BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells
Abstract
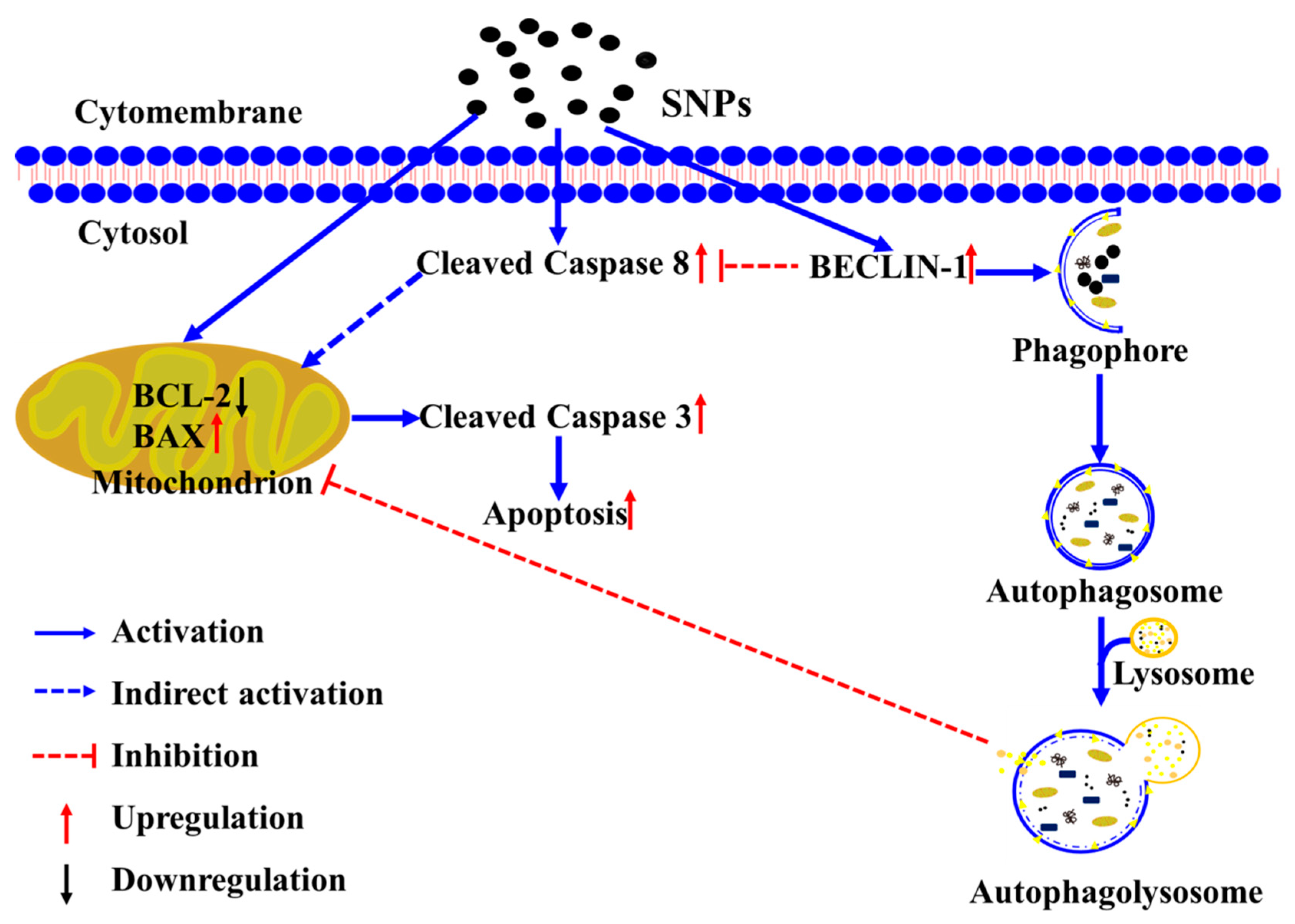
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. SNP Preparation and Characterization
2.3. Animals and Treatment with SNPs
2.4. H&E Staining
2.5. PLC Culture In Vitro and Treatment with SNPs
2.6. Determination of SNP Uptake
2.7. Cell Viability Assay
2.8. Cell Apoptosis Assay
2.9. Construction of Recombinant BECLIN-1 shRNA and Cell Transduction
2.10. Monodansylcadaverine (MDC) Staining Detection
2.11. Western Blotting
2.12. Statistical Analysis
3. Results
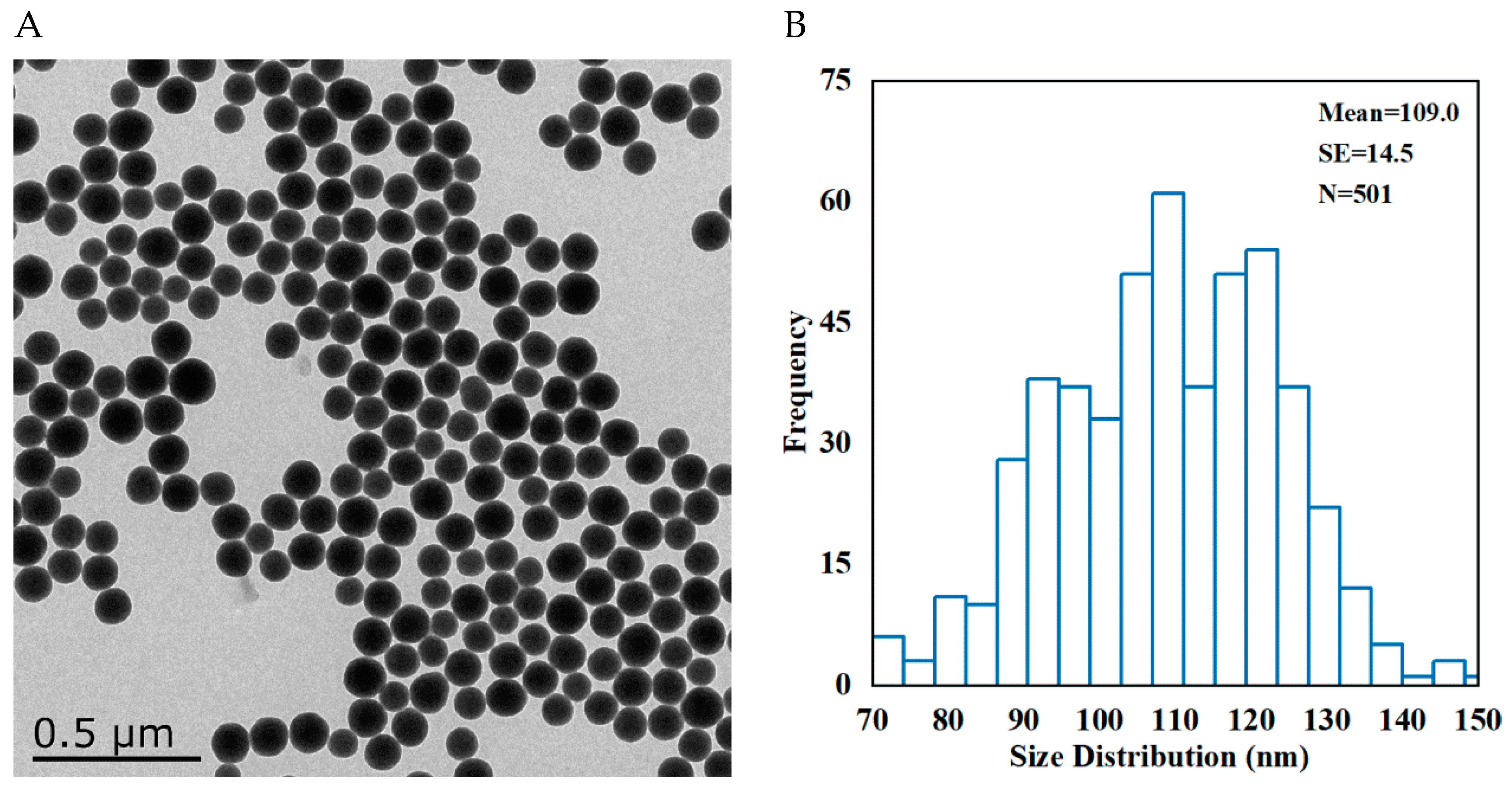
3.1. Characterization of SNPs
3.2. SNPs Change the Histological Structures of Testes In Vivo
3.3. SNP Cellular Uptake in Primary Leydig Cells (PLCs)
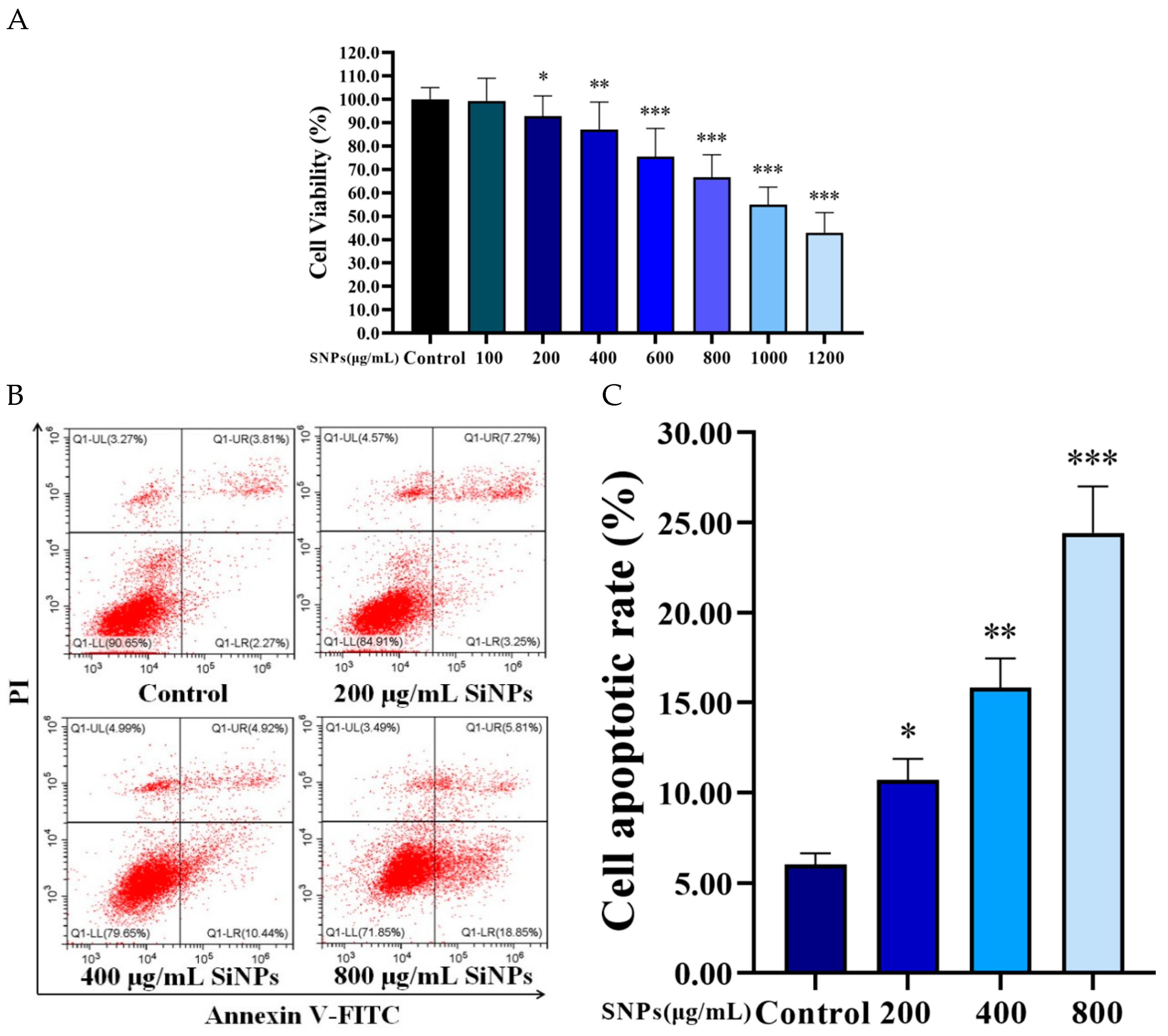
3.4. SNPs Decreased Cell Viability and Increased Cell Apoptosis in the PLCs
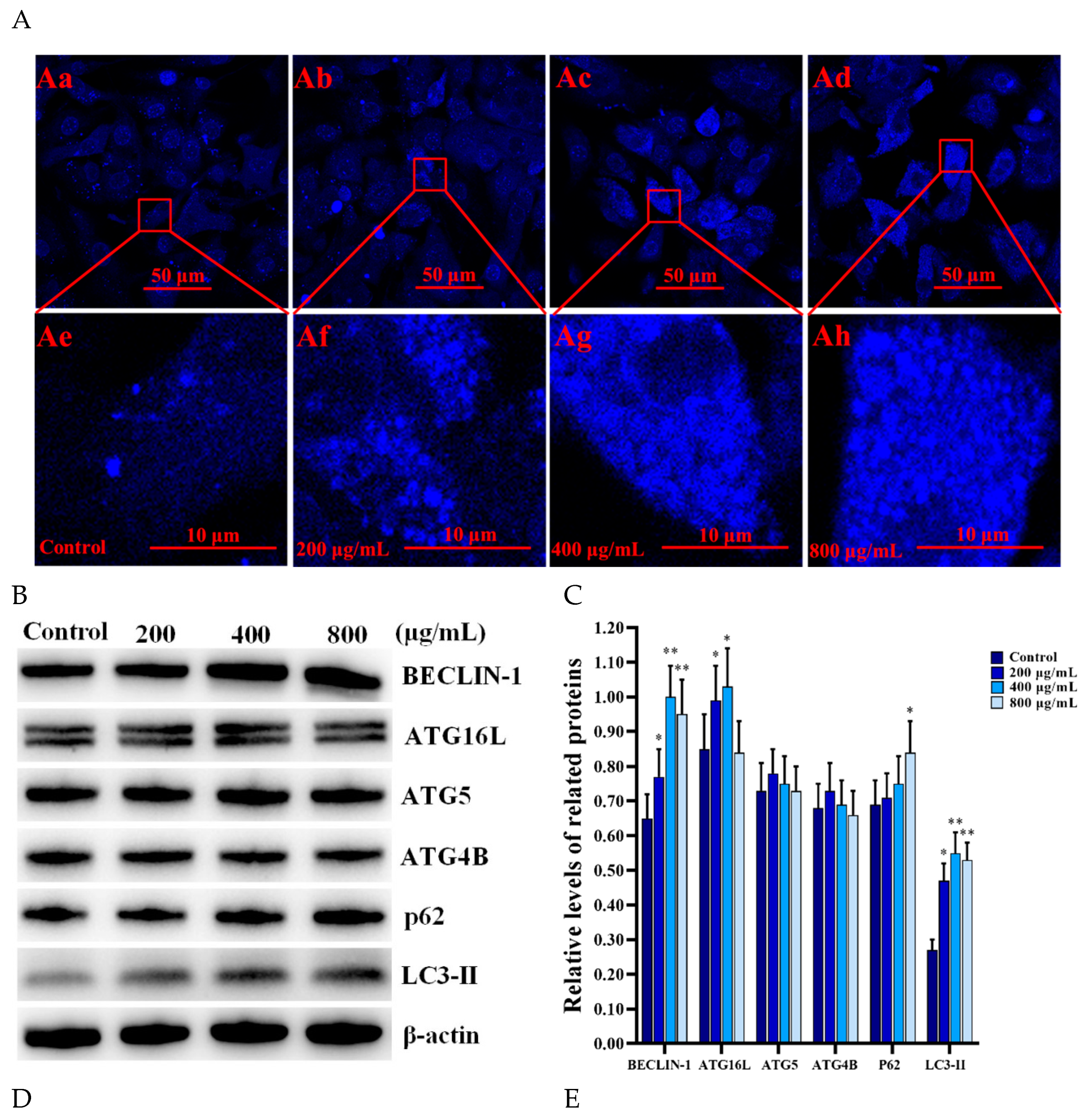
3.5. SNPs Activated Autophagy and the Apoptotic Pathway in the PLCs
3.6. Autophagy Inhibited SNP-Induced Apoptosis in the PLCs
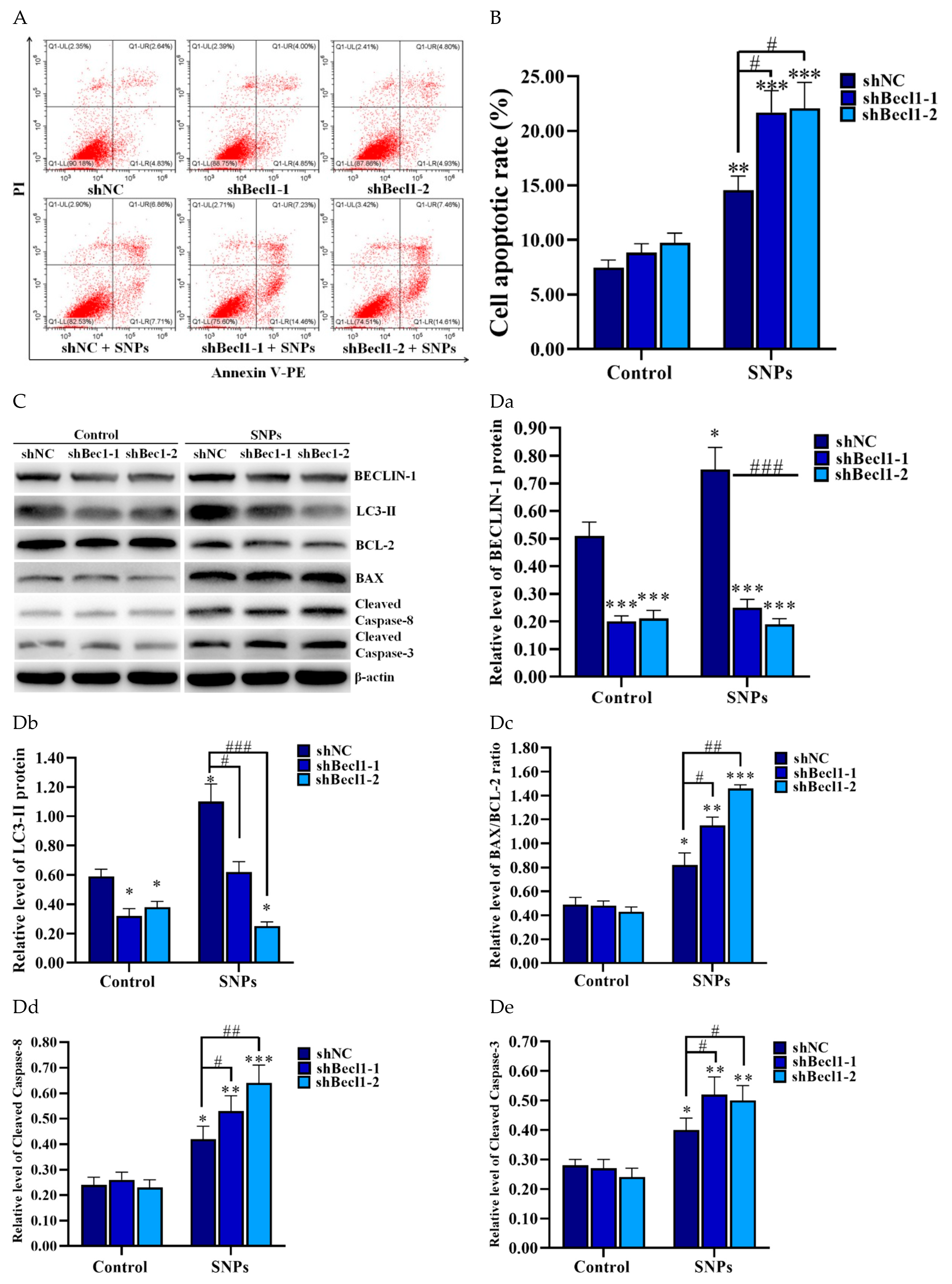
3.7. BECLIN-1 Depletion Increased SNP-Induced Apoptosis in the PLCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murugadoss, S.; Lison, D.; Godderis, L.; Van Den Brule, S.; Mast, J.; Brassinne, F.; Sebaihi, N.; Hoet, P.H. Toxicology of silica nanoparticles: An update. Arch. Toxicol. 2017, 91, 2967–3010. [Google Scholar] [CrossRef]
- Dekkers, S.; Krystek, P.; Peters, R.J.; Lankveld, D.P.; Bokkers, B.G.; van Hoeven-Arentzen, P.H.; Bouwmeester, H.; Oomen, A.G. Presence and risks of nanosilica in food products. Nanotoxicology 2011, 5, 393–405. