be GSCs which maintain their self-renewal capacity outside of the niche. Alternatively, these cells are daughter cells of GSCs, pre-CBs or CBs, which fail to complete their differentiation program. To distinguish between these possibilities, we analyzed the presence of GSC and CB specific molecular markers in the MESR4-RNAi niches. In the wild type, GSCs can be identified by their physical contact with the cap cells and by the presence of phosphorylated Mad (pMad), a reporter of an active dpp pathway. Similar to wild type, in the nosGal4 > MESR4-shRNA niches, two to three GSCs accumulated pMad in their nuclei (Figure 2A,B). NosGal4 > MESR4-RNAi cells located outside the GSC niche lacked nuclear pMad, indicating that these cells are not GSCs.
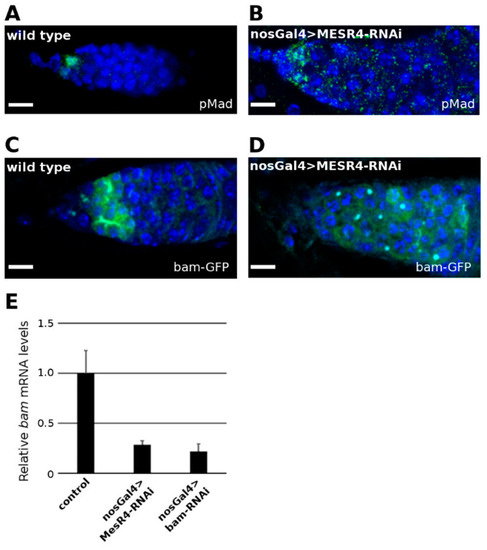
Figure 2.
MESR4 promotes bam expression in the GSC daughter cells. (A,B) Wild type control (A) and nosGal4 > MESR4-RNAi niches (B) stained with pMad (green) and DAPI (blue). Germline-depleted MESR4 niches do not accumulate pMad positive cells. Scale bars: 10 µm. (C,D) Wild type control (C) and nosGal4 > MESR4-RNAi niches (D) expressing GFP under the control of the bam promoter. Niches were stained with GFP (green), and DAPI (blue). In the germline depleted MESR4 niches there are less GFP signals. Scale bars: 10 µm. (E) Quantitation of steady state bam mRNA levels in bgcn1/Df(2R)BSC136 control, nosGal4 > MESR4-RNAi and nosGal4 > bam-RNAi ovaries. Compared to the control (1.0 ± 0.2), bam mRNA levels are decreased in the MESR4 silenced (0.28 ± 0.03) and bam silenced (0.22 ± 0.07) ovaries.
The hallmark of differentiated CBs is the expression of bam which is initiated in the GSC daughter cells that lose physical contact with the cap cells. To monitor bam expression, the bamP-GFP transcriptional reporter line was used which contains the bam promoter region fused with a GFP coding sequence [30]. In wild type niches, strong GFP expression was detected (Figure 2C). In the undifferentiated nosGal4 > MESR4-shRNA germ cells, however, a week bam expression was detected when monitored with the bamP-GFP reporter line (Figure 2D). Consistent with this observation, a significant reduction of bam mRNA levels was found by qRT-PCR in the nosGal4 > MESR4-shRNA ovaries (Figure 2E). The bam mRNA levels of the MESR4-silenced germ cells were similar to these of the tumorous ovaries of nosGal4 > bam-shRNA females, indicating that this degree of reduction in bam mRNA levels is sufficient to prevent differentiation.
Next, we silenced MESR4 with the bamGal4 driver, which is specifically active in the differentiated CBs reflecting the expression pattern of the endogenous bam gene. In the bamGal4 > MESR4-shRNA ovaries, no tumorous niches were found, indicating that MESR4 is not required in bam expressing CBs (Figure S2A,B). Based on these data, we hypothesized that MESR4 functions upstream of bam to promote GSC differentiation. To validate our argument, we performed a genetic rescue experiment using the hs-bam transgenic line. Forced expression of bam from the heat shock inducible hs-bam transgene induced differentiation of the MESR4-RNAi germ cells. Following heat shock, fusome-containing cysts were formed in the nosGal4 > MESR4-shRNA; hs-bam niches (Figure 3B,C). Forced expression of bam resulted in the reduction of the number of GSC-like tumors (37.0% in nosGal4 > MESR4-shRNA; hs-bam, n = 27 vs. 96.3% in nosGal4 > MESR4-shRNA, n = 25) (Figure 3D). In summary, MESR4 regulates the differentiation in the committed GSC daughter cells by promoting bam expression.
