Single Cell/Nucleus Transcriptomics Comparison in Zebrafish and Humans Reveals Common and Distinct Molecular Responses to Alzheimer’s Disease
Abstract
:1. Introduction
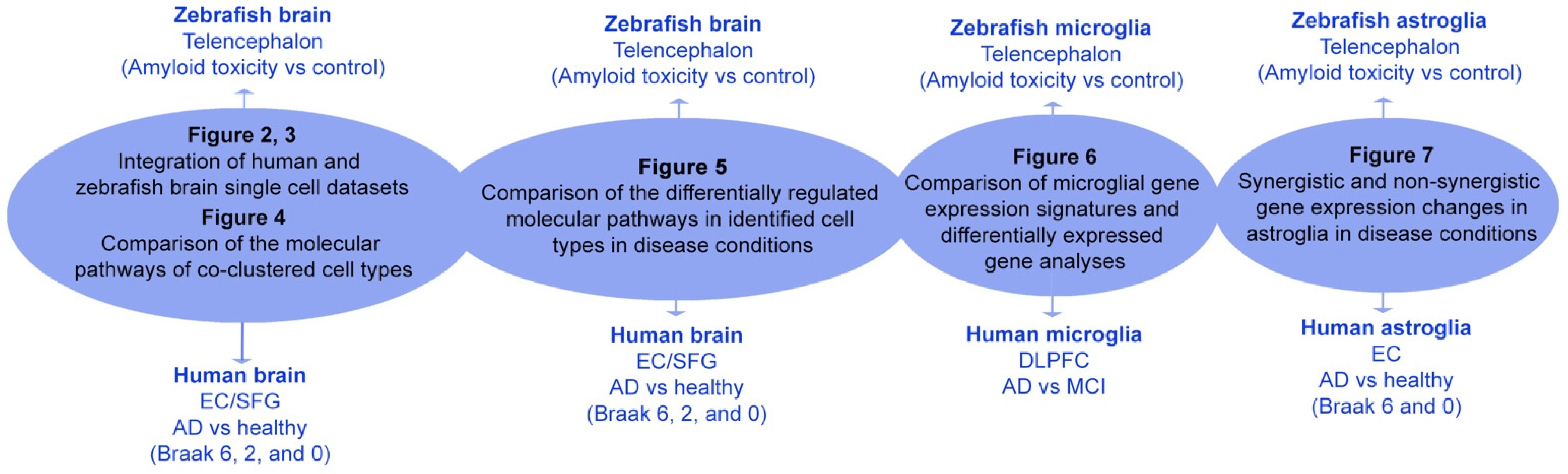
2. Results
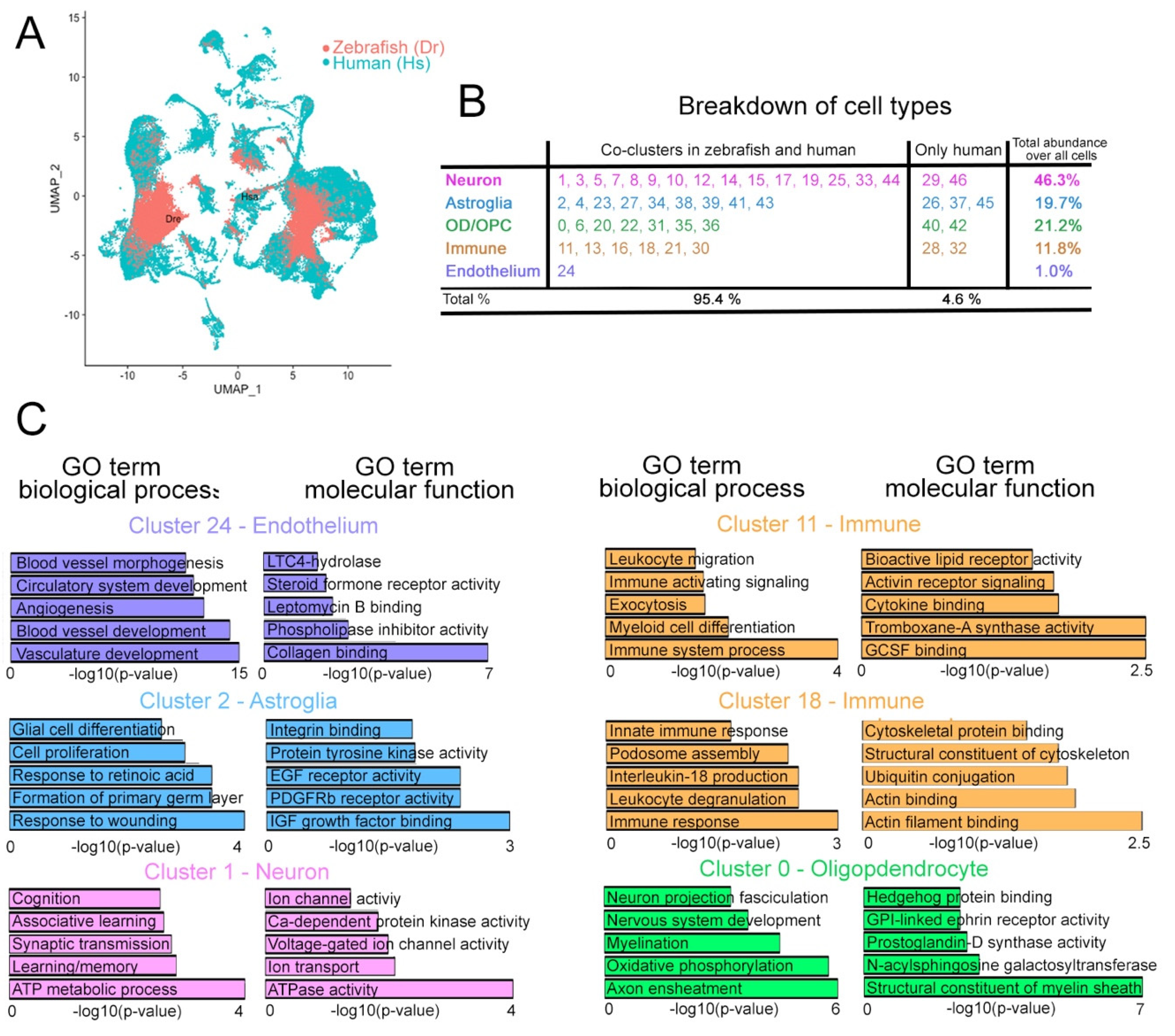
2.1. Integrated Clustering of Zebrafish and Human Cells
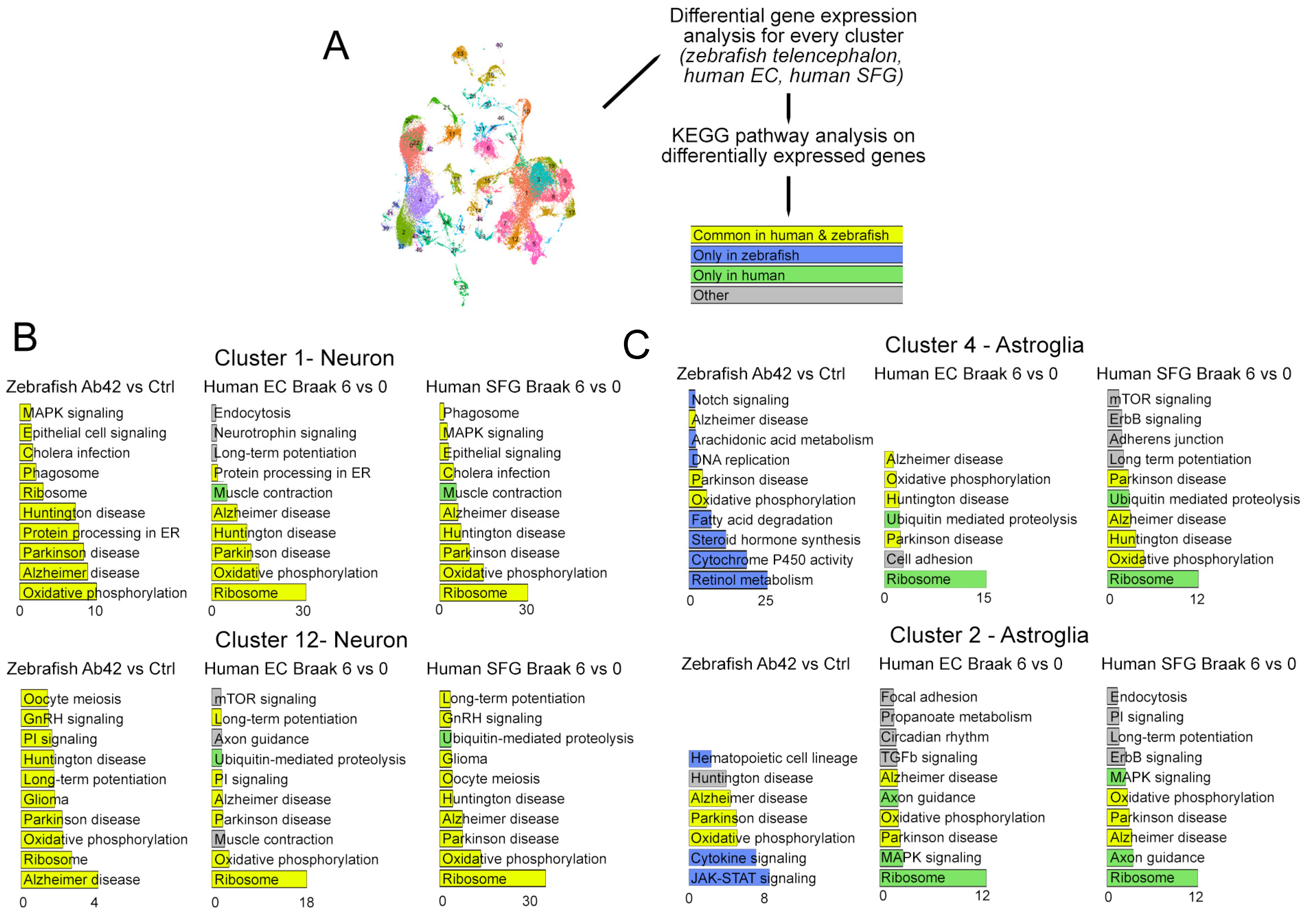
2.2. Differential Expression Analyses between AD and Controls in Zebrafish vs. Human
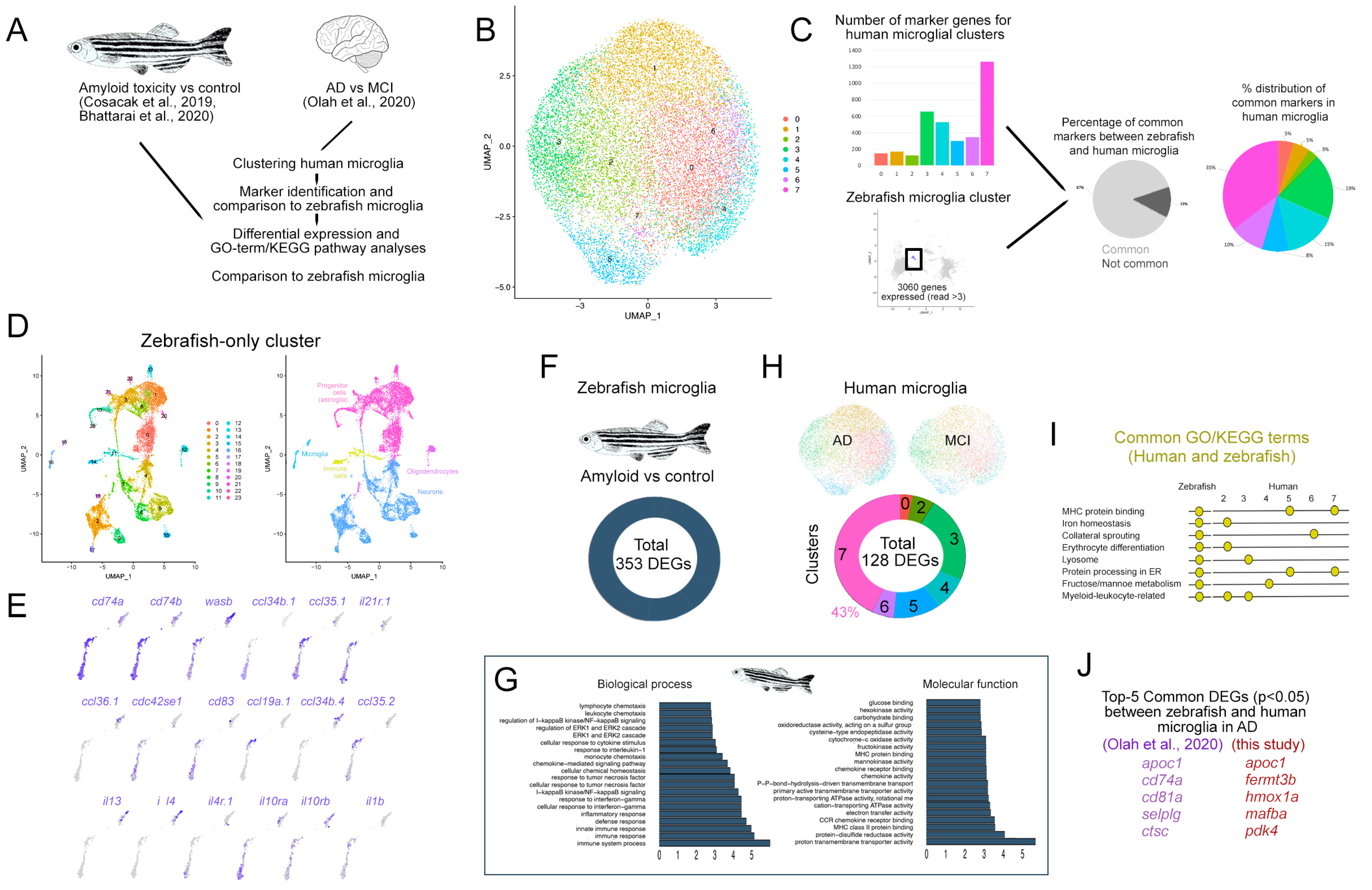
2.3. Comparison of Microglia between Zebrafish and Humans
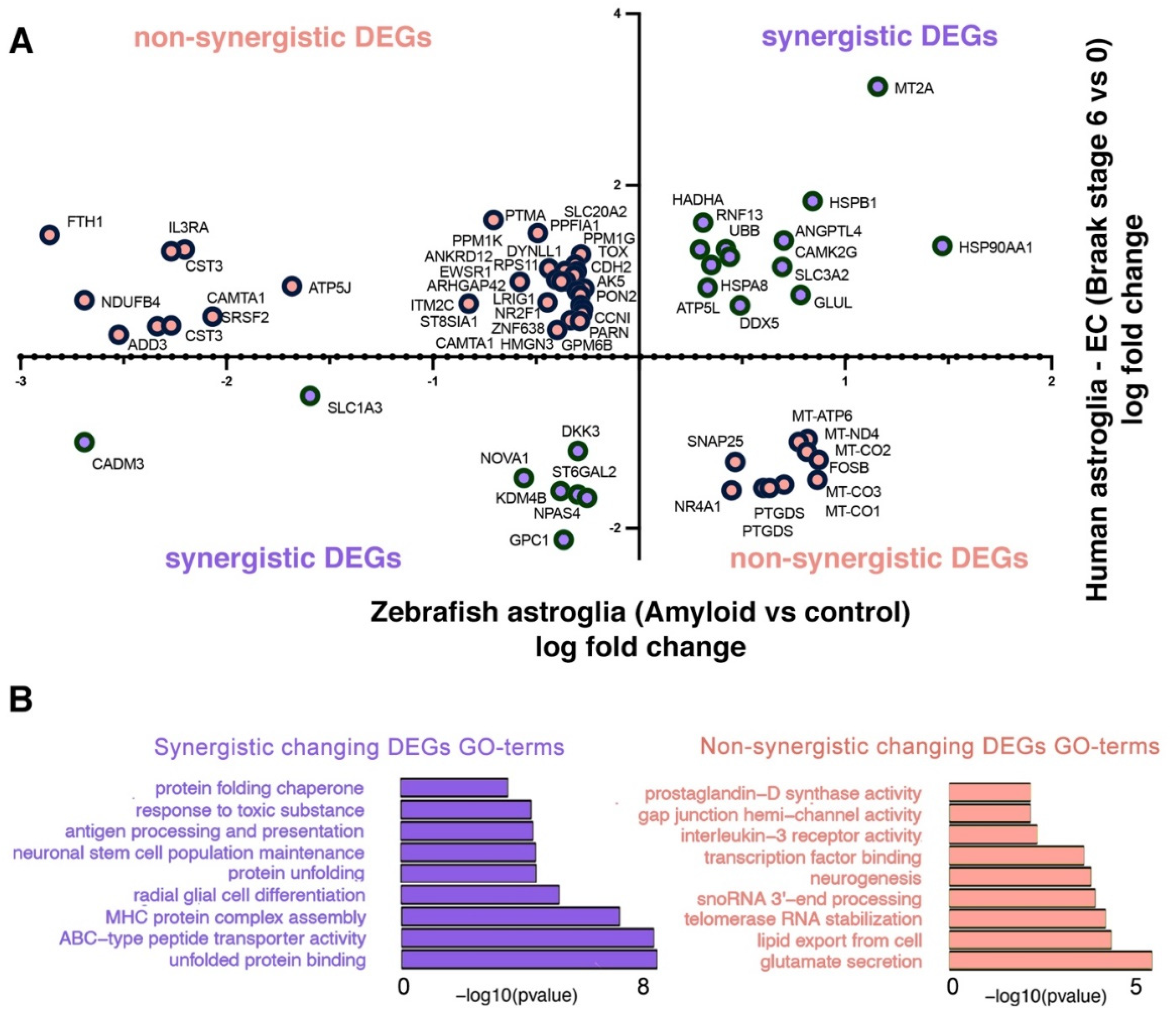
2.4. Comparison of Astroglial Clusters in Human and Zebrafish Brain in AD
2.5. Developing Human Brain versus Zebrafish
3. Discussion
4. Methods
4.1. Single Cell Transcriptomics Data
4.2. Single Cell Data Analyses Using All Genes
4.3. Main Cell Types
4.4. Outcomes
4.5. Orthologous Genes
4.6. Single Cell Data Analyses
4.6.1. Preprocessing of the EC/SFG Datasets
4.6.2. Integrating all Cells from the Zebrafish Telencephalon, Human EC/SFG, and Human Fetal Samples
4.6.3. Marker Genes Analyses
4.6.4. The Effect of Orthologous Genes on Each Dataset
4.6.5. Differentially Expressed Genes and GO Term Analyses
4.6.6. Comparing Human Microglia and Zebrafish Microglia
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- De Strooper, B.; Karran, E. The Cellular Phase of Alzheimer’s Disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef] [Green Version]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Selkoe, D.J. Folding proteins in fatal ways. Nature 2003, 426, 900–904. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s disease is a synaptic failure. Science 2002, 298, 789–791. [Google Scholar] [CrossRef] [Green Version]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef]
- Heppner, F.L.; Ransohoff, R.M.; Becher, B. Immune attack: The role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 2015, 16, 358–372. [Google Scholar] [CrossRef]
- Amor, S.; Puentes, F.; Baker, D.; van der Valk, P. Inflammation in neurodegenerative diseases. Immunology 2010, 129, 154–169. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Bitan, G.; Teplow, D.B. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J. Neurosci. Res. 2002, 69, 567–577. [Google Scholar] [CrossRef] [Green Version]
- Tincer, G.; Mashkaryan, V.; Bhattarai, P.; Kizil, C. Neural stem/progenitor cells in Alzheimer’s disease. Yale J. Biol. Med. 2016, 89, 23–35. [Google Scholar]
- Tong, L.M.; Fong, H.; Huang, Y. Stem cell therapy for Alzheimer’s disease and related disorders: Current status and future perspectives. Exp. Mol. Med. 2015, 47, e151. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.; Zheng, H. Signaling pathways regulating neuron-glia interaction and their implications in Alzheimer’s disease. J. Neurochem. 2016, 136, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Attems, J.; Jellinger, K.A. The overlap between vascular disease and Alzheimer’s disease--lessons from pathology. BMC Med. 2014, 12, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ettle, B.; Schlachetzki, J.C.M.; Winkler, J. Oligodendroglia and Myelin in Neurodegenerative Diseases: More Than Just Bystanders? Mol. Neurobiol. 2016, 53, 3046–3062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartzokis, G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol. Aging 2011, 32, 1341–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobin, M.K.; Musaraca, K.; Disouky, A.; Shetti, A.; Bheri, A.; Honer, W.G.; Kim, N.; Dawe, R.J.; Bennett, D.A.; Arfanakis, K.; et al. Human Hippocampal Neurogenesis Persists in Aged Adults and Alzheimer’s Disease Patients. Cell Stem Cell 2019, 24, 974–982.e973. [Google Scholar] [CrossRef]