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, M.; Song, H.; Yu, C. Silica-Based Nanoparticles for Biomedical Applications: From Nanocarriers to Biomodulators. Acc. Chem. Res. 2020, 53, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Mohammadpour, R.; Yazdimamaghani, M.; Cheney, D.L.; Jedrzkiewicz, J.; Ghandehari, H. Subchronic toxicity of silica nanoparticles as a function of size and porosity. J. Control. Release 2019, 304, 216–232. [Google Scholar] [CrossRef]
- Liu, J.; Yang, M.; Jing, L.; Ren, L.; Wei, J.; Zhang, J.; Zhang, F.; Duan, J.; Zhou, X.; Sun, Z. Silica nanoparticle exposure inducing granulosa cell apoptosis and follicular atresia in female Balb/c mice. Environ. Sci. Pollut. Res. Int. 2018, 25, 3423–3434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.S.; Wei, J.L.; Duan, J.C.; Guo, C.X.; Zhang, J.; Ren, L.H.; Liu, J.H.; Li, Y.B.; Sun, Z.W.; Zhou, X.Q. Silica nanoparticles exacerbates reproductive toxicity development in high-fat diet-treated Wistar rats. J. Hazard. Mater. 2020, 384, 121361. [Google Scholar] [CrossRef] [PubMed]
- Morishita, Y.; Yoshioka, Y.; Satoh, H.; Nojiri, N.; Nagano, K.; Abe, Y.; Kamada, H.; Tsunoda, S.; Nabeshi, H.; Yoshikawa, T.; et al. Distribution and histologic effects of intravenously administered amorphous nanosilica particles in the testes of mice. Biochem. Biophys. Res. Commun. 2012, 420, 297–301. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, N.; Yu, Y.; Li, Y.; Li, Y.B.; Yu, Y.B.; Zhou, X.Q.; Sun, Z.W. Exposure to silica nanoparticles causes reversible damage of the spermatogenic process in mice. PLoS ONE 2014, 9, e101572. [Google Scholar] [CrossRef]