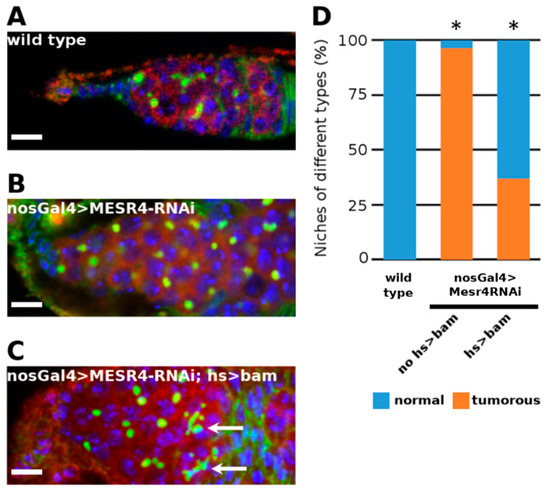
Figure 3.
Rescue of MESR4 silencing by forced bam expression. (A–D) Immunostaining a wild type (A) and a tumorous nosGal4 > MESR4-RNAi niche (B). Rescue of the niche defects in a nosGal4 > MESR4-RNAi; hs > bam niche after heat shock (C). Fusomes of differentiated germ cells are indicated by white arrows. Spectrosomes and fusomes are labelled with HTS (green); germ cells are labelled for Vasa (red); DAPI is blue. Scale bars: 10 µm. (D) Quantification of niche phenotypes of wild type (n = 27), nosGal4 > MESR4-RNAi (n = 25) and nosGal4 > MESR4-RNAi; hs > bam (n = 27) niches. “Normal” niches contain 1 to 6, “tumorous” niches contain more than 6 spectrosome containing germ cells. For statistical analysis, the Chi-square test was used, * p < 0.05.
3.3. MESR4 Is Required for the Transition from the Pre-CB to the CB Stage
An analysis of GSC and CB specific molecular markers of MESR4-silenced germ cells revealed that these cells exit the cells’ GSC state, but do not adopt the differentiated CB fate. Thus, we concluded that the silencing of MESR4 blocks the differentiation process at the pre-CB stage. To test this hypothesis, we investigated the expression of polar granule component (pgc), specifically activated in the pre-CBs prior to the expression of the differentiation factor bam [32]. We used the pgcGFP reporter line expressing EGFP under the control of the endogenous regulatory elements of pgc (Figure 4A). The silencing of bam resulted in the arrest of the differentiation process at the pre-CB stage, as indicated by the accumulation of the pgcGFP-positive pre-CBs in the nosGal4 > bam-shRNA niches (Figure 4B). Similar to bam silencing, the silencing of MESR4 resulted in the accumulation of the pgcGFP-positive pre-CBs in the nosGal4 > MESR4-shRNA niches, indicating that MESR4 promotes the transition from pre-CBs into the CB stage (Figure 4C).
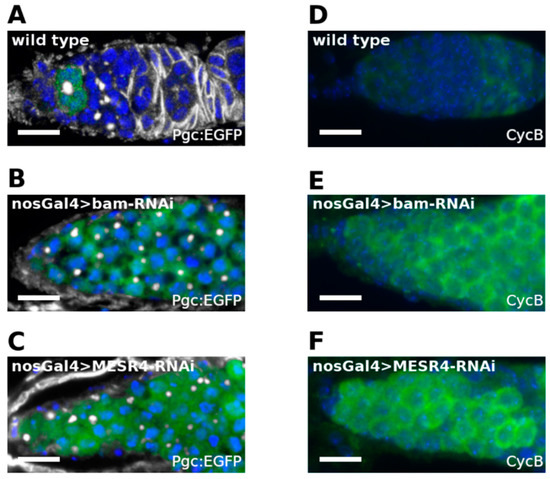
Figure 4.
Analysis of pre-CB specific molecular markers of MESR4-silenced germ cells. (A–C) Immunostaining of niches expressing Pgc:EGFP. Wild type (A), nosGal4 > MESR4-RNAi (B) and nosGal4 > bam-RNAi niches are stained with GFP (green), HTS (white) and DAPI (blue). Niches silenced for MESR4 or bam accumulate a high number of Pgc-positive pre-CBs. (D–F) Immunostaining of niches with CycB (green) and DAPI (blue). Wild type niche (D). NosGal4 > bam-RNAi (E) and nosGal4 > MESR4-RNAi (F) niches accumulate a high number of CycB positive cells.
