Monitoring SARS-CoV-2 Surrogate TGEV Individual Virions Structure Survival under Harsh Physicochemical Environments
Abstract
:1. New concepts
2. Introduction
3. Materials and Methods
3.1. Cell Culture and Virus Purification
3.1.1. Cells and Viruses
3.1.2. Purification of TGEV Virions
3.2. Electron Microscopy
3.3. AFM
3.4. Detergent and Ethanol Time Courses
4. Results
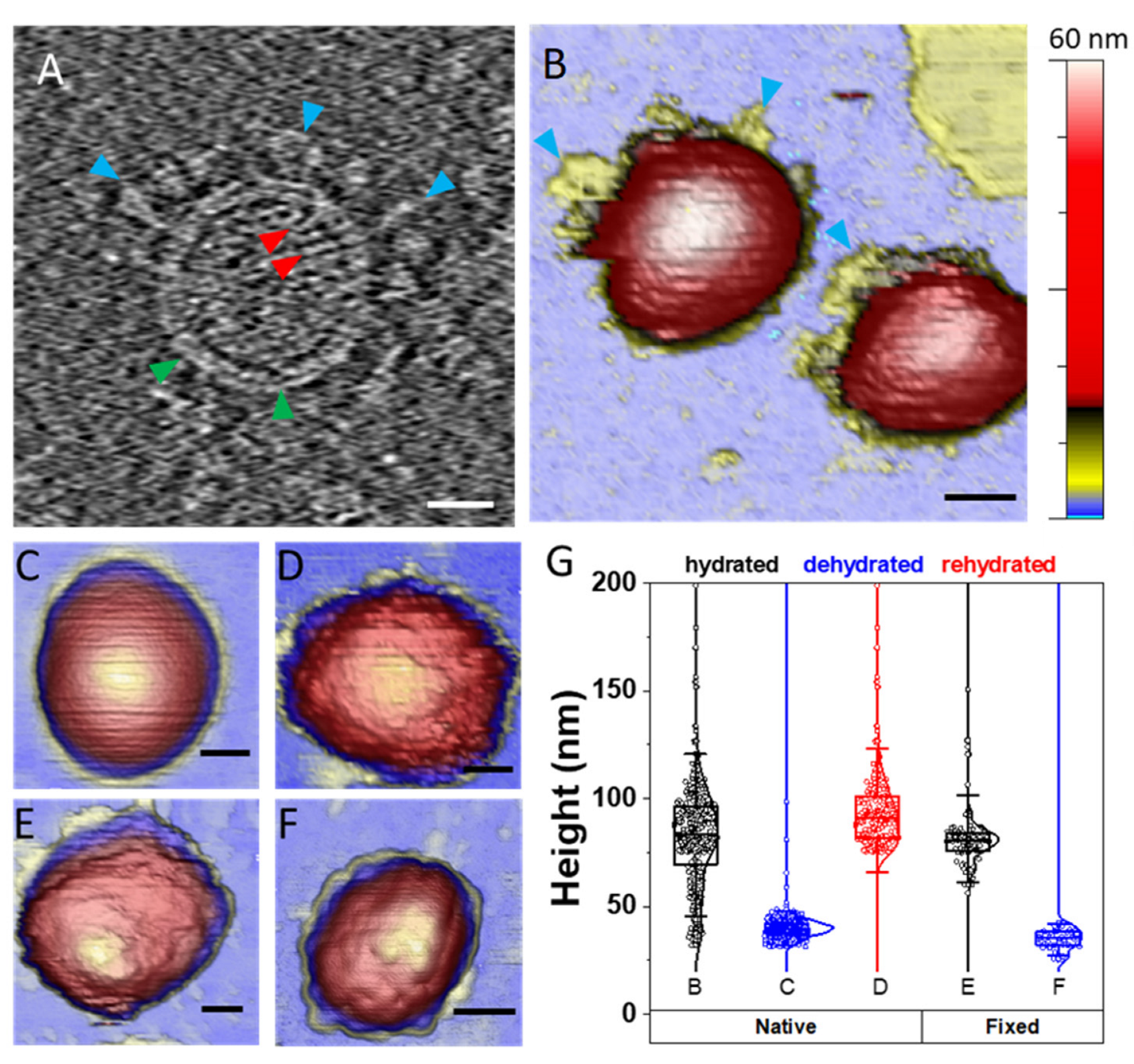
4.1. TGEV Structure Exhibits Different Topographies under Diverse Environments
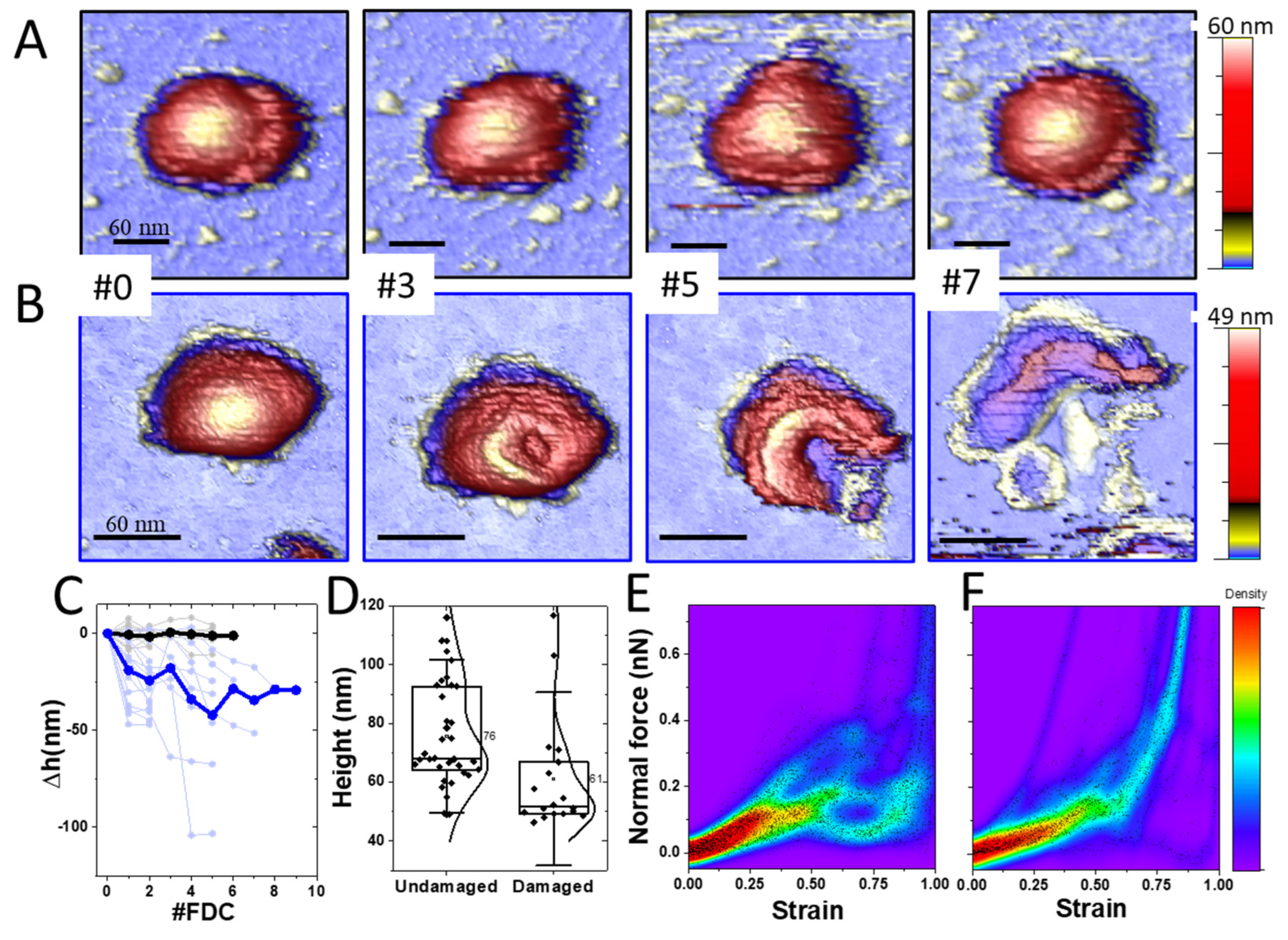
4.2. TGEV Virions Exhibit Two Stability Behaviors under Mechanical Deformation
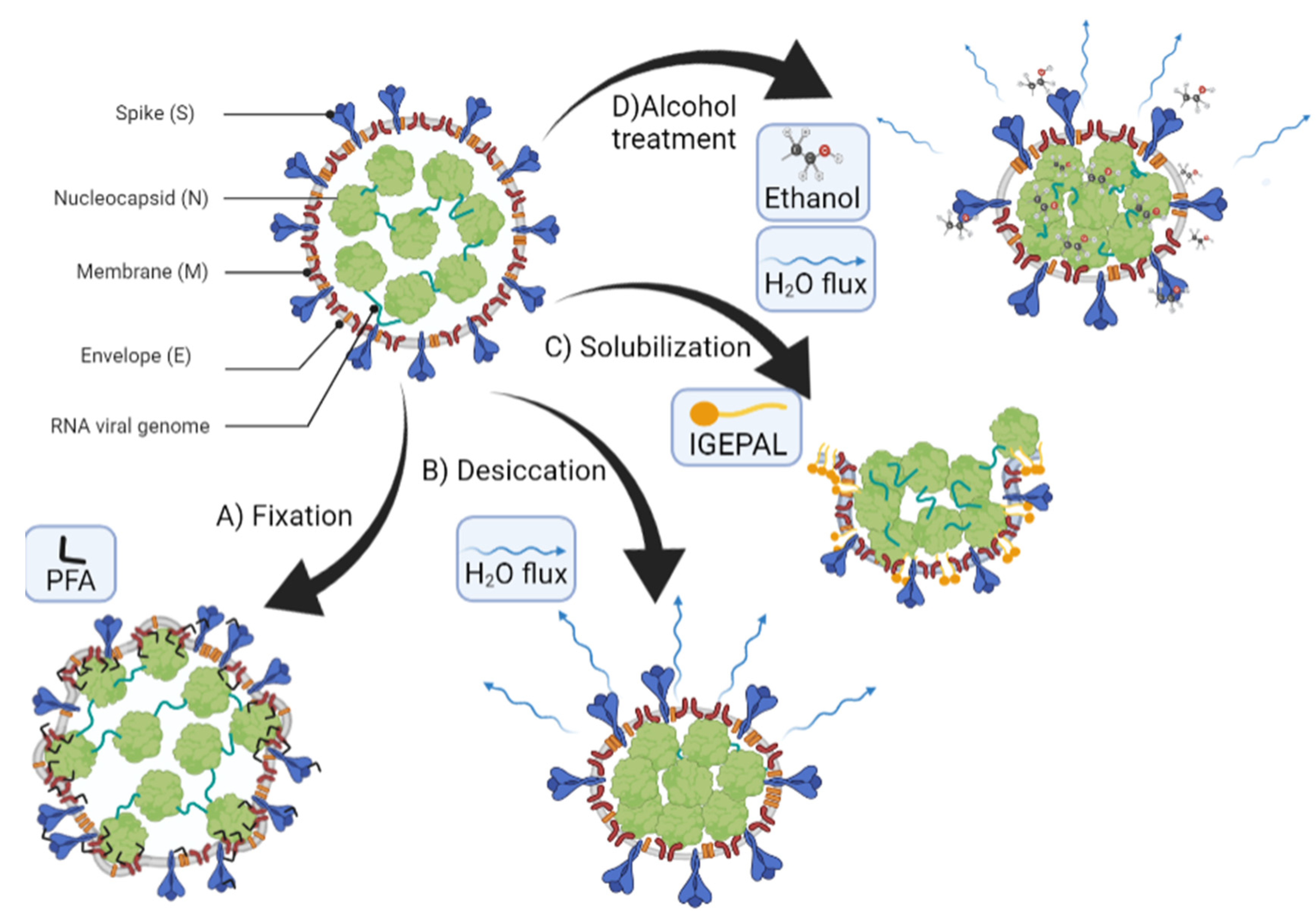
4.3. TGEV Structure Is Differently Affected by Detergents and Ethanol
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flint, S.J.; Racaniello, V.R.; Rall, G.F.; Skalka, A.M.; Enquist, L.W. Principles of Virology; ASM Press: Washington, DC, USA, 2015. [Google Scholar]
- Mateu, M.G. Introduction: The Structural Basis of Virus Function. Struct. Phys. Viruses 2013, 68, 3–51. [Google Scholar]
- Crowe, J.E. Human Respiratory Viruses. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2008; pp. 551–558. ISBN 978-0-12-374410-4. [Google Scholar]
- Gralton, J.; Tovey, E.; McLaws, M.-L.; Rawlinson, W.D. The Role of Particle Size in Aerosolised Pathogen Transmission: A Review. J. Infect. 2011, 62, 1–13. [Google Scholar] [CrossRef]
- Poon, W.C.K.; Brown, A.T.; Direito, S.O.L.; Hodgson, D.J.M.; Nagard, L.L.; Lips, A.; MacPhee, C.E.; Marenduzzo, D.; Royer, J.R.; Silva, A.F.; et al. Soft Matter Science and the COVID-19 Pandemic. Soft Matter 2020, 16, 8310–8324. [Google Scholar] [CrossRef]
