Functional Deficits of 5×FAD Neural Stem Cells Are Ameliorated by Glutathione Peroxidase 4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Isolation of Neural Stem cells NSCs
2.3. Lipid Reactive Oxygen Species (ROS) Assay via Liperfluo Staining
2.4. Protein Expression by Immunoblotting
Autophagy-Lysosomal degradation of GPX4
2.5. Real-Time qRT-PCR
2.6. Cell Proliferation Assay
2.7. Differentiation of Neural Stem Cells (NSCs)
2.8. Immunofluorescence Staining
2.9. Statistical Analysis
3. Results
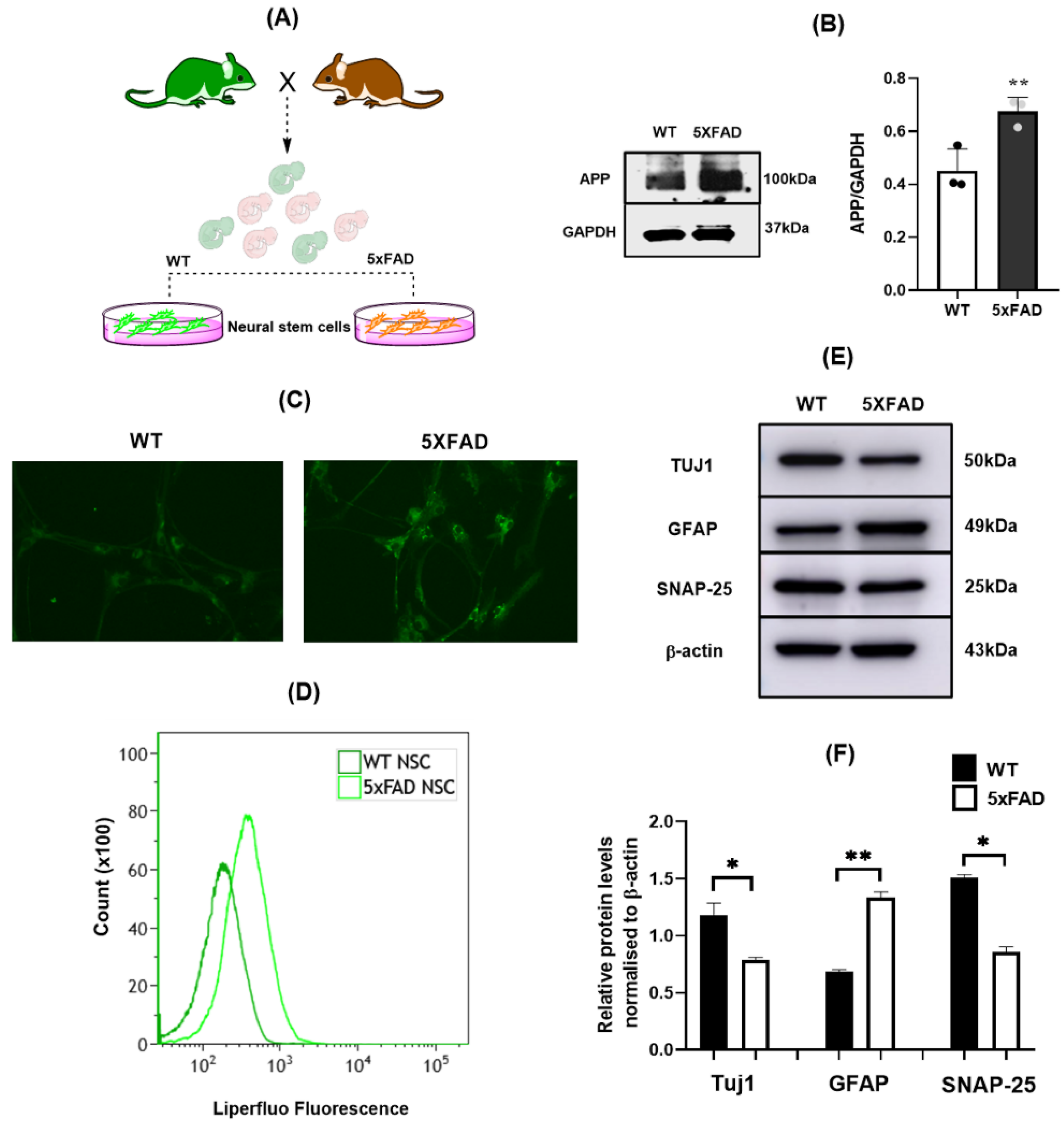
3.1. 5×FAD NSCs Exhibit Elevated Lipid ROS and Reduced Neuronal Differentiation