Cell cycle control is tightly associated with the regulation of the pre-CB-to-CB transition. In the pre-CBs, Pgc indirectly promotes the accumulation of Cyclin B (CycB) at the G2 phase. Then, CycB activates the mechanisms leading to the expression of bam and the transition to the CB stage [32]. We correlated the phase of the cell cycle and the transition state from pre-CBs into the CB stage with the requirement of the MESR4 function. We investigated the expression of the cell cycle regulator CycB, a marker of late G2, in MESR4-silenced germ cells by immunostaining (Figure 4D) [32]. We observed a robust accumulation of CycB-positive germ cells in the nosGal4 > bam-shRNA niches (Figure 4E). Similar to bam silencing, undifferentiated nosGal4 > MESR4-shRNA germ cells were found to express CycB (Figure 4F). The accumulation of late G2 phase pre-CBs in the niches indicates that MESR4 functions downstream of pgc and CycB to promote bam expression. In summary, we conclude that MESR4 is required in the pre-CBs immediately before bam expression to drive the transition of the undifferentiated G2 phase pre-CBs into the differentiated CB state.
3.4. MESR4 Promotes the Transcription of Bam
To identify the molecular mechanisms by which MESR4 regulates GSC differentiation, we first determined the expression pattern of GFP-tagged MESR4 expressed from a genomic bacmid construct (MESR4:GFP) [38]. The tagged MESR4 variant completely rescued the lethal phenotype associated with the MESR479 mutation, indicating that the MESR4:GFP fusion protein is fully functional. We detected ubiquitous MESR4 expression in the somatic and germ cells of the ovaries of MESR4:GFP females. In the cells, MESR4 localized to the nuclei, suggesting a role for MESR4 in the transcriptional regulation of bam (Figure 5A).
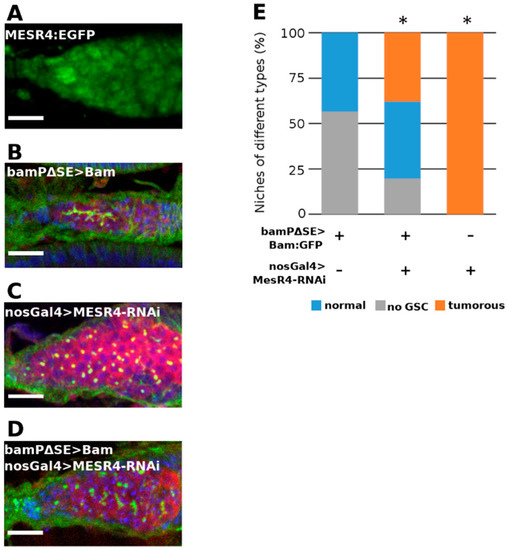
Figure 5.
MESR4 silencing suppresses the GSC-loss phenotype caused by de-repressed bam expression. (A) Immunostaining showing localization of MESR4:EGFP in the niche. MESR4 localizes in the nuclei of the somatic cells and in the nuclei of the germ cells. Scale bar: 10 µm. (B–D) Immunostaining of niches expressing transgenic bam under the control of the bam promoter lacking the pMad responsive SE silencing element. Niches were stained with DAPI (blue), vasa (red) and HTS (green). Scale bars: 10 µm. In bamPΔSE-Bam:GFP niches (B) GSCs were lost; in nosGal4 > MESR4-RNAi (C) niches GSC-like tumors were formed. In bamPΔSE-Bam:GFP; nosGal4 > MESR4-RNAi niches (D) the number of niches without GSCs were reduced. (E) Quantification of germ cell differentiation phenotypes of bamPΔSE-Bam:GFP (n = 99), bamPΔSE-Bam:GFP; nosGal4 > MESR4-RNAi (n = 87), and nosGal4 > MESR4-RNAi (n = 175) niches. Niches of the “no GSC” category lacked spectrosome containing germ cells, “normal” niches contain 1 to 6, “tumorous” niches contain more than 6 spectrosome containing germ cells. For statistical analysis, the Chi-square test was used, * p < 0.05.
The bam transcription has been shown to be negatively regulated by the dpp signaling pathway through the SE element of the bam regulator region. In the bamPΔSE-Bam:GFP transgenic females, the SE element is removed and the GSCs do not respond to the inhibitory pMad-mediated Dpp signal and initiate bam expression from the transgene. Ectopically expressed bam then leads to the differentiation of the GSCs into CBs and cysts, which in turn results in “empty” niches containing no GSCs (Figure 5B). To investigate whether the forced GSC differentiation caused by the de-repressed expression of bam can be rescued by the loss of MESR4, we used bamPΔSE-Bam:GFP transgenic females ectopically expressing bam in the GSCs and simultaneously silenced MESR4 in the germline. We found that in the bamPΔSE-Bam:GFP; nosGal4 > MESR4-shRNA ovaries the number of empty niches was reduced compared with bamPΔSE-Bam:GFP control flies (19.5%, n = 87 vs. 56.6%, n = 99) (Figure 5B–E). Furthermore, bamPΔSE-Bam:GFP; nosGal4 > MESR4-shRNA niches accumulated spectrosome containing single cells (37.9%, n = 87 vs. 0.0%, n = 99) (Figure 5B–E). Taken together, the suppression of GSC-loss phenotype by the silencing of MESR4 indicates that the MESR4 acts as antagonistic to the dpp-mediated bam suppression. In the pre-CBs, the lack of the dpp-mediated suppression is not sufficient, but also a MESR4-mediated positive regulation is required for the initiation of bam expression.