- Xie, X.; Li, Y.; Sun, H.; Liu, L. Exhaled Droplets Due to Talking and Coughing. J. R. Soc. Interface 2009, 6, S703–S714. [Google Scholar] [CrossRef] [Green Version]
- Winther, B.; McCue, K.; Ashe, K.; Rubino, J.R.; Hendley, J.O. Environmental Contamination with Rhinovirus and Transfer to Fingers of Healthy Individuals by Daily Life Activity. J. Med. Virol. 2007, 79, 1606–1610. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of Coronaviruses on Inanimate Surfaces and Their Inactivation with Biocidal Agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-Species Transmission of the Newly Identified Coronavirus 2019-NCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef]
- Rewar, S.; Mirdha, D. Transmission of Ebola Virus Disease: An Overview. Ann. Glob. Health 2015, 80, 444–451. [Google Scholar] [CrossRef]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and Influenza Pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Lin, K.; Liu, M.; Ma, H.; Pan, S.; Qiao, H.; Gao, H. Laboratory Biosafety Emergency Management for SARS-CoV-2. J. Biosaf. Biosecurity 2020, 2, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.; Luque, A.; Hernando-Pérez, M.; Miranda, R.; Carrascosa, J.L.; Serena, P.A.; de Ridder, M.; Raman, A.; Gómez-Herrero, J.; Schaap, I.A.T.; et al. Built-in Mechanical Stress in Viral Shells. Biophys. J. 2011, 100, 1100–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bain, W.; Lee, J.S.; Watson, A.M.; Stitt-Fischer, M.S. Practical Guidelines for Collection, Manipulation and Inactivation of SARS-CoV-2 and COVID-19 Clinical Specimens. Curr. Protoc. Cytom. 2020, 93, e77. [Google Scholar] [CrossRef] [PubMed]