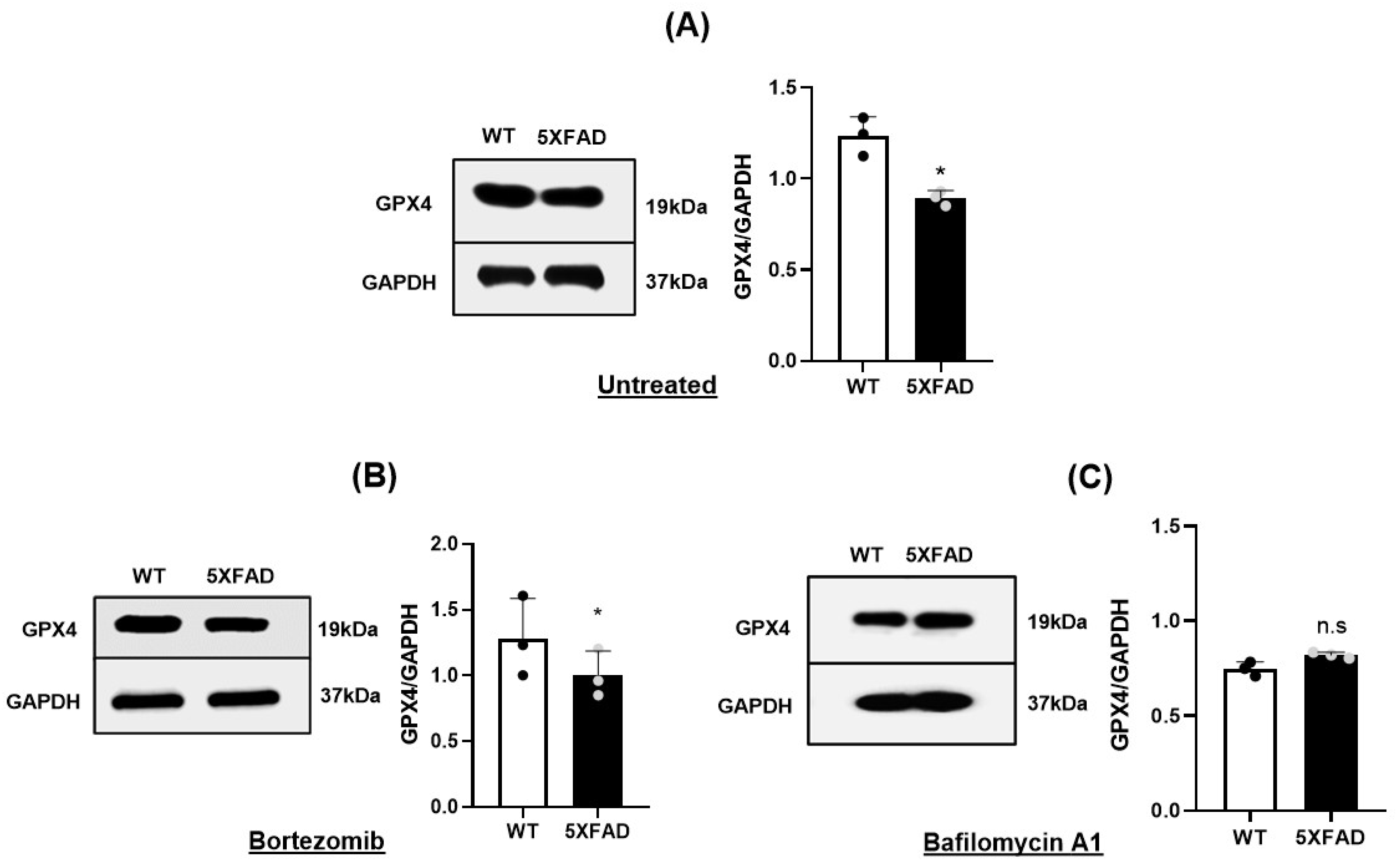
3.2. Reduced Gpx4 in 5×FAD NSCs
3.3. Autophagy-Lysosomal Degradation of GPX4 Was Responsible for Gpx4 Deficit of 5×FAD NSCs
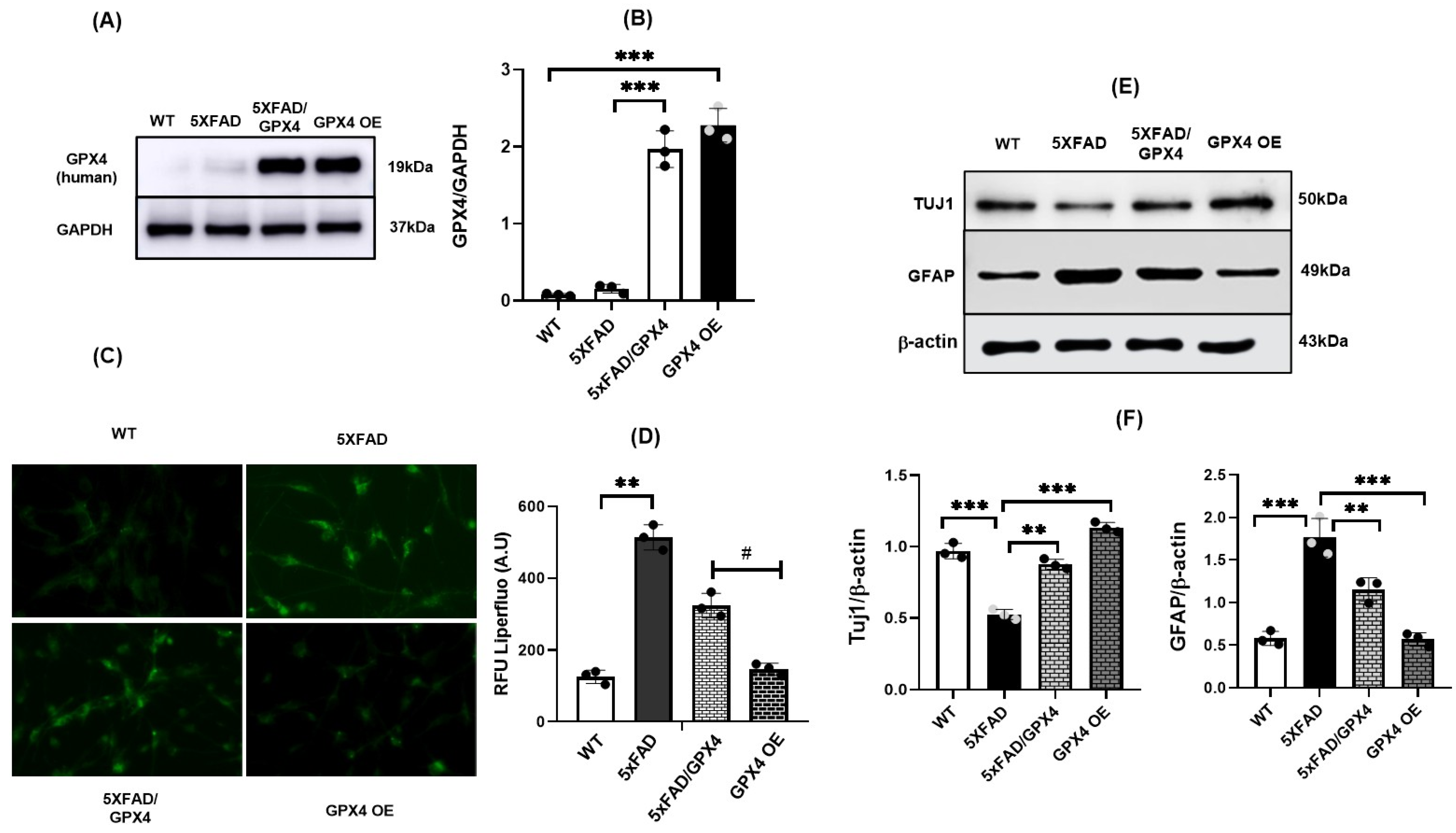
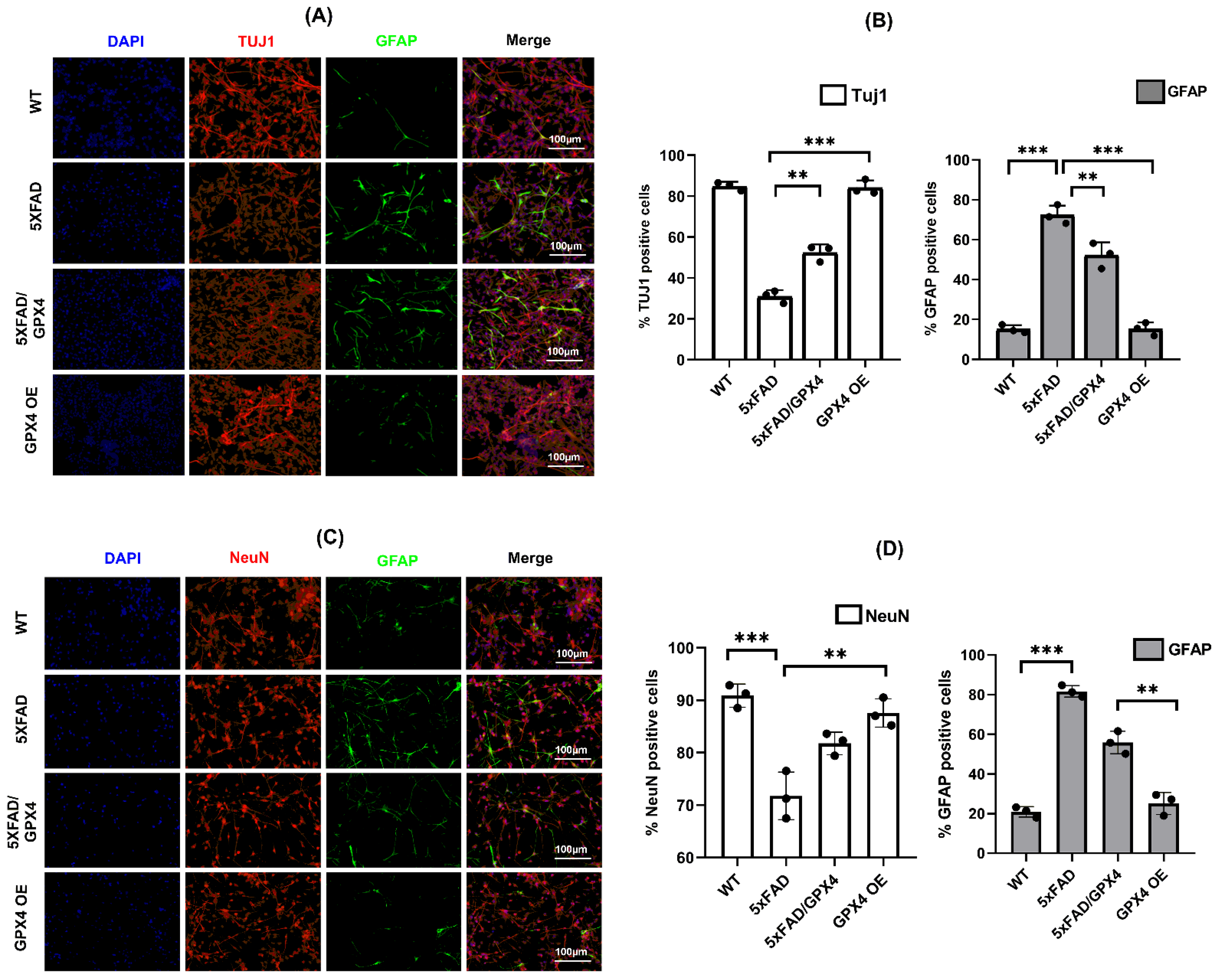
3.4. Overexpression of Gpx4 Restored Differentiation Function of 5×FAD NSCs
4. Discussion
5. Limitation of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- 2020 Alzheimer’s Association Facts and Figures. Available online: https://www.alz.org/media/documents/alzheimers-facts-and-figures.pdf (accessed on 29 December 2021).