3.5. The PHD Domain Is Dispensable for MESR4 Function
An in silico analysis of MESR4 revealed that the MESR4 protein possesses a plant homeodomain (PHD) finger generally involved in chromatin remodeling nine C2H2 type zinc finger (ZF) domains mediating sequence specific DNA–protein interactions [39]. Due to its domain composition, MESR4 can control the bam expression by several, not mutually exclusive, mechanisms. By the PHD finger, it may act as a chromatin remodeling factor and promote bam expression by the formation of a permissive chromatin environment. Alternatively, MESR4 can function as a DNA-binding transcription factor associating directly with cis-regulatory elements of differentiation promoting genes by the ZF domains. To investigate how MESR4 controls the bam expression, we analyzed the activity of the MESR4 PHD finger domain using a CRISPR-based loss-of-function genetic approach.
PHD fingers have been reported to be involved in epigenetic gene regulation by mainly recognizing various unmodified or methylated lysines of H3 histone molecules [40]. To test the biological relevance of the PHD finger of MESR4, we applied CRISPR/Cas9-mediated gene editing to generate MESR4 alleles lacking this domain. This way, we isolated a MESR4ΔPHD allele lacking the C-terminal 106 amino acids of MESR4 covering the entire PHD finger. The homozygous mutant MESRΔPHD animals were viable, indicating that the PHD finger of MESR4 is not required for viability. The homozygous females were fertile, and wild type fusome containing cysts were detected in their niches. A lack of germ cell differentiation defects indicates that the PHD finger is dispensable for the MESR4 function in bam-mediated pre-CB to CB transition.
4. Discussion
The mechanisms behind the decision between self-renewal and differentiation act at multiple levels of gene regulation. Transcriptional control, regulation of protein or RNA turnover and translational regulation of differentiation genes act in concert to ensure the proper temporal and spatial regulation of GSC maintenance and differentiation. Here, we show that MESR4 controls the fate of germ cells in a very narrow developmental window by promoting the pre-CB to CB transition, and functions as a positive transcriptional regulator of the differentiation gene bam.
At the transcriptional level, GSC differentiation can be theoretically triggered by the repression of self-renewal factors, by preventing the expression of differentiation inhibitory factors or by stimulating the expression of differentiation factors. In these processes, epigenetic mechanisms, such as the incorporation of histone variants, posttranslational histone modifications and repositioning of nucleosomes by chromatin remodelers play a key role by establishing a global chromatin landscape to control transcription in GSCs and in their differentiating daughter cells [26]. In the GSCs, epigenetic mechanisms intrinsically suppress the transcription of bam to prevent the differentiation [41,42,43,44,45,46]. In the pre-CBs, however, the chromatin-dependent transcriptional suppression of bam is released to trigger the transition of the pre-CBs to the differentiated CBs. While the mechanisms mediating the transcriptional suppression of bam in the GSCs are well known, the identity of positive transcriptional regulators that drive bam expression to facilitate the differentiation of the GSC daughter pre-CBs remains elusive. We propose that MESR4 is such a factor which functions in the pre-CB to promote bam transcription.
Although MESR4 is expressed in the germ cells and in all somatic cell types of the GSC niche, its function is exclusively required cell-autonomously in the germ cells at the pre-CB stage, as demonstrated by the accumulation of undifferentiated pre-CBs upon germline-specific MESR4 silencing. How the activity of MESR4 is spatially and temporally regulated remains elusive. MESR4 may function as a component of a protein complex specifically assembled in the pre-CBs and may bind a yet undetermined factor, the expression of which is restricted to the pre-CBs. Alternatively, MESR4 may be activated by upstream processes specifically taking place in the pre-CBs. In the pre-CBs, the transcriptional repressor Pgc is transiently expressed mediating a pulse of global transcriptional silencing which prevents the transcription of stem cell fate regulators [32]. This is accompanied by the resetting of chromatin modifications which generates an epigenetic landscape competent to express determinants of a different cell fate. This chromatin environment may permit MESR4 to drive bam expression directly or indirectly.