- Moreno-Jimenez, E.P.; Flor-Garcia, M.; Terreros-Roncal, J.; Rabano, A.; Cafini, F.; Pallas-Bazarra, N.; Avila, J.; Llorens-Martin, M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef]
- Choi, S.H.; Bylykbashi, E.; Chatila, Z.K.; Lee, S.W.; Pulli, B.; Clemenson, G.D.; Kim, E.; Rompala, A.; Oram, M.K.; Asselin, C.; et al. Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer’s mouse model. Science 2018, 361, eaan8821. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.H.; Tanzi, R.E. Is Alzheimer’s Disease a Neurogenesis Disorder? Cell Stem Cell 2019, 25, 7–8. [Google Scholar] [CrossRef]
- Kizil, C.; Bhattarai, P. Is Alzheimer’s Also a Stem Cell Disease?-The Zebrafish Perspective. Front. Cell Dev. Biol. 2018, 6, 159. [Google Scholar] [CrossRef] [Green Version]
- Kizil, C. Mechanisms of Pathology-Induced Neural Stem Cell Plasticity and Neural Regeneration in Adult Zebrafish Brain. Curr. Pathobiol. Rep. 2018, 6, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Cosacak, M.I.; Papadimitriou, C.; Kizil, C. Regeneration, Plasticity, and Induced Molecular Programs in Adult Zebrafish Brain. BioMed Res. Int. 2015, 2015, 769763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishimoto, N.; Shimizu, K.; Sawamoto, K. Neuronal regeneration in a zebrafish model of adult brain injury. Dis. Models Mech. 2012, 5, 200–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kizil, C.; Dudczig, S.; Kyritsis, N.; Machate, A.; Blaesche, J.; Kroehne, V.; Brand, M. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012, 7, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kizil, C.; Kaslin, J.; Kroehne, V.; Brand, M. Adult neurogenesis and brain regeneration in zebrafish. Dev. Neurobiol. 2012, 72, 429–461. [Google Scholar] [CrossRef]
- Kizil, C.; Kyritsis, N.; Dudczig, S.; Kroehne, V.; Freudenreich, D.; Kaslin, J.; Brand, M. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev. Cell 2012, 23, 1230–1237. [Google Scholar] [CrossRef] [Green Version]
- Kyritsis, N.; Kizil, C.; Zocher, S.; Kroehne, V.; Kaslin, J.; Freudenreich, D.; Iltzsche, A.; Brand, M. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 2012, 338, 1353–1356. [Google Scholar] [CrossRef]
- Marz, M.; Schmidt, R.; Rastegar, S.; Strahle, U. Regenerative response following stab injury in the adult zebrafish telencephalon. Dev. Dyn. 2012, 240, 2221–2231. [Google Scholar] [CrossRef]
- Baumgart, E.V.; Barbosa, J.S.; Bally-Cuif, L.; Gotz, M.; Ninkovic, J. Stab wound injury of the zebrafish telencephalon: A model for comparative analysis of reactive gliosis. Glia 2012, 60, 343–357. [Google Scholar] [CrossRef]
- Bhattarai, P.; Thomas, A.K.; Papadimitriou, C.; Cosacak, M.I.; Mashkaryan, V.; Froc, C.; Kurth, T.; Dahl, A.; Zhang, Y.; Kizil, C. IL4/STAT6 signaling activates neural stem cell proliferation and neurogenesis upon Amyloid-β42 aggregation in adult zebrafish brain. Cell Rep. 2016, 17, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Jurisch-Yaksi, N.; Yaksi, E.; Kizil, C. Radial glia in the zebrafish brain: Functional, structural, and physiological comparison with the mammalian glia. Glia 2020, 68, 2451–2470. [Google Scholar] [CrossRef]