- Boldrini, M.; Fulmore, C.A.; Tartt, A.N.; Simeon, L.R.; Pavlova, I.; Poposka, V.; Rosoklija, G.B.; Stankov, A.; Arango, V.; Dwork, A.J. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell 2018, 22, 589–599.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobin, M.K.; Musaraca, K.; Disouky, A.; Shetti, A.; Bheri, A.; Honer, W.G.; Kim, N.; Dawe, R.J.; Bennett, D.A.; Arfanakis, K. Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell 2019, 24, 974–982.e3. [Google Scholar] [CrossRef] [PubMed]
- Briley, D.; Ghirardi, V.; Woltjer, R.; Renck, A.; Zolochevska, O.; Taglialatela, G.; Micci, M.-A. Preserved neurogenesis in non-demented individuals with AD neuropathology. Sci. Rep. 2016, 6, 27812. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Crews, L.; Adame, A.; Patrick, C.; DeLaney, A.; Pham, E.; Rockenstein, E.; Hansen, L.; Masliah, E. Increased BMP6 levels in the brains of Alzheimer’s disease patients and APP transgenic mice are accompanied by impaired neurogenesis. J. Neurosci. 2010, 30, 12252–12262. [Google Scholar] [CrossRef] [PubMed]
- Houben, S.; Homa, M.; Yilmaz, Z.; Leroy, K.; Brion, J.-P.; Ando, K. Tau Pathology and Adult Hippocampal Neurogenesis: What Tau Mouse Models Tell us? Front. Neurol. 2021, 12, 610330. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Tanzi, R.E. Is Alzheimer’s disease a neurogenesis disorder? Cell Stem Cell 2019, 25, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, N.; Niidome, T.; Goto, Y.; Akaike, A.; Kihara, T.; Sugimoto, H. Decreased proliferation of hippocampal progenitor cells in APPswe/PS1dE9 transgenic mice. Neuroreport 2007, 18, 1801–1805. [Google Scholar] [CrossRef]
- Hollands, C.; Tobin, M.K.; Hsu, M.; Musaraca, K.; Yu, T.-S.; Mishra, R.; Kernie, S.G.; Lazarov, O. Depletion of adult neurogenesis exacerbates cognitive deficits in Alzheimer’s disease by compromising hippocampal inhibition. Mol. Neurodegener. 2017, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Baglietto-Vargas, D.; Sánchez-Mejias, E.; Navarro, V.; Jimenez, S.; Trujillo-Estrada, L.; Gómez-Arboledas, A.; Sánchez-Mico, M.; Sánchez-Varo, R.; Vizuete, M.; Dávila, J.C. Dual roles of Aβ in proliferative processes in an amyloidogenic model of Alzheimer’s disease. Sci. Rep. 2017, 7, 10085. [Google Scholar] [CrossRef]
- Clark, J.; Simon, D.K. Transcribe to survive: Transcriptional control of antioxidant defense programs for neuroprotection in Parkinson’s disease. Antioxid. Redox Signal. 2009, 11, 509–528. [Google Scholar] [CrossRef] [PubMed]
- E Abdel Moneim, A. Oxidant/antioxidant imbalance and the risk of Alzheimer’s disease. Curr. Alzheimer Res. 2015, 12, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Angeli, J.P.F.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [Green Version]
- Mattson, M.P. Roles of the lipid peroxidation product 4-hydroxynonenal in obesity, the metabolic syndrome, and associated vascular and neurodegenerative disorders. Exp. Gerontol. 2009, 44, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.-T.; Zou, Y.; Corniola, R. In Oxidative stress and adult neurogenesis—Effects of radiation and superoxide dismutase deficiency. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 738–744. [Google Scholar]