The MESR4 function has been implicated in various developmental processes as a downstream effector of signaling pathways. It is able to suppress the constitutively activated Ras-MAPK pathway [47] or the FGF-signaling [48]. In addition, MESR4 is required for the proper control of EGFR/ERK signaling during embryonic development and wing formation [49]. In the adult female GSC niche, the EGFR/ERK pathway is active in the escort cells, and limits the Dpp gradient to the anterior tip of the niche where the GSCs reside [50]. Several lines of evidence indicate that in this cellular context MESR4 does not function as a component of the EGFR/ERK pathway. Impaired EGFR signaling in the niche results in the accumulation of GSC-like cells induced by the expanded dpp activity, whereas silencing of MESR4 in the germ cells induces the accumulation of pre-CBs. The MESR4 function is dispensable for proper germ cell development in the escort cells where the EGFR pathway is active.
Our results suggest that the molecular mechanism by which MESR4 regulates germ cell development is the transcriptional control of bam. Based on its protein domain composition, MESR4 may promote bam transcription as a chromatin remodeling factor via the PHD finger domain [51,52]. PHD fingers are found in a number of chromatin remodeling factors involved in nucleosome binding where PHD domains interact with modified histones [53,54,55,56]. However, we found that the PHD finger is dispensable for the MESR4 function in germ cell differentiation. Consistent with our observation, previous studies failed to detect the direct association of MESR4 with the modified histones [49]. While we cannot completely exclude that MESR4 is involved in chromatin remodeling, we prefer an alternative hypothesis for the MESR4 function. According to this hypothesis, MESR4 directly interacts with the DNA via its Zn-fingers to specifically control the transcription of key differentiation regulators. An obvious candidate for a target of MESR4 regulation could be bam. This hypothesis is supported by two lines of evidence. We detected decreased bam mRNA levels in MESR4-silenced ovaries. Furthermore, the phenotype caused by the unsilenced bam expression from the bamΔSE transgene can be suppressed by silencing MESR4, thus confirming that MESR4 promotes bam expression. However, the precise mechanisms of bam regulation by MESR4 remain elusive. MESR4 may indirectly affect bam expression through the transcriptional regulation of a yet unknown factor. Alternatively, the transcriptional control of bam by MESR4 may be mediated directly by the association of MESR4 with the bam regulatory sequences. However, further studies are needed to distinguish between these possibilities.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11132056/s1, Figure S1: MESR4 function is required for the early germ cell development exclusively in the niche; Figure S2: MESR4 function is not required in bam expressing CBs.
Author Contributions
Conceptualization, A.B.S.-K., F.J. and M.E.; methodology, A.B.S.-K., M.B., F.J. and M.E.; formal analysis, A.B.S.-K. and F.J.; investigation, A.B.S.-K., Z.T. and F.J; resources, M.E.; writing—original draft preparation, A.B.S.-K., F.J.; writing—review and editing, F.J. and M.E.; visualization, A.B.S.-K., F.J.; supervision, F.J.; project administration, M.E.; funding acquisition, M.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Hungarian Scientific Research Fund (OTKA) (K132384 to M.E. and PD124446 to M.B.), and the National Research, Development, and Innovation Office (NKFIH-469-3/2020 to M.E.). A.B.S.-K. was supported by the Rollin D. Hotchkiss Foundation. M.B. was supported by a János Bolyai Fellowship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon request.
Acknowledgments
We thank R. Lehmann, R.A. Neumüller, L. Gilboa, P. Rangan and D. Chen for fly stocks and reagents.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Morrison, S.J.; Kimble, J. Asymmetric and Symmetric Stem-Cell Divisions in Development and Cancer. Nature 2006, 441, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