- Alunni, A.; Bally-Cuif, L. A comparative view of regenerative neurogenesis in vertebrates. Development 2016, 143, 741–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattarai, P.; Thomas, A.K.; Cosacak, M.I.; Papadimitriou, C.; Mashkaryan, V.; Zhang, Y.; Kizil, C. Modeling Amyloid-β42 Toxicity and Neurodegeneration in Adult Zebrafish Brain. J. Vis. Exp. 2017, 128, e56014. [Google Scholar] [CrossRef]
- Reitz, C.; Mayeux, R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 2014, 88, 640–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaFerla, F.M.; Oddo, S. Alzheimer’s disease: Abeta, tau and synaptic dysfunction. Trends Mol. Med. 2005, 11, 170–176. [Google Scholar] [CrossRef]
- Kizil, C.; Sariya, S.; Kim, Y.A.; Rajabli, F.; Martin, E.; Reyes-Dumeyer, D.; Vardarajan, B.; Maldonado, A.; Haines, J.L.; Mayeux, R.; et al. Admixture Mapping of Alzheimer’s disease in Caribbean Hispanics identifies a new locus on 22q13.1. Mol. Psychiatry 2022. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Raghavan, N.S.; Bhattarai, P.; Siddiqui, T.; Sariya, S.; Reyes-Dumeyer, D.; Flowers, X.E.; Cardoso, S.A.L.; De Jager, P.L.; Bennett, D.A.; et al. FMNL2 regulates gliovascular interactions and is associated with vascular risk factors and cerebrovascular pathology in Alzheimer’s disease. Acta Neuropathol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Leng, K.; Li, E.; Eser, R.; Piergies, A.; Sit, R.; Tan, M.; Neff, N.; Li, S.H.; Rodriguez, R.D.; Suemoto, C.K.; et al. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci. 2021, 24, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Olah, M.; Menon, V.; Habib, N.; Taga, M.F.; Ma, Y.; Yung, C.J.; Cimpean, M.; Khairallah, A.; Coronas-Samano, G.; Sankowski, R.; et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 2020, 11, 6129. [Google Scholar] [CrossRef]
- Eze, U.C.; Bhaduri, A.; Haeussler, M.; Nowakowski, T.J.; Kriegstein, A.R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 2021, 24, 584–594. [Google Scholar] [CrossRef]
- Bhattarai, P.; Cosacak, M.I.; Mashkaryan, V.; Demir, S.; Popova, S.; Govindarajan, N.; Brandt, K.; Zhang, Y.; Chang, W.; Ampatzis, K.; et al. Neuron-glia interaction through Serotonin-BDNF-NGFR axis enables regenerative neurogenesis in Alzheimer’s model of adult zebrafish brain. PLoS Biol. 2020, 18, e3000585. [Google Scholar] [CrossRef] [Green Version]
- Cosacak, M.I.; Bhattarai, P.; Reinhardt, S.; Petzold, A.; Dahl, A.; Zhang, Y.; Kizil, C. Single-Cell Transcriptomics Analyses of Neural Stem Cell Heterogeneity and Contextual Plasticity in a Zebrafish Brain Model of Amyloid Toxicity. Cell Rep. 2019, 27, 1307–1318 e1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Gama, P.P.; Qiu, T.; Cosacak, M.I.; Rayamajhi, D.; Konac, A.; Hansen, J.N.; Ringers, C.; Acuna-Hinrichsen, F.; Hui, S.P.; Olstad, E.W.; et al. Diversity and function of motile ciliated cell types within ependymal lineages of the zebrafish brain. Cell Rep. 2021, 37, 109775. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.; Eckler, M.J.; McKenna, W.L.; McKinsey, G.L.; Rubenstein, J.L.; Chen, B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron 2013, 80, 1167–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakovcevski, I.; Zecevic, N. Olig transcription factors are expressed in oligodendrocyte and neuronal cells in human fetal CNS. J. Neurosci. 2005, 25, 10064–10073. [Google Scholar] [CrossRef]