- Wang, R.; Song, B.; Wu, J.; Zhang, Y.; Chen, A.; Shao, L. Potential adverse effects of nanoparticles on the reproductive system. Int. J. Nanomed. 2018, 13, 8487–8506. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.M.; Ali, H.A.; Saadeldin, I.M.; Ahmed, M.M. Querectin Alleviates Zinc Oxide Nanoreprotoxicity in Male Albino Rats. J. Biochem. Mol. Toxicol. 2016, 30, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, O.A.; Akinloye, O.; Adaramoye, O.A. Cerium oxide nanoparticle elicits oxidative stress, endocrine imbalance and lowers sperm characteristics in testes of balb/c mice. Andrologia 2018, 50, e12920. [Google Scholar] [CrossRef]
- Sharpe, R.M. Testosterone and spermatogenesis. J. Endocrinol. 1987, 113, 1–2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, Z.; Klionsky, D.J. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010, 12, 814–822. [Google Scholar] [CrossRef]
- Tooze, S.A.; Yoshimori, T. The origin of the autophagosomal membrane. Nat. Cell Biol. 2010, 12, 831–835. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, L.; Zheng, X.; Ge, L. Autophagosomal Membrane Origin and Formation. Adv. Exp. Med. Biol. 2021, 1208, 17–42. [Google Scholar]
- He, C.; Levine, B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010, 22, 140–149. [Google Scholar] [CrossRef]
- Oberstein, A.; Jeffrey, P.D.; Shi, Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007, 282, 13123–13132. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Hou, W.; Han, J.; Lu, C.; Goldstein, L.A.; Rabinowich, H. Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy 2010, 6, 891–900. [Google Scholar] [CrossRef]
- Liu, G.; Yuan, Y.; Long, M.; Luo, T.; Bian, J.; Liu, X.; Gu, J.; Zou, H.; Song, R.; Wang, Y.; et al. Beclin-1-mediated Autophagy Protects Against Cadmium-activated Apoptosis via the Fas/FasL Pathway in Primary Rat Proximal Tubular Cell Culture. Sci. Rep. 2017, 7, 977. [Google Scholar] [CrossRef]