- Lin, H. The Stem-Cell Niche Theory: Lessons from Flies. Nat. Rev. Genet. 2002, 3, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Dansereau, D.A.; Lasko, P. The Development of Germline Stem Cells in Drosophila. Methods Mol. Biol. 2008, 450, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R. Germline Stem Cells: Origin and Destiny. Cell Stem Cell 2012, 10, 729–739. [Google Scholar] [CrossRef]
- Spradling, A.; Fuller, M.T.; Braun, R.E.; Yoshida, S. Germline Stem Cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a002642. [Google Scholar] [CrossRef]
- Lin, H.; Spradling, A.C. Germline Stem Cell Division and Egg Chamber Development in Transplanted Drosophila Germaria. Dev. Biol. 1993, 159, 140–152. [Google Scholar] [CrossRef]
- Ohlstein, B.; McKearin, D. Ectopic Expression of the Drosophila Bam Protein Eliminates Oogenic Germline Stem Cells. Development 1997, 124, 3651–3662. [Google Scholar] [CrossRef]
- Song, X.; Wong, M.D.; Kawase, E.; Xi, R.; Ding, B.C.; McCarthy, J.J.; Xie, T. Bmp Signals from Niche Cells Directly Repress Transcription of a Differentiation-Promoting Gene, Bag of Marbles, in Germline Stem Cells in the Drosophila Ovary. Development 2004, 131, 1353–1364. [Google Scholar] [CrossRef]
- Ables, E.T.; Drummond-Barbosa, D. Cyclin E Controls Drosophila Female Germline Stem Cell Maintenance Independently of Its Role in Proliferation by Modulating Responsiveness to Niche Signals. Development 2013, 140, 530–540. [Google Scholar] [CrossRef]
- De Cuevas, M.; Spradling, A.C. Morphogenesis of the Drosophila Fusome and Its Implications for Oocyte Specification. Development 1998, 125, 2781–2789. [Google Scholar] [CrossRef]
- Mathieu, J.; Cauvin, C.; Moch, C.; Radford, S.J.; Sampaio, P.; Perdigoto, C.N.; Schweisguth, F.; Bardin, A.J.; Sunkel, C.E.; McKim, K.; et al. Aurora B and Cyclin B Have Opposite Effects on the Timing of Cytokinesis Abscission in Drosophila Germ Cells and in Vertebrate Somatic Cells. Dev. Cell 2013, 26, 250–265. [Google Scholar] [CrossRef] [PubMed]
- Xie, T. Control of Germline Stem Cell Self-Renewal and Differentiation in the Drosophila Ovary: Concerted Actions of Niche Signals and Intrinsic Factors. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Spradling, A.C. Stem Cells and Niches: Mechanisms That Promote Stem Cell Maintenance throughout Life. Cell 2008, 132, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Jin, Z.; Yu, Y.; Zhang, Y.; Yang, F.; Huang, H.; Cai, T.; Xi, R. A Progressive Somatic Cell Niche Regulates Germline Cyst Differentiation in the Drosophila Ovary. Curr. Biol. 2021, 31, 840–852.e5. [Google Scholar] [CrossRef]
- Tu, R.; Duan, B.; Song, X.; Chen, S.; Scott, A.; Hall, K.; Blanck, J.; DeGraffenreid, D.; Li, H.; Perera, A.; et al. Multiple Niche Compartments Orchestrate Stepwise Germline Stem Cell Progeny Differentiation. Curr. Biol. 2021, 31, 827–839.e3. [Google Scholar] [CrossRef]
- Xie, T.; Spradling, A.C. Decapentaplegic Is Essential for the Maintenance and Division of Germline Stem Cells in the Drosophila Ovary. Cell 1998, 94, 251–260. [Google Scholar] [CrossRef]
- Chen, D.; McKearin, D. Dpp Signaling Silences Bam Transcription Directly to Establish Asymmetric Divisions of Germline Stem Cells. Curr. Biol. 2003, 13, 1786–1791. [Google Scholar] [CrossRef]
- Casanueva, M.O.; Ferguson, E.L. Germline Stem Cell Number in the Drosophila Ovary Is Regulated by Redundant Mechanisms That Control Dpp Signaling. Development 2004, 131, 1881–1890. [Google Scholar] [CrossRef]
- Forbes, A.; Lehmann, R. Nanos and Pumilio Have Critical Roles in the Development and Function of Drosophila Germline Stem Cells. Development 1998, 125, 679–690. [Google Scholar] [CrossRef]