- Menn, B.; Garcia-Verdugo, J.M.; Yaschine, C.; Gonzalez-Perez, O.; Rowitch, D.; Alvarez-Buylla, A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J. Neurosci. 2006, 26, 7907–7918. [Google Scholar] [CrossRef]
- Wenk, G.L. Neuropathologic changes in Alzheimer’s disease: Potential targets for treatment. J. Clin. Psychiatry 2006, 67 (Suppl. 3), 3. [Google Scholar]
- Nowack, A.; Yao, J.; Custer, K.L.; Bajjalieh, S.M. SV2 regulates neurotransmitter release via multiple mechanisms. Am. J. Physiol.-Cell Physiol. 2010, 299, C960–C967. [Google Scholar] [CrossRef] [Green Version]
- Park, D.; Yang, Y.-H.; Bae, D.K.; Lee, S.H.; Yang, G.; Kyung, J.; Kim, D.; Choi, E.-K.; Lee, S.W.; Kim, G.H.; et al. Improvement of cognitive function and physical activity of aging mice by human neural stem cells over-expressing choline acetyltransferase. Neurobiol. Aging 2013, 34, 2639–2646. [Google Scholar] [CrossRef]
- Codega, P.; Silva-Vargas, V.; Paul, A.; Maldonado-Soto, A.R.; Deleo, A.M.; Pastrana, E.; Doetsch, F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 2014, 82, 545–559. [Google Scholar] [CrossRef] [Green Version]
- Mellott, T.J.; Pender, S.M.; Burke, R.M.; Langley, E.A.; Blusztajn, J.K. IGF2 ameliorates amyloidosis, increases cholinergic marker expression and raises BMP9 and neurotrophin levels in the hippocampus of the APPswePS1dE9 Alzheimer’s disease model mice. PLoS ONE 2014, 9, e94287. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Nakajima, K. Role of the Immune System in the Development of the Central Nervous System. Front. Neurosci. 2019, 13, 916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newton, K.; Dixit, V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012, 4, a006049. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.E.; Senger, D.R. Endothelial extracellular matrix: Biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ. Res. 2005, 97, 1093–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillebrand, U.; Hausberg, M.; Lang, D.; Stock, C.; Riethmuller, C.; Callies, C.; Bussemaker, E. How steroid hormones act on the endothelium--insights by atomic force microscopy. Pflügers Arch.-Eur. J. Physiol. 2008, 456, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jebelli, J.; Su, W.; Hopkins, S.; Pocock, J.; Garden, G.A. Glia: Guardians, gluttons, or guides for the maintenance of neuronal connectivity? Ann. N. Y. Acad. Sci. 2015, 1351, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Barker, S.J.; Raju, R.M.; Milman, N.E.P.; Wang, J.; Davila-Velderrain, J.; Gunter-Rahman, F.; Parro, C.C.; Bozzelli, P.L.; Abdurrob, F.; Abdelaal, K.; et al. MEF2 is a key regulator of cognitive potential and confers resilience to neurodegeneration. Sci. Transl. Med. 2021, 13, eabd7695. [Google Scholar] [CrossRef]
- Raghavan, N.S.; Dumitrescu, L.; Mormino, E.; Mahoney, E.R.; Lee, A.J.; Gao, Y.; Bilgel, M.; Goldstein, D.; Harrison, T.; Engelman, C.D.; et al. Association Between Common Variants in RBFOX1, an RNA-Binding Protein, and Brain Amyloidosis in Early and Preclinical Alzheimer Disease. JAMA Neurol. 2020, 77, 1288–1298. [Google Scholar] [CrossRef]
