The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses
Abstract
:1. Introduction
2. Materials and Methods
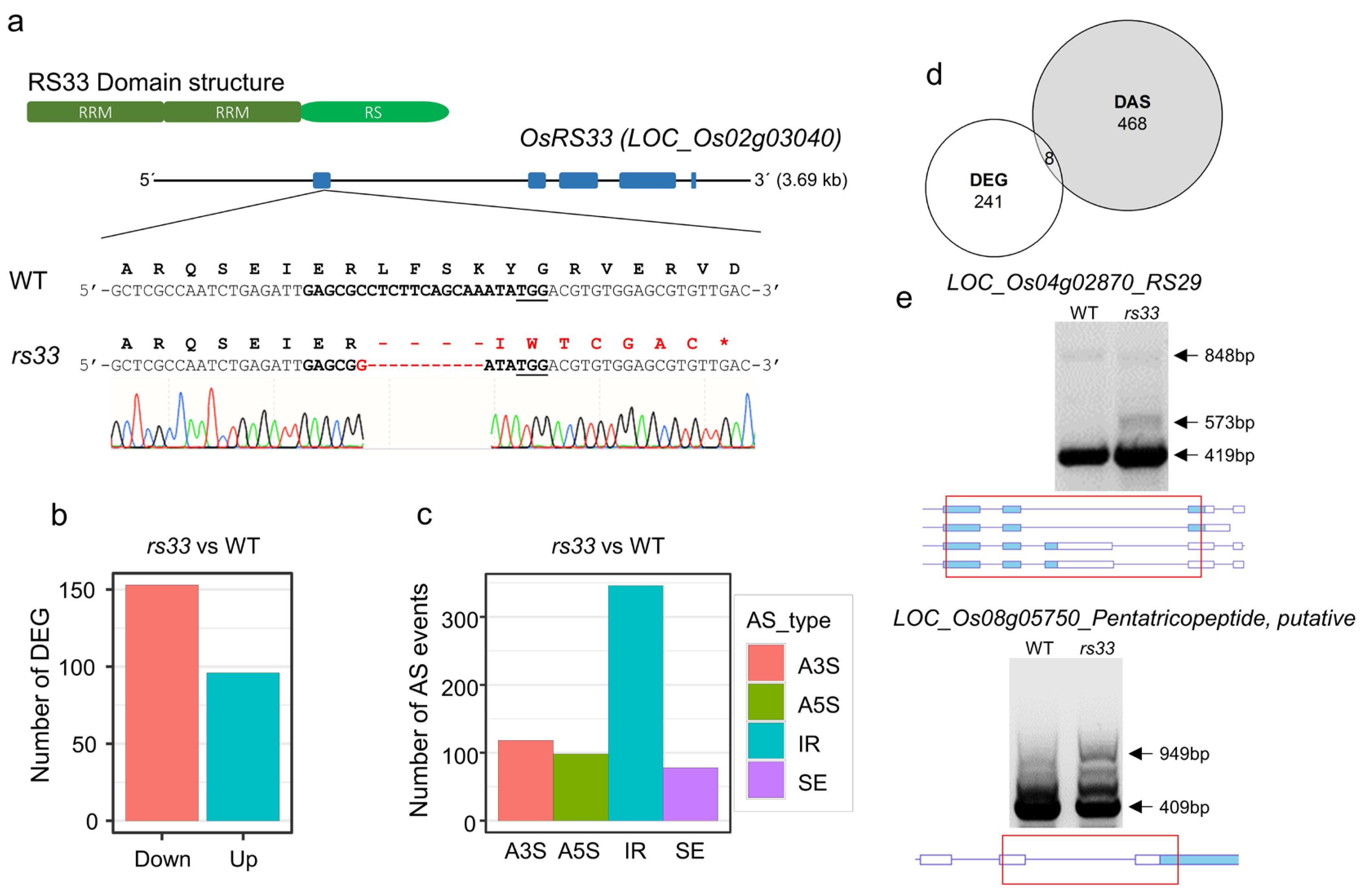
2.1. Plant Materials, Rice Transformation, and Genotyping of the Mutant Plants
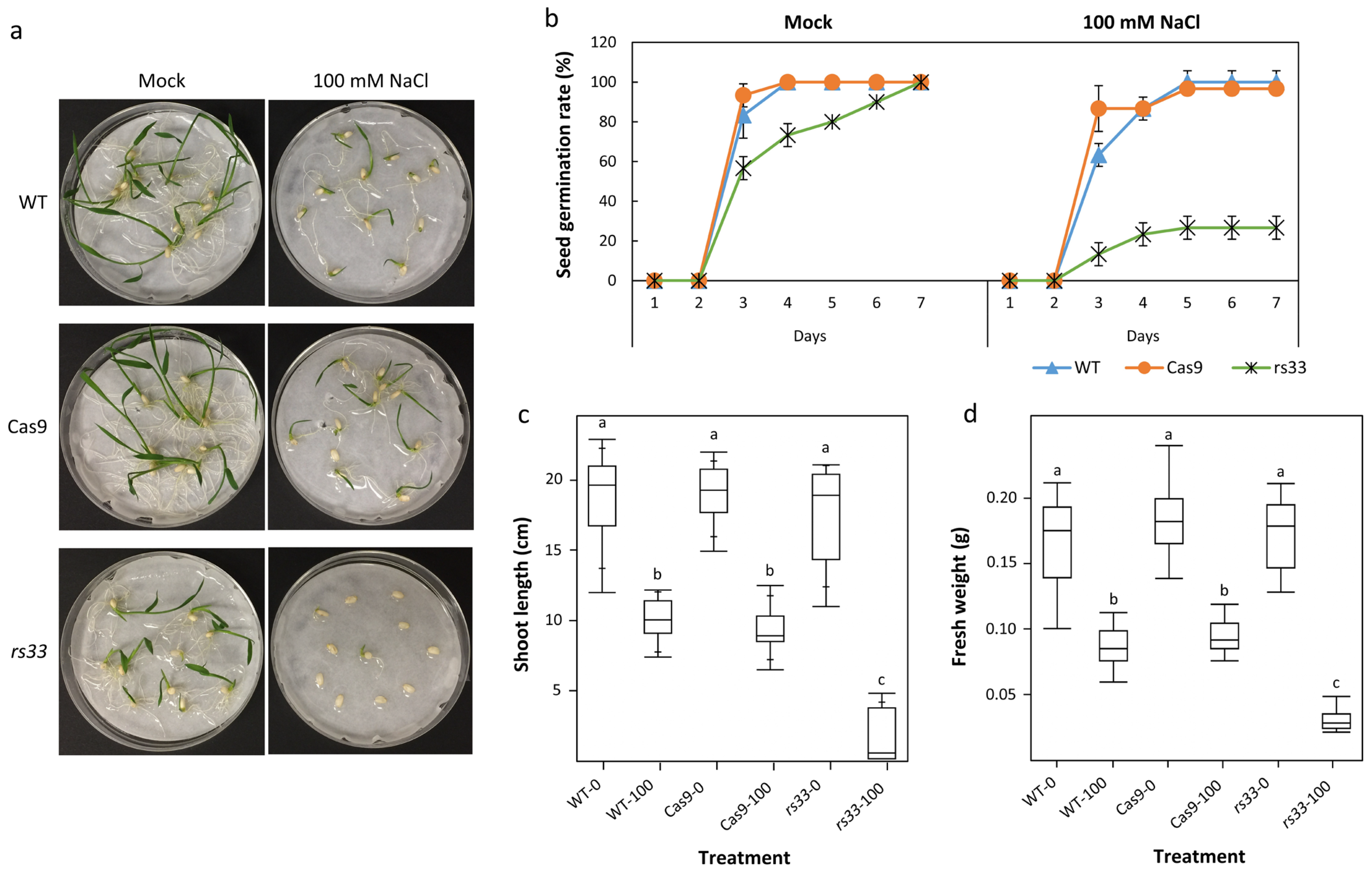
2.2. Phenotypic Analysis
2.3. RNA Isolation and RNA-Seq
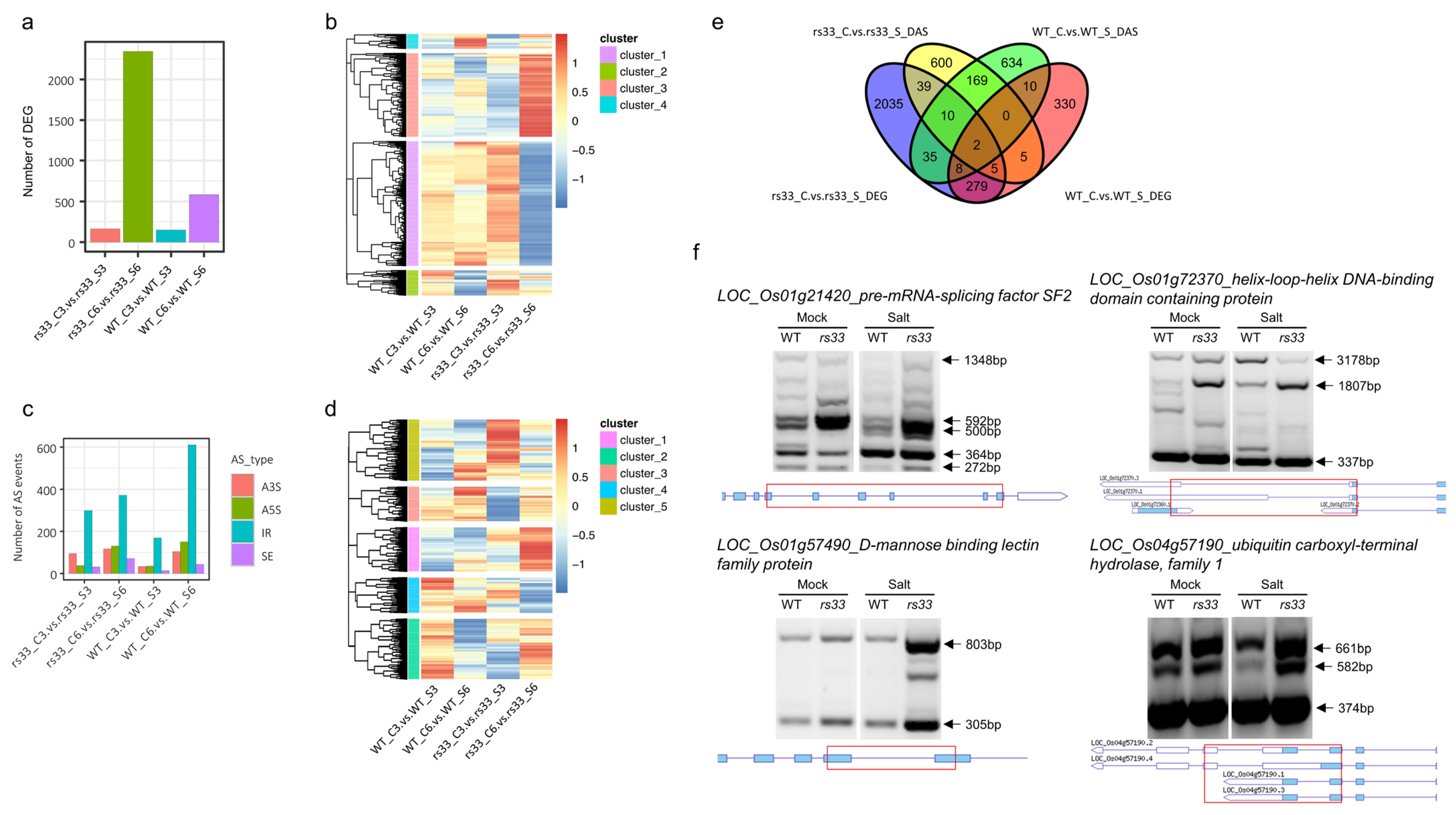
2.4. Analysis of RNA-Seq Data and Gene Functional Classification
2.5. RT-PCR
3. Results
3.1. OsRS33 Regulates Gene Expression and Pre-mRNA Splicing of a Small Set of Genes
3.2. The Osrs33 Mutant Is Hypersensitive to Salt and Cold Stresses
3.3. Loss of OsRS33 Leads to Genome-Wide Splicing Defects and Major Gene Expression Changes under Salt Stress
3.4. Low Temperature Differentially Affects Global Gene Expression and AS in rs33 and WT Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Res 2019, 8, F1000 Faculty Rev-658. [Google Scholar] [CrossRef] [Green Version]
- Chechanovsky, N.; Hovav, R.; Frenkel, R.; Faigenboim, A.; Eselson, Y.; Petreikov, M.; Moy, M.; Shen, S.; Schaffer, A.A. Low temperature upregulates cwp expression and modifies alternative splicing patterns, increasing the severity of cwp-induced tomato fruit cuticular microfissures. Hortic. Res. 2019, 6, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, A.; Nakaminami, K.; Seki, M. Biological Function of Changes in RNA Metabolism in Plant Adaptation to Abiotic Stress. Plant Cell Physiol. 2019, 60, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.A.; Huang, C.K.; Huang, W.S.; Huang, T.S.; Liu, H.Y.; Chen, Y.F. DEAD-Box RNA Helicase 42 Plays a Critical Role in Pre-mRNA Splicing under Cold Stress. Plant Physiol. 2020, 182, 255–271. [Google Scholar] [CrossRef] [Green Version]
- Ganie, S.A.; Reddy, A.S.N. Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance. Biology 2021, 10, 309. [Google Scholar] [CrossRef]
- Yu, H.; Du, Q.; Campbell, M.; Yu, B.; Walia, H.; Zhang, C. Genome-wide discovery of natural variation in pre-mRNA splicing and prioritising causal alternative splicing to salt stress response in rice. New Phytol. 2021, 230, 1273–1287. [Google Scholar] [CrossRef]
- Butt, H.; Eid, A.; Momin, A.A.; Bazin, J.; Crespi, M.; Arold, S.T.; Mahfouz, M.M. CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 2019, 20, 73. [Google Scholar] [CrossRef] [Green Version]
- Butt, H.; Bazin, J.; Alshareef, S.; Eid, A.; Benhamed, M.; Reddy, A.S.N.; Crespi, M.; Mahfouz, M.M. Overlapping roles of spliceosomal components SF3B1 and PHF5A in rice splicing regulation. Commun. Biol. 2021, 4, 529. [Google Scholar] [CrossRef]
