Extracellular Vesicles from Adipose Tissue Could Promote Metabolic Adaptation through PI3K/Akt/mTOR
Abstract
:1. Introduction
2. Biogenesis and Uptake of EVs
3. Functional Role of EVs
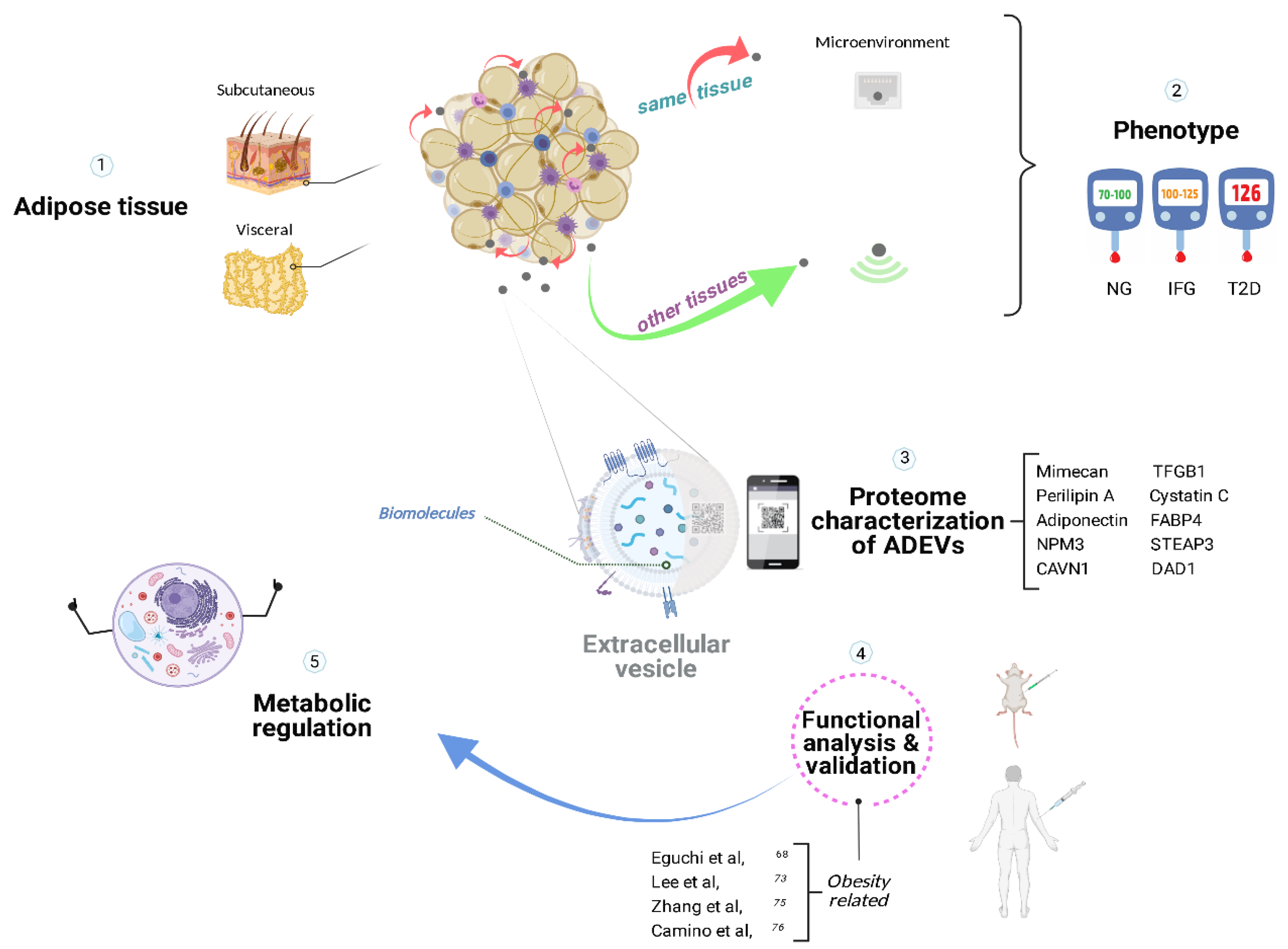
4. Adipose Tissue
5. ADEVs
5.1. Microenvironment in Adipose Tissue
5.2. Distal Intercom from Adipose Tissue
6. PI3K/Akt/mTOR Signaling Cascade
7. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EVs | extracellular vesicles |
| BCAAs | branched-chain amino acids |
| ABCA1 | ATP-binding cassette transporter ABCA1 |
| ABCG1 | ATP-binding cassette sub-family G member 1 |
| ADEVs | adipose-derived extracellular vesicles |
| Akt | “Protein kinase B2” (PKB) |
| AQP7 | Aquaporin-7 |
| AT | adipose tissue |
| ATM | adipose tissue resident macrophages |
| BAT | brown adipose tissue |
| Cav-1 | Caveolin 1 |
| DAD1 | Defender against cell death 1 |
| ESCRT | endosomal sorting complex required for transport |
| FABP4 | fatty acid binding protein 4 |
| GPI | glycosylphosphatidylinositol |
| IL-6 | interleukin 6 |
| IR | insulin resistance |
| IRS-1 | insulin receptor substrate 1 |
| LPL | Lipoprotein lipase |
| MALAT1 | metastasis associated lung adenocarcinoma transcript 1 |
| MCP-1 | monocyte chemoattractant protein-1 |
| MIF | macrophage migration inhibitory factor |
| miRNA | micro ribonucleic acid |
| mRNA | messenger ribonucleic acid |
| mTOR | mammalian target of rapamycin |
| NF-kB | Nuclear factor Kappa B |
| NPM3 | Nucleoplasmin-3 |
| PI3K | Phosphoinositide 3-kinase |
| PM | plasma membrane |
| PPARγ | peroxisome proliferator activated receptor gamma |
| PTEN | Phosphatase and tensin homolog |
| RBP4 | retinol binding protein 4 |
| SHH | sonic hedgehog |
| STEAP3 | Six-transmembrane epithelial antigen of prostate 3 |
| SVF | stromal vascular fraction |
| TFGB1 | transforming growth factor-beta 1 |
| TNF-α | tumor necrosis factor alfa |
| WAT | white adipose tissue |
References
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Saludas, L.; Garbayo, E.; Ruiz-Villalba, A.; Hernández, S.; Vader, P.; Prósper, F.; Blanco-Prieto, M.J. Isolation methods of large and small extracellular vesicles derived from cardiovascular progenitors: A comparative study. Eur. J. Pharm. Biopharm. 2022, 170, 187–196. [Google Scholar] [CrossRef]
- Somiya, M. Where does the cargo go?: Solutions to provide experimental support for the “extracellular vesicle cargo transfer hypothesis”. J. Cell Commun. Signal. 2020, 14, 135–146. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Gandham, S.; Su, X.; Wood, J.; Nocera, A.L.; Alli, S.C.; Milane, L.; Zimmerman, A.; Amiji, M.; Ivanov, A.R. Technologies and Standardization in Research on Extracellular Vesicles. Trends Biotechnol. 2020, 38, 1066–1098. [Google Scholar] [CrossRef]