- Kadowaki, H.; Nishitoh, H.; Urano, F.; Sadamitsu, C.; Matsuzawa, A.; Takeda, K.; Masutani, H.; Yodoi, J.; Urano, Y.; Nagano, T.; et al. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ. 2005, 12, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Beyreuther, K.; Masters, C.L. Alzheimer’s disease. The ins and outs of amyloid-beta. Nature 1997, 389, 677–678. [Google Scholar] [CrossRef]
- Wei, Z.; Koya, J.; Reznik, S.E. Insulin Resistance Exacerbates Alzheimer Disease via Multiple Mechanisms. Front. Neurosci. 2021, 15, 687157. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, A.; Brewerton, S.; Bell, A.; Sargent, S.; Glover, S.; Hardy, C.; Moore, R.; Calley, J.; Ramachandran, D.; Poidinger, M.; et al. An unbiased approach to identifying tau kinases that phosphorylate tau at sites associated with Alzheimer disease. J. Biol. Chem. 2013, 288, 23331–23347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terreros-Roncal, J.; Moreno-Jimenez, E.P.; Flor-Garcia, M.; Rodriguez-Moreno, C.B.; Trinchero, M.F.; Cafini, F.; Rabano, A.; Llorens-Martin, M. Impact of neurodegenerative diseases on human adult hippocampal neurogenesis. Science 2021, 374, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Boldrini, M.; Fulmore, C.A.; Tartt, A.N.; Simeon, L.R.; Pavlova, I.; Poposka, V.; Rosoklija, G.B.; Stankov, A.; Arango, V.; Dwork, A.J.; et al. Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell 2018, 22, 589–599.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrells, S.F.; Paredes, M.F.; Cebrian-Silla, A.; Sandoval, K.; Qi, D.; Kelley, K.W.; James, D.; Mayer, S.; Chang, J.; Auguste, K.I.; et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 2018, 555, 377–381. [Google Scholar] [CrossRef]
- Kempermann, G.; Gage, F.H.; Aigner, L.; Song, H.; Curtis, M.A.; Thuret, S.; Kuhn, H.G.; Jessberger, S.; Frankland, P.W.; Cameron, H.A.; et al. Human Adult Neurogenesis: Evidence and Remaining Questions. Cell Stem Cell 2018, 23, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Berg, D.A.; Zhu, Y.; Shin, J.Y.; Song, J.; Bonaguidi, M.A.; Enikolopov, G.; Nauen, D.W.; Christian, K.M.; Ming, G.L.; et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015, 17, 360–372. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, J.J.; Verkhratsky, A. Neurogenesis in Alzheimer’s disease. J. Anat. 2011, 219, 78–89. [Google Scholar] [CrossRef]
- Knobloch, M.; Pilz, G.A.; Ghesquiere, B.; Kovacs, W.J.; Wegleiter, T.; Moore, D.L.; Hruzova, M.; Zamboni, N.; Carmeliet, P.; Jessberger, S. A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep. 2017, 20, 2144–2155. [Google Scholar] [CrossRef] [Green Version]
- Diotel, N.; Vaillant, C.; Gueguen, M.M.; Mironov, S.; Anglade, I.; Servili, A.; Pellegrini, E.; Kah, O. Cxcr4 and Cxcl12 expression in radial glial cells of the brain of adult zebrafish. J. Comp. Neurol. 2010, 518, 4855–4876. [Google Scholar] [CrossRef]
- Marz, M.; Chapouton, P.; Diotel, N.; Vaillant, C.; Hesl, B.; Takamiya, M.; Lam, C.S.; Kah, O.; Bally-Cuif, L.; Strahle, U. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia 2010, 58, 870–888. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.T.; Wang, L.; Li, S.; Long, Z.Y.; Wu, Y.M.; Liu, Y. Retinoic acid induced the differentiation of neural stem cells from embryonic spinal cord into functional neurons in vitro. Int. J. Clin. Exp. Pathol. 2015, 8, 8129–8135. [Google Scholar]