- AlShareef, S.; Ling, Y.; Butt, H.; Mariappan, K.G.; Benhamed, M.; Mahfouz, M.M. Herboxidiene triggers splicing repression and abiotic stress responses in plants. BMC Genom. 2017, 18, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, Y.; Alshareef, S.; Butt, H.; Lozano-Juste, J.; Li, L.; Galal, A.A.; Moustafa, A.; Momin, A.A.; Tashkandi, M.; Richardson, D.N.; et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 2017, 89, 291–309. [Google Scholar] [CrossRef]
- Wahl, M.C.; Will, C.L.; Luhrmann, R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastner, B.; Will, C.L.; Stark, H.; Luhrmann, R. Structural Insights into Nuclear p e-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb. Perspect. Biol 2019, 11, ARTN a032417. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.G. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Shepard, P.J.; Hertel, K.J. The SR protein family. Genome Biol. 2009, 10, 242. [Google Scholar] [CrossRef] [Green Version]
- Twyffels, L.; Gueydan, C.; Kruys, V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011, 278, 3246–3255. [Google Scholar] [CrossRef]
- Bradley, T.; Cook, M.E.; Blanchette, M. SR proteins control a complex network of RNA-processing events. Rna 2015, 21, 75–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, J.M.; Sanford, J.R. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 2015, 6, 93–110. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S. SR Proteins: Binders, Regulators, and Connectors of RNA. Mol. Cells 2017, 40, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, H.; Piatek, A.; Li, L.; Reddy, A.S.N.; Mahfouz, M.M. Multiplex CRISPR Mutagenesis of the Serine/Arginine-Rich (SR) Gene Family in Rice. Genes 2019, 10, 596. [Google Scholar] [CrossRef] [Green Version]
- Barta, A.; Kalyna, M.; Reddy, A.S.N. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef]
- Morton, M.; AlTamimi, N.; Butt, H.; Reddy, A.S.N.; Mahfouz, M. Serine/Arginine-rich protein family of splicing regulators: New approaches to study splice isoform functions. Plant Sci. 2019, 283, 127–134. [Google Scholar] [CrossRef]