- Iraci, N.; Leonardi, T.; Gessler, F.; Vega, B.; Pluchino, S. Focus on Extracellular Vesicles: Physiological Role and Signalling Properties of Extracellular Membrane Vesicles. Int. J. Mol. Sci. 2016, 17, 171. [Google Scholar] [CrossRef] [Green Version]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.E.; Sneider, A.; Witwer, K.W.; Bergese, P.; Bhattacharyya, S.N.; Cocks, A.; Cocucci, E.; Erdbrügger, U.; Falcon-Perez, J.M.; Freeman, D.W.; et al. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. J. Extracell. Vesicles 2019, 8, 1684862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavík, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef] [Green Version]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedgwick, A.E.; D’Souza-Schorey, C. The biology of extracellular microvesicles. Traffic 2018, 19, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Buzás, E.I.; Tóth, E.; Sódar, B.W.; Szabó-Taylor, K. Molecular interactions at the surface of extracellular vesicles. Semin. Immunopathol. 2018, 40, 453–464. [Google Scholar] [CrossRef] [Green Version]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef] [Green Version]
- Roche, P.A.; Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015, 15, 203–216. [Google Scholar] [CrossRef]
- Tung, S.L.; Boardman, D.A.; Sen, M.; Letizia, M.; Peng, Q.; Cianci, N.; Dioni, L.; Carlin, L.M.; Lechler, R.; Bollati, V.; et al. Regulatory T cell-derived extracellular vesicles modify dendritic cell function. Sci. Rep. 2018, 8, 6065. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Pan, Y.; Zhang, L.; Shen, C.; Qin, G.; Ashraf, M.; Weintraub, N.; Ma, G.; Tang, Y. Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2013, 431, 566–571. [Google Scholar] [CrossRef] [Green Version]
- Rhee, J.S.; Black, M.; Schubert, U.; Fischer, S.; Morgenstern, E.; Hammes, H.P.; Preissner, K.T. The functional role of blood platelet components in angiogenesis. Thromb. Haemost. 2004, 92, 394–402. [Google Scholar] [PubMed]
- Oggero, S.; Austin-Williams, S.; Norling, L.V. The Contrasting Role of Extracellular Vesicles in Vascular Inflammation and Tissue Repair. Front. Pharmacol. 2019, 10, 1479. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tan, C. miRNAs in Adipocyte-Derived Extracellular Vesicles: Multiple Roles in Development of Obesity-Associated Disease. Front. Mol. Biosci. 2020, 7, 171. [Google Scholar] [CrossRef] [PubMed]
- Kita, S.; Maeda, N.; Shimomura, I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J. Clin. Investig. 2019, 129, 4041–4049. [Google Scholar] [CrossRef] [Green Version]
- Akbar, N.; Azzimato, V.; Choudhury, R.P.; Aouadi, M. Extracellular vesicles in metabolic disease. Diabetologia 2019, 62, 2179–2187. [Google Scholar] [CrossRef] [Green Version]
- Guay, C.; Regazzi, R. Exosomes as new players in metabolic organ cross-talk. Diabetes Obes. Metab. 2017, 19 (Suppl. 1), 137–146. [Google Scholar] [CrossRef]
- Esteve Ràfols, M. Adipose tissue: Cell heterogeneity and functional diversity. Endocrinol. Nutr. 2014, 61, 100–112. [Google Scholar] [CrossRef]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Olefsky, J.M.; Glass, C.K. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 2010, 72, 219–246. [Google Scholar] [CrossRef]