- Valdivia, L.E.; Lamb, D.B.; Horner, W.; Wierzbicki, C.; Tafessu, A.; Williams, A.M.; Gestri, G.; Krasnow, A.M.; Vleeshouwer-Neumann, T.S.; Givens, M.; et al. Antagonism between Gdf6a and retinoic acid pathways controls timing of retinal neurogenesis and growth of the eye in zebrafish. Development 2016, 143, 1087–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gongal, P.A.; March, L.D.; Holly, V.L.; Pillay, L.M.; Berry-Wynne, K.M.; Kagechika, H.; Waskiewicz, A.J. Hmx4 regulates Sonic hedgehog signaling through control of retinoic acid synthesis during forebrain patterning. Dev. Biol. 2011, 355, 55–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alunni, A.; Krecsmarik, M.; Bosco, A.; Galant, S.; Pan, L.; Moens, C.B.; Bally-Cuif, L. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development 2013, 140, 3335–3347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turolo, S.; Edefonti, A.; Mazzocchi, A.; Syren, M.L.; Morello, W.; Agostoni, C.; Montini, G. Role of Arachidonic Acid and Its Metabolites in the Biological and Clinical Manifestations of Idiopathic Nephrotic Syndrome. Int. J. Mol. Sci. 2021, 22, 5452. [Google Scholar] [CrossRef]
- Cosacak, M.I.; Bhattarai, P.; Kizil, C. Alzheimer’s disease, neural stem cells and neurogenesis: Cellular phase at single-cell level. Neural Regen. Res. 2020, 15, 824–827. [Google Scholar] [CrossRef]
- Mashkaryan, V.; Siddiqui, T.; Popova, S.; Cosacak, M.I.; Bhattarai, P.; Brandt, K.; Govindarajan, N.; Petzold, A.; Reinhardt, S.; Dahl, A.; et al. Type 1 Interleukin-4 signaling obliterates mouse astroglia in vivo but not in vitro. Front. Cell Dev. Biol. 2020, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Celikkaya, H.; Cosacak, M.I.; Papadimitriou, C.; Popova, S.; Bhattarai, P.; Biswas, S.N.; Siddiqui, T.; Wistorf, S.; Nevado-Alcalde, I.; Naumann, L.; et al. GATA3 Promotes the Neural Progenitor State but Not Neurogenesis in 3D Traumatic Injury Model of Primary Human Cortical Astrocytes. Front. Cell Neurosci. 2019, 13, 23. [Google Scholar] [CrossRef]
- Papadimitriou, C.; Celikkaya, H.; Cosacak, M.I.; Mashkaryan, V.; Bray, L.; Bhattarai, P.; Brandt, K.; Hollak, H.; Chen, X.; He, S.; et al. 3D Culture Method for Alzheimer’s Disease Modeling Reveals Interleukin-4 Rescues Abeta42-Induced Loss of Human Neural Stem Cell Plasticity. Dev. Cell 2018, 46, 85–101.e8. [Google Scholar] [CrossRef] [Green Version]
- Arber, C.; Lovejoy, C.; Harris, L.; Willumsen, N.; Alatza, A.; Casey, J.M.; Lines, G.; Kerins, C.; Mueller, A.K.; Zetterberg, H.; et al. Familial Alzheimer’s Disease Mutations in PSEN1 Lead to Premature Human Stem Cell Neurogenesis. Cell Rep. 2021, 34, 108615. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Nicola, D.; Suzzi, S.; Vargas-Caballero, M.; Fransen, N.L.; Al-Malki, H.; Cebrian-Silla, A.; Garcia-Verdugo, J.M.; Riecken, K.; Fehse, B.; Perry, V.H. Temporal dynamics of hippocampal neurogenesis in chronic neurodegeneration. Brain 2014, 137, 2312–2328. [Google Scholar] [CrossRef] [PubMed]
- Leissring, M.A.; Farris, W.; Chang, A.Y.; Walsh, D.M.; Wu, X.; Sun, X.; Frosch, M.P.; Selkoe, D.J. Enhanced proteolysis of beta-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron 2003, 40, 1087–1093. [Google Scholar] [CrossRef] [Green Version]
- Carson, J.A.; Turner, A.J. Beta-amyloid catabolism: Roles for neprilysin (NEP) and other metallopeptidases? J. Neurochem. 2002, 81, 1–8. [Google Scholar] [CrossRef]