- Melo, J.P.; Kalyna, M.; Duque, P. Current Challenges in Studying Alternative Splicing in Plants: The Case of Physcomitrella patens SR Proteins. Front. Plant Sci. 2020, 11, 286. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente van Bentem, S.; Anrather, D.; Roitinger, E.; Djamei, A.; Hufnagl, T.; Barta, A.; Csaszar, E.; Dohnal, I.; Lecourieux, D.; Hirt, H. Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res. 2006, 34, 3267–3278. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Szakonyi, D.; Simpson, C.G.; Barbosa, I.C.; Brown, J.W.; Baena-Gonzalez, E.; Duque, P. The Arabidopsis SR45 Splicing Factor, a Negative Regulator of Sugar Signaling, Modulates SNF1-Related Protein Kinase 1 Stability. Plant Cell 2016, 28, 1910–1925. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Steitz, J.A. SRprises along a messenger’s journey. Mol. Cell 2005, 17, 613–615. [Google Scholar] [CrossRef]
- Wu, H.; Sun, S.Y.; Tu, K.; Gao, Y.; Xie, B.; Krainer, A.R.; Zhu, J. A Splicing-Independent Function of SF2/ASF in MicroRNA Processing. Mol. Cell 2010, 38, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, S.; Jabre, I.; Reddy, A.S.N.; Staiger, D.; Syed, N.H. Perspective on Alternative Splicing and Proteome Complexity in Plants. Trends Plant Sci. 2019, 24, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.C.; Cheng, S.C. Functional roles of protein splicing factors. Biosci. Rep. 2012, 32, 345–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [Green Version]
- Shang, X.D.; Cao, Y.; Ma, L.G. Alternative Splicing in Plant Genes: A Means of Regulating the Environmental Fitness of Plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Szakonyi, D.; Duque, P. Alternative Splicing as a Regulator of Early Plant Development. Front. Plant Sci. 2018, 9, 1174. [Google Scholar] [CrossRef] [Green Version]
- Jabre, I.; Reddy, A.S.N.; Kalyna, M.; Chaudhary, S.; Khokhar, W.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res. 2019, 47, 2716–2726. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Carvalho, S.D.; Duque, P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010, 154, 772–783. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013, 9, e1003875. [Google Scholar] [CrossRef] [Green Version]
- Cruz, T.M.; Carvalho, R.F.; Richardson, D.N.; Duque, P. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 2014, 15, 17541–17564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, D.; Wang, Y.; Hamilton, M.; Ben-Hur, A.; Reddy, A.S. Transcriptome-Wide Identification of RNA Targets of Arabidopsis SERINE/ARGININE-RICH45 Uncovers the Unexpected Roles of This RNA Binding Protein in RNA Processing. Plant Cell 2015, 27, 3294–3308. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, M.; Tsumoto, A.; Shimamoto, K. The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 2006, 18, 146–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S.N. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Albaqami, M.; Laluk, K.; Reddy, A.S.N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative Splicing Plays a Critical Role in Maintaining Mineral Nutrient Homeostasis in Rice (Oryza sativa). Plant Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Du, B.; Liu, D.; Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 2014, 455, 312–317. [Google Scholar] [CrossRef]
- Xin, R.; Kathare, P.K.; Huq, E. Coordinated Regulation of Pre-mRNA Splicing by the SFPS-RRC1 Complex to Promote Photomorphogenesis. Plant Cell 2019, 31, 2052–2069. [Google Scholar] [CrossRef] [Green Version]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010, 329, 1355–1358. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, H.; Eid, A.; Ali, Z.; Atia, M.A.M.; Mokhtar, M.M.; Hassan, N.; Lee, C.M.; Bao, G.; Mahfouz, M.M. Efficient CRISPR/Cas9-Mediated Genome Editing Using a Chimeric Single-Guide RNA Molecule. Front. Plant Sci. 2017, 8, 1441. [Google Scholar] [CrossRef] [Green Version]
- Butt, H.; Rao, G.S.; Sedeek, K.; Aman, R.; Kamel, R.; Mahfouz, M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020, 18, 2370. [Google Scholar] [CrossRef]
- Butt, H.; Jamil, M.; Wang, J.Y.; Al-Babili, S.; Mahfouz, M. Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 2018, 18, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, H.; Zaidi, S.S.; Hassan, N.; Mahfouz, M. CRISPR-Based Directed Evolution for Crop Improvement. Trends Biotechnol. 2020, 38, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Rao, G.S.; Jiang, W.; Mahfouz, M. Synthetic directed evolution in plants: Unlocking trait engineering and improvement. Synth. Biol. 2021, 6, ysab025. [Google Scholar] [CrossRef]
- Horan, K.; Jang, C.; Bailey-Serres, J.; Mittler, R.; Shelton, C.; Harper, J.F.; Zhu, J.K.; Cushman, J.C.; Gollery, M.; Girke, T. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 2008, 147, 41–57. [Google Scholar] [CrossRef] [Green Version]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010, 24, 1073–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, G.S.; Palusa, S.G.; Golovkin, M.; Prasad, J.; Manley, J.L.; Reddy, A.S.N. Regulation of Plant Developmental Processes by a Novel Splicing Factor. PLoS ONE 2007, 2, e471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.N.; Mount, S.M. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009, 150, 1450–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, D.; Prasad, A.M.; Srinivasan, R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 2007, 45, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Manna, S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie 2015, 113, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrillo, E.; Godoy Herz, M.A.; Fuchs, A.; Reifer, D.; Fuller, J.; Yanovsky, M.J.; Simpson, C.; Brown, J.W.; Barta, A.; Kalyna, M.; et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science 2014, 344, 427–430. [Google Scholar] [CrossRef] [Green Version]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Ding, F.; Cui, P.; Wang, Z.Y.; Zhang, S.D.; Ali, S.; Xiong, L.M. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, N.; Yoshimura, K.; Kimura, A.; Yabuta, Y.; Shigeoka, S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 2007, 48, 1036–1049. [Google Scholar] [CrossRef] [Green Version]
- Kan, C.C.; Chung, T.Y.; Juo, Y.A.; Hsieh, M.H. Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genom. 2015, 16, 731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Li, L.; Ying, Y.; Wang, J.; Shao, J.F.; Yamaji, N.; Whelan, J.; Ma, J.F.; Shou, H. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice. New Phytol. 2020, 225, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ye, J.; Ma, Y.; Wang, T.; Shou, H.; Zheng, L. OsIRO3 Plays an Essential Role in Iron Deficiency Responses and Regulates Iron Homeostasis in Rice. Plants 2020, 9, 1095. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Itai, R.N.; Kobayashi, T.; Aung, M.S.; Nakanishi, H.; Nishizawa, N.K. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 2011, 75, 593–605. [Google Scholar] [CrossRef]
- Blanco, F.; Salinas, P.; Cecchini, N.M.; Jordana, X.; Van Hummelen, P.; Alvarez, M.E.; Holuigue, L. Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 2009, 70, 79–102. [Google Scholar] [CrossRef] [Green Version]
- Naithani, S.; Dikeman, D.; Garg, P.; Al-Bader, N.; Jaiswal, P. Beyond gene ontology (GO): Using biocuration approach to improve the gene nomenclature and functional annotation of rice S-domain kinase subfamily. PeerJ 2021, 9, e11052. [Google Scholar] [CrossRef]
- Wang, D.H.; Song, W.; Wei, S.W.; Zheng, Y.F.; Chen, Z.S.; Han, J.D.; Zhang, H.T.; Luo, J.C.; Qin, Y.M.; Xu, Z.H.; et al. Characterization of the Ubiquitin C-Terminal Hydrolase and Ubiquitin-Specific Protease Families in Rice (Oryza sativa). Front. Plant Sci. 2018, 9, 1636. [Google Scholar] [CrossRef]