- Chait, A.; den Hartigh, L.J. Adipose Tissue Distribution, Inflammation and Its Metabolic Consequences, Including Diabetes and Cardiovascular Disease. Front. Cardiovasc. Med. 2020, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Kajimura, S. Metabolic adaptation and maladaptation in adipose tissue. Nat. Metab. 2019, 1, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Schenk, S.; Saberi, M.; Olefsky, J.M. Insulin sensitivity: Modulation by nutrients and inflammation. J. Clin. Investig. 2008, 118, 2992–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Shi, L.; Mei, H.; Zhang, J.; Zhu, Y.; Han, X.; Zhu, D. Inflamed macrophage microvesicles induce insulin resistance in human adipocytes. Nutr. Metab. 2015, 12, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yu, M.; Dai, M.; Chen, C.; Tang, Q.; Jing, W.; Wang, H.; Tian, W. miR-450a-5p within rat adipose tissue exosome-like vesicles promotes adipogenic differentiation by targeting WISP2. J. Cell Sci. 2017, 130, 1158–1168. [Google Scholar] [CrossRef] [Green Version]
- Son, Y.H.; Ka, S.; Kim, A.Y.; Kim, J.B. Regulation of Adipocyte Differentiation via MicroRNAs. Endocrinol. Metab. 2014, 29, 122–135. [Google Scholar] [CrossRef] [Green Version]
- Uranga, R.M.; Keller, J.N. The Complex Interactions between Obesity, Metabolism and the Brain. Front. Neurosci. 2019, 13, 513. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Liu, M. Adipose tissue in control of metabolism. J. Endocrinol. 2016, 231, R77–R99. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.B.; Poliakov, A.; Hardy, R.W.; Clements, R.; Liu, C.; Liu, Y.; Wang, J.; Xiang, X.; Zhang, S.; Zhuang, X.; et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 2009, 58, 2498–2505. [Google Scholar] [CrossRef] [Green Version]
- Dahik, V.D.; Frisdal, E.; Le Goff, W. Rewiring of Lipid Metabolism in Adipose Tissue Macrophages in Obesity: Impact on Insulin Resistance and Type 2 Diabetes. Int. J. Mol. Sci. 2020, 21, 5505. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Dietze, D.; Koenen, M.; Röhrig, K.; Horikoshi, H.; Hauner, H.; Eckel, J. Impairment of insulin signaling in human skeletal muscle cells by co-culture with human adipocytes. Diabetes 2002, 51, 2369–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, N.; Jin-no, S.; Nakagawa, Y.; Asai, N.; Arakawa, E.; Tamura, N.; Tamura, T.; Matsuda, T. Identification and characterization of microvesicles secreted by 3T3-L1 adipocytes: Redox- and hormone-dependent induction of milk fat globule-epidermal growth factor 8-associated microvesicles. Endocrinology 2007, 148, 3850–3862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kratz, M.; Coats, B.R.; Hisert, K.B.; Hagman, D.; Mutskov, V.; Peris, E.; Schoenfelt, K.Q.; Kuzma, J.N.; Larson, I.; Billing, P.S.; et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014, 20, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, R.; Tanaka, C.; Sato, M.; Nagasaki, H.; Sugimura, K.; Okumura, K.; Nakagawa, Y.; Aoki, N. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem. Biophys. Res. Commun. 2010, 398, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2018, 19, 808. [Google Scholar] [CrossRef] [Green Version]
- Müller, G.; Schneider, M.; Biemer-Daub, G.; Wied, S. Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell. Signal. 2011, 23, 1207–1223. [Google Scholar] [CrossRef]