- Kiss, B.; Kis, Z.; Pályi, B.; Kellermayer, M.S.Z. Topography, Spike Dynamics, and Nanomechanics of Individual Native SARS-CoV-2 Virions. Nano Lett. 2021, 21, 2675–2680. [Google Scholar] [CrossRef] [PubMed]
- Lyonnais, S.; Hénaut, M.; Neyret, A.; Merida, P.; Cazevieille, C.; Gros, N.; Chable-Bessia, C.; Muriaux, D. Atomic Force Microscopy Analysis of Native Infectious and Inactivated SARS-CoV-2 Virions. Sci. Rep. 2021, 11, 11885. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Preece, B.; Swann, H.; Fan, X.; McKenney, R.J.; Ori-McKenney, K.M.; Saffarian, S.; Vershinin, M.D. Structural Stability of SARS-CoV-2 Virus like Particles Degrades with Temperature. Biochem. Biophys. Res. Commun. 2021, 534, 343–346. [Google Scholar] [CrossRef]
- Risco, C.; Antón, I.M.; Enjuanes, L.; Carrascosa, J.L. The Transmissible Gastroenteritis Coronavirus Contains a Spherical Core Shell Consisting of M and N Proteins. J. Virol. 1996, 70, 4773–4777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of Surrogate Coronaviruses in Water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef]
- Neuman, B.W.; Buchmeier, M.J. Supramolecular Architecture of the Coronavirus Particle. Adv. Virus Res. 2016, 96, 1–27. [Google Scholar] [CrossRef]
- Casanova, L.M.; Jeon, S.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Effects of Air Temperature and Relative Humidity on Coronavirus Survival on Surfaces. Appl. Environ. Microbiol. 2010, 76, 2712–2717. [Google Scholar] [CrossRef] [Green Version]
- Hatanaka, N.; Yasugi, M.; Sato, T.; Mukamoto, M.; Yamasaki, S. Hypochlorous Acid Solution Is a Potent Antiviral Agent against SARS-CoV-2. J. Appl. Microbiol. 2022, 132, 1496–1502. [Google Scholar] [CrossRef] [PubMed]