- Vardarajan, B.N.; Zhang, Y.; Lee, J.H.; Cheng, R.; Bohm, C.; Ghani, M.; Reitz, C.; Reyes-Dumeyer, D.; Shen, Y.; Rogaeva, E.; et al. Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 2015, 77, 215–227. [Google Scholar] [CrossRef] [Green Version]
- Reitz, C.; Mayeux, R. Use of genetic variation as biomarkers for Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2009, 1180, 75–96. [Google Scholar] [CrossRef] [Green Version]
- Scheltens, P.; Blennow, K.; Breteler, M.M.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Arendt, T. Synaptic degeneration in Alzheimer’s disease. Acta Neuropathol. 2009, 118, 167–179. [Google Scholar] [CrossRef]
- Caramillo, E.M.; Echevarria, D.J. Alzheimer’s disease in the zebrafish: Where can we take it? Behav. Pharmacol. 2017, 28, 179–186. [Google Scholar] [CrossRef]
- Santana, S.; Rico, E.P.; Burgos, J.S. Can zebrafish be used as animal model to study Alzheimer’s disease? Am. J. Neurodegener. Dis. 2012, 1, 32–48. [Google Scholar]
- DeCarolis, N.A.; Mechanic, M.; Petrik, D.; Carlton, A.; Ables, J.L.; Malhotra, S.; Bachoo, R.; Gotz, M.; Lagace, D.C.; Eisch, A.J. In vivo contribution of nestin- and GLAST-lineage cells to adult hippocampal neurogenesis. Hippocampus 2013, 23, 708–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, D.J.; Fort, P.E. Heat Shock Proteins Regulatory Role in Neurodevelopment. Front. Neurosci. 2018, 12, 821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choy, F.C.; Klaric, T.S.; Koblar, S.A.; Lewis, M.D. The Role of the Neuroprotective Factor Npas4 in Cerebral Ischemia. Int. J. Mol. Sci. 2015, 16, 29011–29028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tai, H.C.; Schuman, E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008, 9, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.J.; An, J.; Jou, I.; Park, S.M.; Joe, E.H. A Parkinson’s disease gene, DJ-1, regulates anti-inflammatory roles of astrocytes through prostaglandin D2 synthase expression. Neurobiol. Dis. 2019, 127, 482–491. [Google Scholar] [CrossRef]
- Ekdahl, C.T.; Kokaia, Z.; Lindvall, O. Brain inflammation and adult neurogenesis: The dual role of microglia. Neuroscience 2009, 158, 1021–1029. [Google Scholar] [CrossRef]
- Yutsudo, N.; Kamada, T.; Kajitani, K.; Nomaru, H.; Katogi, A.; Ohnishi, Y.H.; Ohnishi, Y.N.; Takase, K.; Sakumi, K.; Shigeto, H.; et al. fosB-null mice display impaired adult hippocampal neurogenesis and spontaneous epilepsy with depressive behavior. Neuropsychopharmacology 2013, 38, 895–906. [Google Scholar] [CrossRef]
- Kalebic, N.; Gilardi, C.; Stepien, B.; Wilsch-Brauninger, M.; Long, K.R.; Namba, T.; Florio, M.; Langen, B.; Lombardot, B.; Shevchenko, A.; et al. Neocortical Expansion Due to Increased Proliferation of Basal Progenitors Is Linked to Changes in Their Morphology. Cell Stem Cell 2019, 24, 535–550.e9. [Google Scholar] [CrossRef] [Green Version]
- Pirttila, T.J.; Lukasiuk, K.; Hakansson, K.; Grubb, A.; Abrahamson, M.; Pitkanen, A. Cystatin C modulates neurodegeneration and neurogenesis following status epilepticus in mouse. Neurobiol. Dis. 2005, 20, 241–253. [Google Scholar] [CrossRef]