- Mori, M.A.; Thomou, T.; Boucher, J.; Lee, K.Y.; Lallukka, S.; Kim, J.K.; Torriani, M.; Yki-Järvinen, H.; Grinspoon, S.K.; Cypess, A.M.; et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J. Clin. Investig. 2014, 124, 3339–3351. [Google Scholar] [CrossRef] [Green Version]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef]
- Zhang, Y.; Mei, H.; Chang, X.; Chen, F.; Zhu, Y.; Han, X. Adipocyte-derived microvesicles from obese mice induce M1 macrophage phenotype through secreted miR-155. J. Mol. Cell Biol. 2016, 8, 505–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, F.; Yu, Y.; Feng, L.; Li, J.; Zhang, M.; Lan, X.; Yan, X.; Liu, Y.; Guan, F.; Zhang, M.; et al. Adipogenic miR-27a in adipose tissue upregulates macrophage activation via inhibiting PPARγ of insulin resistance induced by high-fat diet-associated obesity. Exp. Cell Res. 2017, 355, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, S.Y.; Leng, Y.; Wang, Z.X.; Xiao, X.; Zhang, X.; Wen, T.; Gong, H.Z.; Hong, A.; Ma, Y. Exosomal transfer of obesity adipose tissue for decreased miR-141-3p mediate insulin resistance of hepatocytes. Int. J. Biol. Sci. 2019, 15, 351–368. [Google Scholar] [CrossRef]
- Wang, Y.C.; Li, Y.; Wang, X.Y.; Zhang, D.; Zhang, H.; Wu, Q.; He, Y.Q.; Wang, J.Y.; Zhang, L.; Xia, H.; et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 2013, 56, 2275–2285. [Google Scholar] [CrossRef]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-Derived Exosomal MiR-27a Induces Insulin Resistance in Skeletal Muscle Through Repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef]
- Gao, J.; Li, X.; Wang, Y.; Cao, Y.; Yao, D.; Sun, L.; Qin, L.; Qiu, H.; Zhan, X. Adipocyte-derived extracellular vesicles modulate appetite and weight through mTOR signalling in the hypothalamus. Acta Physiol. 2020, 228, e13339. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, X.; Liu, X.; Du, H.; Sun, C.; Shao, X.; Tian, J.; Gu, X.; Wang, H.; Tian, J.; et al. Adipose-Derived Exosomes Exert Proatherogenic Effects by Regulating Macrophage Foam Cell Formation and Polarization. J. Am. Heart Assoc. 2018, 7, e007442. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, K.; Xu, T.; Du, W.; Yu, B.; Liu, Y.; Nie, H. Exosomal microRNA-29a mediates cardiac dysfunction and mitochondrial inactivity in obesity-related cardiomyopathy. Endocrine 2019, 63, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Kranendonk, M.E.; Visseren, F.L.; van Herwaarden, J.A.; Nolte-’t Hoen, E.N.; de Jager, W.; Wauben, M.H.; Kalkhoven, E. Effect of extracellular vesicles of human adipose tissue on insulin signaling in liver and muscle cells. Obesity 2014, 22, 2216–2223. [Google Scholar] [CrossRef]
- Song, M.; Han, L.; Chen, F.F.; Wang, D.; Wang, F.; Zhang, L.; Wang, Z.H.; Zhong, M.; Tang, M.X.; Zhang, W. Adipocyte-Derived Exosomes Carrying Sonic Hedgehog Mediate M1 Macrophage Polarization-Induced Insulin Resistance via Ptch and PI3K Pathways. Cell. Physiol. Biochem. 2018, 48, 1416–1432. [Google Scholar] [CrossRef] [PubMed]
- Mleczko, J.; Ortega, F.J.; Falcon-Perez, J.M.; Wabitsch, M.; Fernandez-Real, J.M.; Mora, S. Extracellular Vesicles from Hypoxic Adipocytes and Obese Subjects Reduce Insulin-Stimul.lated Glucose Uptake. Mol. Nutr. Food Res. 2018, 62, 1700917. [Google Scholar] [CrossRef] [Green Version]