- Keller, N.; Mares, J.; Zerbe, O.; Grutter, M.G. Structural and Biochemical Studies on Procaspase-8: New Insights on Initiator Caspase Activation. Structure 2009, 17, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Voicu, S.N.P.; Dinu, D.; Sima, C.; Hermenean, A.; Ardelean, A.; Codrici, E.; Stan, M.S.; Zarnescu, O.; Dinischiotu, A. Silica Nanoparticles Induce Oxidative Stress and Autophagy but Not Apoptosis in the MRC-5 Cell Line. Int. J. Mol. Sci. 2015, 16, 29398–29416. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, C.; Fritsch-Decker, S.; Al-Rawi, M.; Diabate, S.; Weiss, C. Autophagy induced by silica nanoparticles protects RAW264.7 macrophages from cell death. Toxicology 2017, 379, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Zhou, J.; Du, S.; Peng, S. Autophagy upregulation promotes macrophages to escape mesoporous silica nanoparticle (MSN)-induced NF-kappaB-dependent inflammation. Inflamm. Res. 2016, 65, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Duan, J.; Yu, Y.; Li, Y.; Liu, X.; Zhou, X.; Ho, K.F.; Tian, L.; Sun, Z. Silica nanoparticles induce autophagy and autophagic cell death in HepG2 cells triggered by reactive oxygen species. J. Hazard. Mater. 2014, 270, 176–186. [Google Scholar] [CrossRef]
- Zhang, J.; Ye, R.; Grunberger, J.W.; Jin, J.; Zhang, Q.; Mohammadpour, R.; Khurana, N.; Xu, X.; Ghandehari, H.; Chen, F. Activation of Autophagy by Low-Dose Silica Nanoparticles Enhances Testosterone Secretion in Leydig Cells. Int. J. Mol. Sci. 2022, 23, 3104. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, Y.; Liu, Q.; Hu, J.; Jin, J.; Ma, Z.; Zhang, J. ERO1alpha promotes testosterone secretion in hCG-stimulated mouse Leydig cells via activation of the PI3K/AKT/mTOR signaling pathway. J. Cell. Physiol. 2020, 235, 5666–5678. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Liu, J.; Zhang, J.; Wang, J.; Wei, J.; Li, Y.; Guo, C.; Sun, Z.; Zhou, X. Silica nanoparticles induce spermatocyte cell autophagy through microRNA-494 targeting AKT in GC-2spd cells. Environ. Pollut. 2019, 255 Pt 1, 113172. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, N.; Ito, Y.; Ramdhan, D.H.; Yanagiba, Y.; Hayashi, Y.; Wang, D.; Li, C.M.; Taneda, S.; Suzuki, A.K.; Taya, K.; et al. Effect of nanoparticle-rich diesel exhaust on testicular and hippocampus steroidogenesis in male rats. Inhal. Toxicol. 2012, 24, 459–467. [Google Scholar] [CrossRef]
- Ozgur, M.E.; Ulu, A.; Ozcan, I.; Balcioglu, S.; Ates, B.; Koytepe, S. Investigation of toxic effects of amorphous SiO2 nanoparticles on motility and oxidative stress markers in rainbow trout sperm cells. Environ. Sci. Pollut. Res. Int. 2019, 26, 15641–15652. [Google Scholar] [CrossRef] [PubMed]
- Bara, N.; Eshwarmoorthy, M.; Subaharan, K.; Kaul, G. Mesoporous silica nanoparticle is comparatively safer than zinc oxide nanoparticle which can cause profound steroidogenic effects on pregnant mice and male offspring exposed in utero. Toxicol. Ind. Health 2018, 34, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, L.; Klein, J.P.; Forest, V.; Boudard, D.; Martini, M.; Pourchez, J.; Blanchin, M.G.; Cottier, M. Testicular biodistribution of silica-gold nanoparticles after intramuscular injection in mice. Biomed. Microdevices 2015, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int J. Biochem. Cell B 2004, 36, 2503–2518. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Myeku, N.; Figueiredo-Pereira, M.E. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: Association with sequestosome 1/p62. J. Biol. Chem. 2011, 286, 22426–22440. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Mansilla, A.; Menzies, F.M.; Rubinsztein, D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 2009, 33, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.X.; Yang, M.; Jing, L.; Wang, J.; Yu, Y.; Li, Y.; Duan, J.C.; Zhou, X.Q.; Li, Y.B.; Sun, Z.W. Amorphous silica nanoparticles trigger vascular endothelial cell injury through apoptosis and autophagy via reactive oxygen species-mediated MAPK/Bcl-2 and PI3K/Akt/mTOR signaling. Int. J. Nanomed. 2016, 11, 5257–5276. [Google Scholar] [CrossRef]
- Chou, C.C.; Chen, W.; Hung, Y.; Mou, C.Y. Molecular Elucidation of Biological Response to Mesoporous Silica Nanoparticles in Vitro and in Vivo. ACS Appl. Mater. Interfaces 2017, 9, 22235–22251. [Google Scholar] [CrossRef]
- Wei, F.; Wang, Y.; Luo, Z.; Li, Y.; Duan, Y. New findings of silica nanoparticles induced ER autophagy in human colon cancer cell. Sci. Rep. 2017, 7, 42591. [Google Scholar] [CrossRef]
- Ha, S.W.; Weitzmann, M.N.; Beck, G.R., Jr. Bioactive silica nanoparticles promote osteoblast differentiation through stimulation of autophagy and direct association with LC3 and p62. ACS Nano 2014, 8, 5898–5910. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Duan, J.C.; Yang, M.; Yu, Y.; Feng, L.; Yang, X.Z.; Zhou, X.Q.; Zhao, Z.D.; Sun, Z.W. Silica nanoparticles induce autophagosome accumulation via activation of the EIF2AK3 and ATF6 UPR pathways in hepatocytes. Autophagy 2018, 14, 1185–1200. [Google Scholar] [CrossRef]
- Kretowski, R.; Kusaczuk, M.; Naumowicz, M.; Kotynska, J.; Szynaka, B.; Cechowska-Pasko, M. The Effects of Silica Nanoparticles on Apoptosis and Autophagy of Glioblastoma Cell Lines. Nanomaterials 2017, 7, 230. [Google Scholar] [CrossRef] [PubMed]