- Hayama, R.; Yang, P.; Valverde, F.; Mizoguchi, T.; Furutani-Hayama, I.; Vierstra, R.D.; Coupland, G. Ubiquitin carboxyl-terminal hydrolases are required for period maintenance of the circadian clock at high temperature in Arabidopsis. Sci. Rep. 2019, 9, 17030. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Wang, X.; Zhao, Y.; Khan, N.U.; Zhao, Z.; Zhang, Y.; Wen, X.; Tang, F.; Wang, F.; Li, Z. Genetic basis and identification of candidate genes for salt tolerance in rice by GWAS. Sci. Rep. 2020, 10, 9958. [Google Scholar] [CrossRef]
- Mao, C.; Ding, J.; Zhang, B.; Xi, D.; Ming, F. OsNAC2 positively affects salt-induced cell death and binds to the OsAP37 and OsCOX11 promoters. Plant J. 2018, 94, 454–468. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Morales, S.; Gomez-Merino, F.C.; Trejo-Tellez, L.I. NAC transcription factor expression, amino acid concentration and growth of elite rice cultivars upon salt stress. Acta Physiol. Plant 2014, 36, 1927–1936. [Google Scholar] [CrossRef] [Green Version]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef]
- Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 2014, 80, 93–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.L.; Guo, Z.J.; Wang, H.H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z. Alternative Splicing of Rice WRKY62 and WRKY76 Transcription Factor Genes in Pathogen Defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Xu, J.; Meng, X.; Fang, X.; Xia, M.; Zhang, J.; Cao, S.; Fan, T. Linker histone variant HIS1-3 and WRKY1 Oppositely Regulate Salt Stress Tolerance in Arabidopsis. Plant Physiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Assuncao, A.G.; Herrero, E.; Lin, Y.F.; Huettel, B.; Talukdar, S.; Smaczniak, C.; Immink, R.G.; van Eldik, M.; Fiers, M.; Schat, H.; et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 10296–10301. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Humbert, S.; Liu, J.X.; Srivastava, R.; Rothstein, S.J.; Howell, S.H. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7247–7252. [Google Scholar] [CrossRef] [Green Version]
- Nagashima, Y.; Mishiba, K.; Suzuki, E.; Shimada, Y.; Iwata, Y.; Koizumi, N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 2011, 1, 29. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.; Gong, Z.; Rock, C.D.; Subramanian, S.; Guo, Y.; Xu, W.; Galbraith, D.; Zhu, J.K. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 2001, 1, 771–781. [Google Scholar] [CrossRef] [Green Version]