- Camino, T.; Lago-Baameiro, N.; Bravo, S.B.; Sueiro, A.; Couto, I.; Santos, F.; Baltar, J.; Casanueva, F.F.; Pardo, M. Vesicles Shed by Pathological Murine Adipocytes Spread Pathology: Characterization and Functional Role of Insulin Resistant/Hypertrophied Adiposomes. Int. J. Mol. Sci. 2020, 21, 2252. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Shang, Q.; Pan, Z.; Bai, Y.; Li, Z.; Zhang, H.; Zhang, Q.; Guo, C.; Zhang, L.; Wang, Q. Exosomes from Adipose-Derived Stem Cells Attenuate Adipose Inflammation and Obesity through Polarizing M2 Macrophages and Beiging in White Adipose Tissue. Diabetes 2018, 67, 235–247. [Google Scholar] [CrossRef] [Green Version]
- Eguchi, A.; Lazic, M.; Armando, A.M.; Phillips, S.A.; Katebian, R.; Maraka, S.; Quehenberger, O.; Sears, D.D.; Feldstein, A.E. Circulating adipocyte-derived extracellular vesicles are novel markers of metabolic stress. J. Mol. Med. 2016, 94, 1241–1253. [Google Scholar] [CrossRef]
- Connolly, K.D.; Guschina, I.A.; Yeung, V.; Clayton, A.; Draman, M.S.; Von Ruhland, C.; Ludgate, M.; James, P.E.; Rees, D.A. Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J. Extracell. Vesicles 2015, 4, 29159. [Google Scholar] [CrossRef]
- Durcin, M.; Fleury, A.; Taillebois, E.; Hilairet, G.; Krupova, Z.; Henry, C.; Truchet, S.; Trötzmüller, M.; Köfeler, H.; Mabilleau, G.; et al. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1305677. [Google Scholar] [CrossRef]
- Crewe, C.; Joffin, N.; Rutkowski, J.M.; Kim, M.; Zhang, F.; Towler, D.A.; Gordillo, R.; Scherer, P.E. An Endothelial-to-Adipocyte Extracellular Vesicle Axis Governed by Metabolic State. Cell 2018, 175, 695–708.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehr, S.; Hartwig, S.; Sell, H. Adipokines: A treasure trove for the discovery of biomarkers for metabolic disorders. Proteom. Clin. Appl. 2012, 6, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Moon, P.G.; Lee, I.K.; Baek, M.C. Proteomic Analysis of Extracellular Vesicles Released by Adipocytes of Otsuka Long-Evans Tokushima Fatty (OLETF) Rats. Protein J. 2015, 34, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, S.; De Filippo, E.; Göddeke, S.; Knebel, B.; Kotzka, J.; Al-Hasani, H.; Roden, M.; Lehr, S.; Sell, H. Exosomal proteins constitute an essential part of the human adipose tissue secretome. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, M.; Dong, J.; Wu, Y.; Tian, W. Identification of Novel Adipokines through Proteomic Profiling of Small Extracellular Vesicles Derived from Adipose Tissue. J. Proteome Res. 2020, 19, 3130–3142. [Google Scholar] [CrossRef] [PubMed]
- Camino, T.; Lago-Baameiro, N.; Bravo, S.B.; Molares-Vila, A.; Sueiro, A.; Couto, I.; Baltar, J.; Casanueva, E.F.; Pardo, M. Human obese white adipose tissue sheds depot-specific extracellular vesicles and reveals candidate biomarkers for monitoring obesity and its comorbidities. Transl. Res. 2021, 239, 85–102. [Google Scholar] [CrossRef]
- Kranendonk, M.E.; Visseren, F.L.; van Balkom, B.W.; Nolte-’t Hoen, E.N.; van Herwaarden, J.A.; de Jager, W.; Schipper, H.S.; Brenkman, A.B.; Verhaar, M.C.; Wauben, M.H.M. Human adipocyte extracellular vesicles in reciprocal signaling between adipocytes and macrophages. Obesity 2014, 22, 1296–1308. [Google Scholar] [CrossRef]