- Ciechomska, I.A.; Goemans, G.C.; Skepper, J.N.; Tolkovsky, A.M. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene 2009, 28, 2128–2141. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Gonzalez-Polo, R.A.; Casares, N.; Perfettini, J.L.; Dessen, P.; Larochette, N.; Metivier, D.; Meley, D.; Souquere, S.; Yoshimori, T.; et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005, 25, 1025–1040. [Google Scholar] [CrossRef] [PubMed]
- Bohensky, J.; Shapiro, I.M.; Leshinsky, S.; Terkhorn, S.P.; Adams, C.S.; Srinivas, V. HIF-1 regulation of chondrocyte apoptosis: Induction of the autophagic pathway. Autophagy 2007, 3, 207–214. [Google Scholar] [CrossRef]
- Shi, M.; Cheng, L.; Zhang, Z.; Liu, Z.; Mao, X. Ferroferric oxide nanoparticles induce prosurvival autophagy in human blood cells by modulating the Beclin 1/Bcl-2/VPS34 complex. Int. J. Nanomed. 2015, 10, 207–216. [Google Scholar]
- Ren, L.; Zhang, J.; Wang, J.; Wei, J.; Liu, J.; Li, X.; Zhu, Y.; Li, Y.; Guo, C.; Duan, J.; et al. Silica nanoparticles induce spermatocyte cell apoptosis through microRNA-2861 targeting death receptor pathway. Chemosphere 2019, 228, 709–720. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, J.; Zou, Y.; Zhang, L.; Wei, J.; Shi, Z.; Li, Y.; Guo, C.; Sun, Z.; Zhou, X. Silica nanoparticles induce reversible damage of spermatogenic cells via RIPK1 signal pathways in C57 mice. Int. J. Nanomed. 2016, 11, 2251–2264. [Google Scholar]
- Jalmar, O.; Francois-Moutal, L.; Garcia-Saez, A.J.; Perry, M.; Granjon, T.; Gonzalvez, F.; Gottlieb, E.; Ayala-Sanmartin, J.; Klosgen, B.; Schwille, P.; et al. Caspase-8 binding to cardiolipin in giant unilamellar vesicles provides a functional docking platform for bid. PLoS ONE 2013, 8, e55250. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, R.; Moosavi, M.A.; Tavakol, S.; Vardar, D.O.; Hosseini, A.; Rahmati, M.; Dini, L.; Hussain, S.; Mandegary, A.; Klionsky, D.J. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 2019, 15, 4–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, Y.; Lu, K.; Yang, M.; Li, Y.; Zhou, X.; Sun, Z. Silica nanoparticles induce autophagy dysfunction via lysosomal impairment and inhibition of autophagosome degradation in hepatocytes. Int. J. Nanomed. 2017, 12, 809–825. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, J.; Dong, S.; Cai, X.; Simaiti, A.; Yang, X.; Zhu, X.; Luo, J.; Jiang, L.H.; Du, B.; et al. Silica nanoparticles induce lung inflammation in mice via ROS/PARP/TRPM2 signaling-mediated lysosome impairment and autophagy dysfunction. Part. Fibre Toxicol. 2020, 17, 23. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Grunberger, J.W.; Khurana, N.; Zhou, X.; Xu, X.; Ghandehari, H.; Chen, F. BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells. Cells 2022, 11, 1863. https://doi.org/10.3390/cells11121863
Zhang Q, Grunberger JW, Khurana N, Zhou X, Xu X, Ghandehari H, Chen F. BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells. Cells. 2022; 11(12):1863. https://doi.org/10.3390/cells11121863
Chicago/Turabian StyleZhang, Qianru, Jason William Grunberger, Nitish Khurana, Xin Zhou, Xianyu Xu, Hamidreza Ghandehari, and Fenglei Chen. 2022. "BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells" Cells 11, no. 12: 1863. https://doi.org/10.3390/cells11121863
APA StyleZhang, Q., Grunberger, J. W., Khurana, N., Zhou, X., Xu, X., Ghandehari, H., & Chen, F. (2022). BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells. Cells, 11(12), 1863. https://doi.org/10.3390/cells11121863