- Gilboa, L.; Lehmann, R. Repression of Primordial Germ Cell Differentiation Parallels Germ Line Stem Cell Maintenance. Curr. Biol. 2004, 14, 981–986. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, H. Nanos Maintains Germline Stem Cell Self-Renewal by Preventing Differentiation. Science 2004, 303, 2016–2019. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Xie, T. Dcr-1 Maintains Drosophila Ovarian Stem Cells. Curr. Biol. 2007, 17, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Liu, X.; Strauss, T.J.; McKearin, D.M.; Liu, Q. The MiRNA Pathway Intrinsically Controls Self-Renewal of Drosophila Germline Stem Cells. Curr. Biol. 2007, 17, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, D.; Duan, R.; Xia, L.; Wang, J.; Qurashi, A.; Jin, P.; Chen, D. Argonaute 1 Regulates the Fate of Germline Stem Cells in Drosophila. Development 2007, 134, 4265–4272. [Google Scholar] [CrossRef]
- Sanchez, C.G.; Teixeira, F.K.; Czech, B.; Preall, J.B.; Zamparini, A.L.; Seifert, J.R.K.; Malone, C.D.; Hannon, G.J.; Lehmann, R. Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell 2016, 18, 276–290. [Google Scholar] [CrossRef]
- Flora, P.; McCarthy, A.; Upadhyay, M.; Rangan, P. Role of Chromatin Modifications in Drosophila Germline Stem Cell Differentiation. In Signaling-Mediated Control of Cell Division; Results and Problems in Cell Differentiation, 59; Springer: New York, NY, USA, 2017; pp. 1–30. [Google Scholar] [CrossRef]
- Vidaurre, V.; Chen, X. Epigenetic Regulation of Drosophila Germline Stem Cell Maintenance and Differentiation. Dev. Biol. 2021, 473, 105–118. [Google Scholar] [CrossRef]
- Sardi, J.; Bener, M.B.; Simao, T.; Descoteaux, A.E.; Slepchenko, B.M.; Inaba, M. Mad Dephosphorylation at the Nuclear Pore Is Essential for Asymmetric Stem Cell Division. Proc. Natl. Acad. Sci. USA 2021, 118, e2006786118. [Google Scholar] [CrossRef]
- Banisch, T.U.; Slaidina, M.; Gupta, S.; Ho, M.; Gilboa, L.; Lehmann, R. A Transitory Signaling Center Controls Timing of Primordial Germ Cell Differentiation. Dev. Cell 2021, 56, 1742–1755.e4. [Google Scholar] [CrossRef]
- Chen, D.; McKearin, D.M. A Discrete Transcriptional Silencer in the Bam Gene Determines Asymmetric Division of the Drosophila Germline Stem Cell. Development 2003, 130, 1159–1170. [Google Scholar] [CrossRef]
- Wissel, S.; Kieser, A.; Yasugi, T.; Duchek, P.; Roitinger, E.; Gokcezade, J.; Steinmann, V.; Gaul, U.; Mechtler, K.; Förstemann, K.; et al. A Combination of CRISPR/Cas9 and Standardized RNAi as a Versatile Platform for the Characterization of Gene Function. G3 Genes Genomes Genet. 2016, 6, 2467–2478. [Google Scholar] [CrossRef]
- Flora, P.; Schowalter, S.; Wong-Deyrup, S.; DeGennaro, M.; Nasrallah, M.A.; Rangan, P. Transient Transcriptional Silencing Alters the Cell Cycle to Promote Germline Stem Cell Differentiation in Drosophila. Dev. Biol. 2018, 434, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Van Doren, M.; Williamson, A.L.; Lehmann, R. Regulation of Zygotic Gene Expression in Drosophila Primordial Germ Cells. Curr. Biol. 1998, 8, 243–246. [Google Scholar] [CrossRef]
- Port, F.; Chen, H.-M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas Tools for Efficient Germline and Somatic Genome Engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [PubMed]
- Jankovics, F.; Bence, M.; Sinka, R.; Faragó, A.; Bodai, L.; Pettkó-Szandtner, A.; Ibrahim, K.; Takács, Z.; Szarka-Kovács, A.B.; Erdélyi, M. Drosophila Small Ovary Gene Is Required for Transposon Silencing and Heterochromatin Organization, and Ensures Germline Stem Cell Maintenance and Differentiation. Development 2018, 145, dev170639. [Google Scholar] [CrossRef]
- Jankovics, F.; Henn, L.; Bujna, Á.; Vilmos, P.; Spirohn, K.; Boutros, M.; Erdélyi, M. Functional Analysis of the Drosophila Embryonic Germ Cell Transcriptome by RNA Interference. PLoS ONE 2014, 9, e98579. [Google Scholar] [CrossRef]
- Lavoie, C.A.; Ohlstein, B.; McKearin, D.M. Localization and Function of Bam Protein Require the Benign Gonial Cell Neoplasm Gene Product. Dev. Biol. 1999, 212, 405–413. [Google Scholar] [CrossRef]
- Kudron, M.M.; Victorsen, A.; Gevirtzman, L.; Hillier, L.W.; Fisher, W.W.; Vafeados, D.; Kirkey, M.; Hammonds, A.S.; Gersch, J.; Ammouri, H.; et al. The ModERN Resource: Genome-Wide Binding Profiles for Hundreds of Drosophila and Caenorhabditis Elegans Transcription Factors. Genetics 2018, 208, 937–949. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Jain, K.; Fraser, C.S.; Marunde, M.R.; Parker, M.M.; Sagum, C.; Burg, J.M.; Hall, N.; Popova, I.K.; Rodriguez, K.L.; Vaidya, A.; et al. Characterization of the Plant Homeodomain (PHD) Reader Family for Their Histone Tail Interactions. Epigenetics Chromatin 2020, 13, 3. [Google Scholar] [CrossRef]