- Liu, T.; Sun, Y.-C.; Cheng, P.; Shao, H.-G. Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochem. Biophys. Res. Commun. 2019, 515, 352–358. [Google Scholar] [CrossRef]
- Gesmundo, I.; Pardini, B.; Gargantini, E.; Gamba, G.; Birolo, G.; Fanciulli, A.; Banfi, D.; Congiusta, N.; Favaro, E.; Deregibus, M.C. Adipocyte-derived extracellular vesicles regulate survival and function of pancreatic β cells. JCI Insight 2021, 6, e141962. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Eguchi, A.; Tempaku, M.; Honda, T.; Togashi, K.; Iwasa, M.; Hasegawa, H.; Takei, Y.; Sumida, Y.; Taguchi, O. Circulating extracellular vesicles are associated with lipid and insulin metabolism. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E574–E582. [Google Scholar] [CrossRef]
- Afrisham, R.; Sadegh-Nejadi, S.; Meshkani, R.; Emamgholipour, S.; Paknejad, M.J.D. Effect of circulating exosomes derived from normal-weight and obese women on gluconeogenesis, glycogenesis, lipogenesis and secretion of FGF21 and fetuin A in HepG2 cells. Diabetol. Metab. Syndr. 2020, 12, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarado, H.M.; Munguia, B.C.; Millan, E.C.; Hernandez, M.H.; Teros, V.L.; Moreno, B.P.; Duran, L.R.; Garcia, H.A. Morphometric Characterization of Extracellular Vesicles in Adults with Obesity. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Huang, X.; Liu, G.; Guo, J.; Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef] [Green Version]
- Schultze, S.M.; Hemmings, B.A.; Niessen, M.; Tschopp, O. PI3K/AKT, MAPK and AMPK signalling: Protein kinases in glucose homeostasis. Expert Rev. Mol. Med. 2012, 14, e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Z.; Zhang, W. Role of mTOR in Glucose and Lipid Metabolism. Int. J. Mol. Sci 2018, 19, 2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva Rosa, S.C.; Nayak, N.; Caymo, A.M.; Gordon, J.W. Mechanisms of muscle insulin resistance and the cross-talk with liver and adipose tissue. Physiol. Rep. 2020, 8, e14607. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.L.; Jung, S.M.; Guertin, D.A. The Complex Roles of Mechanistic Target of Rapamycin in Adipocytes and Beyond. Trends Endocrinol. Metab. 2017, 28, 319–339. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Dong, L.Q.; Liu, F. Recent Advances in Adipose mTOR Signaling and Function: Therapeutic Prospects. Trends Pharmacol. Sci. 2016, 37, 303–317. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, F.; Krebs, M.; Dombrowski, L.; Brehm, A.; Bernroider, E.; Roth, E.; Nowotny, P.; Waldhäusl, W.; Marette, A.; Roden, M.J.D. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes 2005, 54, 2674–2684. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Yoon, S.-O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edick, A.M.; Auclair, O.; Burgos, S.A. Role of Grb10 in mTORC1-dependent regulation of insulin signaling and action in human skeletal muscle cells. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E173–E183. [Google Scholar] [CrossRef] [PubMed]