- Cui, P.; Zhang, S.; Ding, F.; Ali, S.; Xiong, L. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 2014, 15, R1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, S.; Zhang, Y.; Wang, X.; Li, D.; Li, Q.; Yue, M.; Li, Q.; Zhang, Y.E.; Xu, Y.; et al. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 2011, 23, 396–411. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Xia, Z.; Luo, Y.; Jiang, X.; Qian, B.; Xie, H.; Zhu, J.K.; Xiong, L.; Zhu, J.; Wang, Z.Y. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 2018, 46, 1777–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.F.; Miki, D.; Tang, K.; Zhou, H.R.; Zheng, Z.; Chen, W.; Ma, Z.Y.; Yang, L.; Zhang, H.; Liu, R.; et al. A Pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013, 9, e1003779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.L.; Zhang, S.W.; Huang, H.W.; Cai, T.; Li, L.; Chen, S.; He, X.J. The Splicing Factor PRP31 Is Involved in Transcriptional Gene Silencing and Stress Response in Arabidopsis. Mol. Plant 2015, 8, 1053–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.H.; Kapoor, A.; Zhu, J.; Zhu, J.K. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 2006, 18, 1736–1749. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.D.; Cho, Y.H.; Lee, B.H.; Yoo, S.D. STABILIZED1 Modulates Pre-mRNA Splicing for Thermotolerance. Plant Physiol. 2017, 173, 2370–2382. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butt, H.; Bazin, J.; Prasad, K.V.S.K.; Awad, N.; Crespi, M.; Reddy, A.S.N.; Mahfouz, M.M. The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses. Cells 2022, 11, 1796. https://doi.org/10.3390/cells11111796
Butt H, Bazin J, Prasad KVSK, Awad N, Crespi M, Reddy ASN, Mahfouz MM. The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses. Cells. 2022; 11(11):1796. https://doi.org/10.3390/cells11111796
Chicago/Turabian StyleButt, Haroon, Jeremie Bazin, Kasavajhala V. S. K. Prasad, Nourelislam Awad, Martin Crespi, Anireddy S. N. Reddy, and Magdy M. Mahfouz. 2022. "The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses" Cells 11, no. 11: 1796. https://doi.org/10.3390/cells11111796
APA StyleButt, H., Bazin, J., Prasad, K. V. S. K., Awad, N., Crespi, M., Reddy, A. S. N., & Mahfouz, M. M. (2022). The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses. Cells, 11(11), 1796. https://doi.org/10.3390/cells11111796