- Xi, R.; Xie, T. Stem Cell Self-Renewal Controlled by Chromatin Remodeling Factors. Science 2005, 310, 1487–1489. [Google Scholar] [CrossRef]
- Buszczak, M.; Paterno, S.; Spradling, A.C. Drosophila Stem Cells Share a Common Requirement for the Histone H2B Ubiquitin Protease Scrawny. Science 2009, 323, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pan, L.; Wang, S.; Zhou, J.; McDowell, W.; Park, J.; Haug, J.; Staehling, K.; Tang, H.; Xie, T. Histone H3K9 Trimethylase Eggless Controls Germline Stem Cell Maintenance and Differentiation. PLoS Genet. 2011, 7, e1002426. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Xuan, T.; Tan, J.; Li, M.; Zhao, G.; Li, M. The Drosophila Putative Histone Acetyltransferase Enok Maintains Female Germline Stem Cells through Regulating Bruno and the Niche. Dev. Biol. 2013, 384, 1–12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xuan, T.; Xin, T.; He, J.; Tan, J.; Gao, Y.; Feng, S.; He, L.; Zhao, G.; Li, M. DBre1/DSet1-Dependent Pathway for Histone H3K4 Trimethylation Has Essential Roles in Controlling Germline Stem Cell Maintenance and Germ Cell Differentiation in the Drosophila Ovary. Dev. Biol. 2013, 379, 167–181. [Google Scholar] [CrossRef]
- Sun, J.; Wei, H.-M.; Xu, J.; Chang, J.-F.; Yang, Z.; Ren, X.; Lv, W.-W.; Liu, L.-P.; Pan, L.-X.; Wang, X.; et al. Histone H1-Mediated Epigenetic Regulation Controls Germline Stem Cell Self-Renewal by Modulating H4K16 Acetylation. Nat. Commun. 2015, 6, 8856. [Google Scholar] [CrossRef]
- Huang, A.M.; Rubin, G.M. A Misexpression Screen Identifies Genes That Can Modulate RAS1 Pathway Signaling in Drosophila Melanogaster. Genetics 2000, 156, 1219–1230. [Google Scholar] [CrossRef]
- Zhu, M.Y.; Wilson, R.; Leptin, M. A Screen for Genes That Influence Fibroblast Growth Factor Signal Transduction in Drosophila. Genetics 2005, 170, 767–777. [Google Scholar] [CrossRef]
- Seong, K.-H.; Tsuda, M.; Tsuda-Sakurai, K.; Aigaki, T. The Plant Homeodomain Finger Protein MESR4 Is Essential for Embryonic Development in Drosophila. Genesis 2015, 53, 701–708. [Google Scholar] [CrossRef]
- Matsuoka, S.; Hiromi, Y.; Asaoka, M. Egfr Signaling Controls the Size of the Stem Cell Precursor Pool in the Drosophila Ovary. Mech. Dev. 2013, 130, 241–253. [Google Scholar] [CrossRef]
- Morra, R.; Lee, B.M.; Shaw, H.; Tuma, R.; Mancini, E.J. Concerted Action of the PHD, Chromo and Motor Domains Regulates the Human Chromatin Remodelling ATPase CHD4. FEBS Lett. 2012, 586, 2513–2521. [Google Scholar] [CrossRef]
- Watson, A.A.; Mahajan, P.; Mertens, H.D.T.; Deery, M.J.; Zhang, W.; Pham, P.; Du, X.; Bartke, T.; Zhang, W.; Edlich, C.; et al. The PHD and Chromo Domains Regulate the ATPase Activity of the Human Chromatin Remodeler CHD4. J. Mol. Biol. 2012, 422, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Bienz, M. The PHD Finger, a Nuclear Protein-Interaction Domain. Trends Biochem. Sci. 2006, 31, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Peña, P.V.; Davrazou, F.; Shi, X.; Walter, K.L.; Verkhusha, V.V.; Gozani, O.; Zhao, R.; Kutateladze, T.G. Molecular Mechanism of Histone H3K4me3 Recognition by Plant Homeodomain of ING2. Nature 2006, 442, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Kachirskaia, I.; Walter, K.L.; Kuo, J.-H.A.; Lake, A.; Davrazou, F.; Chan, S.M.; Martin, D.G.E.; Fingerman, I.M.; Briggs, S.D.; et al. Proteome-Wide Analysis in Saccharomyces Cerevisiae Identifies Several PHD Fingers as Novel Direct and Selective Binding Modules of Histone H3 Methylated at Either Lysine 4 or Lysine 36. J. Biol. Chem. 2007, 282, 2450–2455. [Google Scholar] [CrossRef]
- Mansfield, R.E.; Musselman, C.A.; Kwan, A.H.; Oliver, S.S.; Garske, A.L.; Davrazou, F.; Denu, J.M.; Kutateladze, T.G.; Mackay, J.P. Plant Homeodomain (PHD) Fingers of CHD4 Are Histone H3-Binding Modules with Preference for Unmodified H3K4 and Methylated H3K9. J. Biol. Chem. 2011, 286, 11779–11791. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).