- Um, S.H.; D’Alessio, D.; Thomas, G.J. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006, 3, 393–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanazawa, T.; Taneike, I.; Akaishi, R.; Yoshizawa, F.; Furuya, N.; Fujimura, S.; Kadowaki, M.J. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J. Biol. Chem. 2004, 279, 8452–8459. [Google Scholar] [CrossRef] [Green Version]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e5. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Z.; Qi, M.; Zhao, P.; Duan, Y.; Yang, G.; Yuan, L. Brown adipose tissue-derived exosomes mitigate the metabolic syndrome in high fat diet mice. Theranostics 2020, 10, 8197–8210. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Ye, Y.; Li, D.; Liu, Y.; Lee, E.; Zhang, M.; Dai, X.; Zhang, X.; Wang, S.; et al. Pancreatic β cells control glucose homeostasis via the secretion of exosomal miR-29 family. J. Extracell. Vesicles 2021, 10, e12055. [Google Scholar] [CrossRef]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, M.F.; Jiang, S.; Wu, J.; Liu, J.; Yuan, X.W.; Shen, D.; Zhang, J.Z.; Zhou, N.; He, J.; et al. Liver governs adipose remodelling via extracellular vesicles in response to lipid overload. Nat. Commun. 2020, 11, 719. [Google Scholar] [CrossRef] [Green Version]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef]
- Lorente-Cebrián, S.; Mejhert, N.; Kulyté, A.; Laurencikiene, J.; Åström, G.; Hedén, P.; Rydén, M.; Arner, P. MicroRNAs regulate human adipocyte lipolysis: Effects of miR-145 are linked to TNF-α. PLoS ONE 2014, 9, e86800. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Meng, X.; Che, H.; Shen, N.; Xiao, D.; Song, X.; Liang, M.; Fu, X.; Ju, J.; Li, Y.; et al. Regulation of Insulin Resistance by Multiple MiRNAs via Targeting the GLUT4 Signalling Pathway. Cell Physiol. Biochem. 2016, 38, 2063–2078. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Igata, M.; Kondo, T.; Kitano, S.; Takaki, Y.; Hanatani, S.; Sakaguchi, M.; Goto, R.; Senokuchi, T.; Kawashima, J.; et al. Identification of microRNA that represses IRS-1 expression in liver. PLoS ONE 2018, 13, e0191553. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ai, H.; Liu, L.; Zhang, X.; Gao, F.; Zheng, L.; Yi, J.; Sun, L.; Yu, C.; Zhao, H.; et al. Micro-RNA-27a/b negatively regulates hepatic gluconeogenesis by targeting FOXO1. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E911–E924. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a promotes insulin resistance and mediates glucose metabolism by targeting PPAR-γ-mediated PI3K/AKT signaling. Aging 2019, 11, 7510–7524. [Google Scholar] [CrossRef]
- Kukreti, H.; Amuthavalli, K. MicroRNA-34a causes ceramide accumulation and effects insulin signaling pathway by targeting ceramide kinase (CERK) in aging skeletal muscle. J. Cell. Biochem. 2020, 121, 3070–3089. [Google Scholar] [CrossRef]
- Gómez-Serrano, M.; Ponath, V.; Preußer, C.; Pogge von Strandmann, E. Beyond the Extracellular Vesicles: Technical Hurdles, Achieved Goals and Current Challenges When Working on Adipose Cells. Int. J. Mol. Sci. 2021, 22, 3362. [Google Scholar] [CrossRef]
| Source * | Content | Role | Refs |
|---|---|---|---|