- Hamilton, L.K.; Dufresne, M.; Joppé, S.E.; Petryszyn, S.; Aumont, A.; Calon, F.; Barnabé-Heider, F.; Furtos, A.; Parent, M.; Chaurand, P. Aberrant lipid metabolism in the forebrain niche suppresses adult neural stem cell proliferation in an animal model of Alzheimer’s disease. Cell Stem Cell 2015, 17, 397–411. [Google Scholar] [CrossRef] [Green Version]
- Blurton-Jones, M.; Kitazawa, M.; Martinez-Coria, H.; Castello, N.A.; Müller, F.-J.; Loring, J.F.; Yamasaki, T.R.; Poon, W.W.; Green, K.N.; LaFerla, F.M. Neural stem cells improve cognition via BDNF in a transgenic model of Alzheimer disease. Proc. Natl. Acad. Sci. USA 2009, 106, 13594–13599. [Google Scholar] [CrossRef] [Green Version]
- McGinley, L.M.; Kashlan, O.N.; Bruno, E.S.; Chen, K.S.; Hayes, J.M.; Kashlan, S.R.; Raykin, J.; Johe, K.; Murphy, G.G.; Feldman, E.L. Human neural stem cell transplantation improves cognition in a murine model of Alzheimer’s disease. Sci. Rep. 2018, 8, 14776. [Google Scholar] [CrossRef]
- Espósito, M.S.; Piatti, V.C.; Laplagne, D.A.; Morgenstern, N.A.; Ferrari, C.C.; Pitossi, F.J.; Schinder, A.F. Neuronal differentiation in the adult hippocampus recapitulates embryonic development. J. Neurosci. 2005, 25, 10074–10086. [Google Scholar] [CrossRef]
- Hayashi, Y.; Lin, H.-T.; Lee, C.-C.; Tsai, K.-J. Effects of neural stem cell transplantation in Alzheimer’s disease models. J. Biomed. Sci. 2020, 27, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesler, S.R.; Acton, P.; Rao, V.; Ray, W.J. Functional and structural connectome properties in the 5XFAD transgenic mouse model of Alzheimer’s disease. Netw. Neurosci. 2018, 2, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Ran, Q.; Liang, H.; Gu, M.; Qi, W.; Walter, C.A.; Roberts, L.J.; Herman, B.; Richardson, A.; Van Remmen, H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004, 279, 55137–55146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, B.A.; Liyanage, V.R.; Zachariah, R.M.; Olson, C.O.; Bailey, M.A.; Rastegar, M. Dynamic expression of MEIS1 homeoprotein in E14. 5 forebrain and differentiated forebrain-derived neural stem cells. Ann. Anat.-Anat. Anz. 2013, 195, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, V.R.; Zachariah, R.M.; Rastegar, M. Decitabine alters the expression of Mecp2 isoforms via dynamic DNA methylation at the Mecp2 regulatory elements in neural stem cells. Mol. Autism 2013, 4, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, N.J.; Satti, N.K.; Dutt, P.; Hamid, A.; Ahmad, M. Attenuation of glutamate-induced excitotoxicity by withanolide-A in neuron-like cells: Role for PI3K/Akt/MAPK signaling pathway. Mol. Neurobiol. 2018, 55, 2725–2739. [Google Scholar] [CrossRef]
- Hambright, W.S.; Fonseca, R.S.; Chen, L.; Na, R.; Ran, Q. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol. 2017, 12, 8–17. [Google Scholar] [CrossRef]
- Lone, A.M.; Dar, N.J.; Hamid, A.; Shah, W.A.; Ahmad, M.; Bhat, B.A. Promise of retinoic acid-triazolyl derivatives in promoting differentiation of neuroblastoma cells. ACS Chem. Neurosci. 2016, 7, 82–89. [Google Scholar] [CrossRef]