- Svetoni, F.; De Paola, E.; La Rosa, P.; Mercatelli, N.; Caporossi, D.; Sette, C.; Paronetto, M.P. Post-transcriptional regulation of FUS and EWS protein expression by miR-141 during neural differentiation. Hum. Mol. Genet. 2017, 26, 2732–2746. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Gim, J.; Lee, J.; Gunasekaran, T.I.; Choi, K.Y.; Lee, J.J.; Seo, E.H.; Ko, P.W.; Chung, J.Y.; Choi, S.M.; et al. Potential Novel Genes for Late-Onset Alzheimer’s Disease in East-Asian Descent Identified by APOE-Stratified Genome-Wide Association Study. J. Alzheimer’s Dis. 2021, 82, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Gur, G.; Rubin, C.; Katz, M.; Amit, I.; Citri, A.; Nilsson, J.; Amariglio, N.; Henriksson, R.; Rechavi, G.; Hedman, H.; et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004, 23, 3270–3281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, H.; Hodges, A.K.; Curtis, C.; Lee, S.H.; Troakes, C.; Dobson, R.J.B.; Newhouse, S.J. Transcriptomic analysis of probable asymptomatic and symptomatic alzheimer brains. Brain Behav. Immun. 2019, 80, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902 e1821. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, A.; Andrews, M.G.; Mancia Leon, W.; Jung, D.; Shin, D.; Allen, D.; Jung, D.; Schmunk, G.; Haeussler, M.; Salma, J.; et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature 2020, 578, 142–148. [Google Scholar] [CrossRef]


| Human | Human | Human | Zebrafish | |
|---|---|---|---|---|
| Region | Adult entorhinal cortex (EC) and superior frontal gyrus (SFG) | Adult dorsolateral prefrontal cortex (DLPFC) | Embryonic telencephalon and cortex | Adult telencephalon (pallium and subpalium) |
| Sequencing type | Single nuclear RNA sequencing—mix | Single nuclear RNA sequencing—microglia | Single cell RNA sequencing—mix | Single cell RNA sequencing—mix |
| Number of individuals | 10 | 48 | 1st trimester—10 | 12 |
| Number of cells | 41,578 (EC), 62,086 (SFG) | 16,172 microglia nuclei | 6665 | 15,447 |
| Comparison | Alzheimer’s disease (Braak stage 2 and 6) vs. control (Braak stage 0) | Alzheimer’s disease (Braak stage 2 and 6) vs. control (Braak stage 0) | Developmental stages | Amyloid-injected versus control |
| References | [37] | [38] | [39] | [40,41,42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cosacak, M.I.; Bhattarai, P.; De Jager, P.L.; Menon, V.; Tosto, G.; Kizil, C. Single Cell/Nucleus Transcriptomics Comparison in Zebrafish and Humans Reveals Common and Distinct Molecular Responses to Alzheimer’s Disease. Cells 2022, 11, 1807. https://doi.org/10.3390/cells11111807
Cosacak MI, Bhattarai P, De Jager PL, Menon V, Tosto G, Kizil C. Single Cell/Nucleus Transcriptomics Comparison in Zebrafish and Humans Reveals Common and Distinct Molecular Responses to Alzheimer’s Disease. Cells. 2022; 11(11):1807. https://doi.org/10.3390/cells11111807
Chicago/Turabian StyleCosacak, Mehmet Ilyas, Prabesh Bhattarai, Philip L. De Jager, Vilas Menon, Giuseppe Tosto, and Caghan Kizil. 2022. "Single Cell/Nucleus Transcriptomics Comparison in Zebrafish and Humans Reveals Common and Distinct Molecular Responses to Alzheimer’s Disease" Cells 11, no. 11: 1807. https://doi.org/10.3390/cells11111807
APA StyleCosacak, M. I., Bhattarai, P., De Jager, P. L., Menon, V., Tosto, G., & Kizil, C. (2022). Single Cell/Nucleus Transcriptomics Comparison in Zebrafish and Humans Reveals Common and Distinct Molecular Responses to Alzheimer’s Disease. Cells, 11(11), 1807. https://doi.org/10.3390/cells11111807