| AT | RBP4 | Activate bone marrow macrophages | [40] |
| Adipocyte | mRNA (Adiponectin, resistin, PPARγ2) | Adipogenesis | [46] |
| Adipocyte | GPI | Lipogenesis | [49] |
| Adipocyte | miR-155 | M1 macrophage polarization; AT IR | [52] |
| AT | miR-27a | Activate macrophage; IR, AT IR | [53] |
| Adipocyte | miR-34a | M1 macrophage polarization; adipose inflammation | [54] |
| ATM | miR-155 | Insulin sensitivity | [55] |
| Plasma | miR-34a, miR-122, miR-192 | Glucose and lipid metabolism | [56] |
| AT | miR-141-3p | Hepatic IR | [57] |
| Adipocyte | miR-27a | Skeletal muscle IR | [59] |
| Adipocyte | MALAT1 | Increase food intake | [60] |
| AT | ABCA1, ABCG1 | Macrophage foam cell generation and M1 polarization | [61] |
| Plasma | miR-29a | ATP production cardiomyopathy | [62] |
| AT | IL-6, MCP-1, MIF, RBP-4, Resistin | Hepatic IR | [63] |
| Adipocyte | SHH | M1 macrophage polarization; AT IR | [64] |
| Adipocyte | PTEN | AT IR | [65] |
| Adipocyte | Mimecan, Perilipin A, FABP4, TGB1, Cystatin C | AT Inflammation & IR; Biomarker | [66] |
| ADSCs | Arginase-1, Tyrosine Hidroxylase | M2 macrophage polarization; AT insulin sensitivity | [67] |
| Plasma | Perilipin A | DIO mice/Obese Humans Biomarker | [68] |
| Adipocyte | Adiponectin, FABP4 | Adipogenesis Biomarker | [69] |
| Adipocyte | Specific cholesterol enrichment (sEVs)/fosfatidil serine (lEVs) | Small & Large EVs Biomarker | [70] |
| Adipocyte-Endothelial | Cav-1 | Nutrient state Biomarker | [71] |
| Adipocyte | Cav-1, LPL, AQP7 | Obesity Biomarker | [73] |
| Adipocyte | AT secretome | Exosomal adipokines footprint | [74] |
| AT | NPM3, STEAP3, DAD1 | Obesity & Adipogenesis Biomarker | [75] |
| ADSCs | |||
| Serum | |||
| AT | TGFB1, Cav-1, FABP4, Mimecan | Obesity Biomarker | [76] |
| Plasma | |||
| Adipocyte | IL-6, TNFα, MCP-1 | AT & Systemic IR | [77] |
| AT | |||
| ATM | miR-29a | IR (AT, myocyte and hepatocyte) | [78] |
| Adipocyte | miR-125a-5p, miR-296-3p, miR-298-5p, miR-351-5p | Insulin secretion & β cell function | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgadillo-Velázquez, J.; Mendivil-Alvarado, H.; Coronado-Alvarado, C.D.; Astiazaran-Garcia, H. Extracellular Vesicles from Adipose Tissue Could Promote Metabolic Adaptation through PI3K/Akt/mTOR. Cells 2022, 11, 1831. https://doi.org/10.3390/cells11111831
Delgadillo-Velázquez J, Mendivil-Alvarado H, Coronado-Alvarado CD, Astiazaran-Garcia H. Extracellular Vesicles from Adipose Tissue Could Promote Metabolic Adaptation through PI3K/Akt/mTOR. Cells. 2022; 11(11):1831. https://doi.org/10.3390/cells11111831
Chicago/Turabian StyleDelgadillo-Velázquez, Jaime, Herminia Mendivil-Alvarado, Carlos Daniel Coronado-Alvarado, and Humberto Astiazaran-Garcia. 2022. "Extracellular Vesicles from Adipose Tissue Could Promote Metabolic Adaptation through PI3K/Akt/mTOR" Cells 11, no. 11: 1831. https://doi.org/10.3390/cells11111831
APA StyleDelgadillo-Velázquez, J., Mendivil-Alvarado, H., Coronado-Alvarado, C. D., & Astiazaran-Garcia, H. (2022). Extracellular Vesicles from Adipose Tissue Could Promote Metabolic Adaptation through PI3K/Akt/mTOR. Cells, 11(11), 1831. https://doi.org/10.3390/cells11111831