- Dar, N.J.; Bhat, J.A.; Satti, N.K.; Sharma, P.R.; Hamid, A.; Ahmad, M. Withanone, an active constituent from Withania somnifera, affords protection against NMDA-induced excitotoxicity in neuron-like cells. Mol. Neurobiol. 2017, 54, 5061–5073. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Castegna, A.; Lauderback, C.M.; Drake, J. Evidence that amyloid beta-peptide-induced lipid peroxidation and its sequelae in Alzheimer’s disease brain contribute to neuronal death. Neurobiol. Aging 2002, 23, 655–664. [Google Scholar] [CrossRef]
- Cutler, R.G.; Kelly, J.; Storie, K.; Pedersen, W.A.; Tammara, A.; Hatanpaa, K.; Troncoso, J.C.; Mattson, M.P. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2004, 101, 2070–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratico, D.; Uryu, K.; Leight, S.; Trojanoswki, J.Q.; Lee, V.M.-Y. Increased lipid peroxidation precedes amyloid plaque formation in an animal model of Alzheimer amyloidosis. J. Neurosci. 2001, 21, 4183–4187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzel, S.E.; Tyurina, Y.Y.; Zhao, J.; Croix, C.M.S.; Dar, H.H.; Mao, G.; Tyurin, V.A.; Anthonymuthu, T.S.; Kapralov, A.A.; Amoscato, A.A. PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell 2017, 171, 628–641.e26. [Google Scholar] [CrossRef] [Green Version]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- Scopa, C.; Marrocco, F.; Latina, V.; Ruggeri, F.; Corvaglia, V.; La Regina, F.; Ammassari-Teule, M.; Middei, S.; Amadoro, G.; Meli, G. Impaired adult neurogenesis is an early event in Alzheimer’s disease neurodegeneration, mediated by intracellular Aβ oligomers. Cell Death Differ. 2020, 27, 934–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, K.; Saito, Y.; Sakiyama, J.; Ohuchi, Y.; Oseto, F.; Noguchi, N. A novel fluorescent probe with high sensitivity and selective detection of lipid hydroperoxides in cells. RSC Adv. 2012, 2, 7894–7900. [Google Scholar] [CrossRef]
- Blurton-Jones, M.; Spencer, B.; Michael, S.; Castello, N.A.; Agazaryan, A.A.; Davis, J.L.; Müller, F.-J.; Loring, J.F.; Masliah, E.; LaFerla, F.M. Neural stem cells genetically-modified to express neprilysin reduce pathology in Alzheimer transgenic models. Stem Cell Res. Ther. 2014, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-Q.; Cai, Q.; Shen, Y.-Y.; Wang, P.-Y.; Li, M.-H.; Teng, G.-Y. Neural stem cell transplantation improves spatial learning and memory via neuronal regeneration in amyloid-β precursor protein/presenilin 1/tau triple transgenic mice. Am. J. Alzheimer’s Dis. Other Dement. 2014, 29, 142–149. [Google Scholar] [CrossRef]
- Dráberová, E.; Del Valle, L.; Gordon, J.; Marková, V.; Šmejkalová, B.; Bertrand, L.; de Chadarévian, J.-P.; Agamanolis, D.P.; Legido, A.; Khalili, K. Class III β-tubulin is constitutively coexpressed with glial fibrillary acidic protein and nestin in midgestational human fetal astrocytes: Implications for phenotypic identity. J. Neuropathol. Exp. Neurol. 2008, 67, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Eng, L.F.; Ghirnikar, R.S.; Lee, Y.L. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000). Neurochem. Res. 2000, 25, 1439–1451. [Google Scholar] [CrossRef]