- Tatzber, F.; Wonisch, W.; Balka, G.; Marosi, A.; Rusvai, M.; Resch, U.; Lindschinger, M.; Moerkl, S.; Cvirn, G. Coating with Hypertonic Saline Improves Virus Protection of Filtering Facepiece Manyfold—Benefit of Salt Impregnation in Times of Pandemic. Int. J. Environ. Res. Public Health 2021, 18, 7406. [Google Scholar] [CrossRef] [PubMed]
- Shah, Y.; Kurelek, J.W.; Peterson, S.D.; Yarusevych, S. Experimental Investigation of Indoor Aerosol Dispersion and Accumulation in the Context of COVID-19: Effects of Masks and Ventilation. Phys. Fluids 2021, 33, 073315. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kolloli, A.; Subbian, S. Inactivation and Elimination of SARS-CoV-2 in Biosamples Using Simple Fixatives and Ultrafiltration. Methods Protoc. 2021, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- de Pablo, P.J. Atomic Force Microscopy of Virus Shells. Semin. Cell Dev. Biol. 2018, 73, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Zaragoza, M.; Yubero, M.P.; Martín-Forero, E.; Castón, J.R.; Reguera, D.; Luque, D.; de Pablo, P.J.; Rodríguez, J.M. Biophysical Properties of Single Rotavirus Particles Account for the Functions of Protein Shells in a Multilayered Virus. eLife 2018, 7, e37295. [Google Scholar] [CrossRef]
- Hernando-Pérez, M.; Cartagena-Rivera, A.X.; Božič, A.L.; Carrillo, P.J.P.; Martín, C.S.; Mateu, M.G.; Raman, A.; Podgornik, R.; Pablo, P.J. de Quantitative Nanoscale Electrostatics of Viruses. Nanoscale 2015, 7, 17289–17298. [Google Scholar] [CrossRef] [Green Version]
- Ivanovska, I.L.; de Pablo, P.J.; Ibarra, B.; Sgalari, G.; MacKintosh, F.C.; Carrascosa, J.L.; Schmidt, C.F.; Wuite, G.J.L. Bacteriophage Capsids: Tough Nanoshells with Complex Elastic Properties. Proc. Natl. Acad. Sci. USA 2004, 101, 7600–7605. [Google Scholar] [CrossRef] [Green Version]