- Nazir, F.H.; Becker, B.; Brinkmalm, A.; Höglund, K.; Sandelius, Å.; Bergström, P.; Satir, T.M.; Öhrfelt, A.; Blennow, K.; Agholme, L. Expression and secretion of synaptic proteins during stem cell differentiation to cortical neurons. Neurochem. Int. 2018, 121, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.A.; Lahiri, D.K. Neuronal Differentiation Is Accompanied by Increased Levels of SNAP-25 Protein in Fetal Rat Primary Cortical Neurons: Implications in Neuronal Plasticity and Alzheimer’s Disease. Ann. N. Y. Acad. Sci. 2006, 1086, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Demars, M.; Hu, Y.S.; Gadadhar, A.; Lazarov, O. Impaired neurogenesis is an early event in the etiology of familial Alzheimer’s disease in transgenic mice. J. Neurosci. Res. 2010, 88, 2103–2117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña-Bautista, C.; Baquero, M.; Vento, M.; Cháfer-Pericás, C. Free radicals in Alzheimer’s disease: Lipid peroxidation biomarkers. Clin. Chim. Acta 2019, 491, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Clémot, M.; Sênos Demarco, R.; Jones, D.L. Lipid mediated regulation of adult stem cell behavior. Front. Cell Dev. Biol. 2020, 8, 115. [Google Scholar] [CrossRef] [Green Version]
- Albadri, S.; Naso, F.; Thauvin, M.; Gauron, C.; Parolin, C.; Duroure, K.; Vougny, J.; Fiori, J.; Boga, C.; Vriz, S. Redox signaling via lipid peroxidation regulates retinal progenitor cell differentiation. Dev. Cell 2019, 50, 73–89.e6. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hambright, W.S.; Na, R.; Ran, Q. Ablation of the ferroptosis inhibitor glutathione peroxidase 4 in neurons results in rapid motor neuron degeneration and paralysis. J. Biol. Chem. 2015, 290, 28097–28106. [Google Scholar] [CrossRef] [Green Version]
- Raciti, M.; Ceccatelli, S. Redox unbalance modifies neurogenic potential. Oncotarget 2017, 8, 3762. [Google Scholar] [CrossRef] [Green Version]
- Peng, G.; Tang, Z.; Xiang, Y.; Chen, W. Glutathione peroxidase 4 maintains a stemness phenotype, oxidative homeostasis and regulates biological processes in Panc-1 cancer stem-like cells. Oncol. Rep. 2019, 41, 1264–1274. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Geng, Y.; Lu, X.; Shi, Y.; Wu, G.; Zhang, M.; Shan, B.; Pan, H.; Yuan, J. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. USA 2019, 116, 2996–3005. [Google Scholar] [CrossRef] [Green Version]
- Orr, M.E.; Oddo, S. Autophagic/lysosomal dysfunction in Alzheimer’s disease. Alzheimer’s Res. Ther. 2013, 5, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nixon, R.A. Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer’s disease: Inseparable partners in a multifactorial disease. FASEB J. 2017, 31, 2729–2743. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dar, N.J.; Na, R.; Ran, Q. Functional Deficits of 5×FAD Neural Stem Cells Are Ameliorated by Glutathione Peroxidase 4. Cells 2022, 11, 1770. https://doi.org/10.3390/cells11111770
Dar NJ, Na R, Ran Q. Functional Deficits of 5×FAD Neural Stem Cells Are Ameliorated by Glutathione Peroxidase 4. Cells. 2022; 11(11):1770. https://doi.org/10.3390/cells11111770
Chicago/Turabian StyleDar, Nawab John, Ren Na, and Qitao Ran. 2022. "Functional Deficits of 5×FAD Neural Stem Cells Are Ameliorated by Glutathione Peroxidase 4" Cells 11, no. 11: 1770. https://doi.org/10.3390/cells11111770
APA StyleDar, N. J., Na, R., & Ran, Q. (2022). Functional Deficits of 5×FAD Neural Stem Cells Are Ameliorated by Glutathione Peroxidase 4. Cells, 11(11), 1770. https://doi.org/10.3390/cells11111770