- Hernando-Pérez, M.; Lambert, S.; Nakatani-Webster, E.; Catalano, C.E.; de Pablo, P.J. Cementing Proteins Provide Extra Mechanical Stabilization to Viral Cages. Nat. Commun. 2014, 5, 4520. [Google Scholar] [CrossRef] [Green Version]
- Llauró, A.; Luque, D.; Edwards, E.; Trus, B.L.; Avera, J.; Reguera, D.; Douglas, T.; de Pablo, P.J.; Castón, J.R. Cargo–Shell and Cargo–Cargo Couplings Govern the Mechanics of Artificially Loaded Virus-Derived Cages. Nanoscale 2016, 8, 9328–9336. [Google Scholar] [CrossRef]
- Kellermayer, M.S.Z.; Vörös, Z.; Csík, G.; Herényi, L. Forced Phage Uncorking: Viral DNA Ejection Triggered by a Mechanically Sensitive Switch. Nanoscale 2018, 10, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.; Castellanos, M.; de Pablo, P.J.; Mateu, M.G. Manipulation of the Mechanical Properties of a Virus by Protein Engineering. Proc. Natl. Acad. Sci. USA 2008, 105, 4150–4155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Eghiaian, F.; Sieben, C.; Herrmann, A.; Schaap, I.A.T. Bending and Puncturing the Influenza Lipid Envelope. Biophys. J. 2011, 100, 637–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín-González, N.; Hernando-Pérez, M.; Condezo, G.N.; Pérez-Illana, M.; Šiber, A.; Reguera, D.; Ostapchuk, P.; Hearing, P.; San Martín, C.; de Pablo, P.J. Adenovirus Major Core Protein Condenses DNA in Clusters and Bundles, Modulating Genome Release and Capsid Internal Pressure. Nucleic Acids Res. 2019, 47, 9231–9242. [Google Scholar] [CrossRef] [Green Version]
- Mertens, J.; Casado, S.; Mata, C.P.; Hernando-Pérez, M.; de Pablo, P.J.; Carrascosa, J.L.; Castón, J.R. A Protein with Simultaneous Capsid Scaffolding and DsRNA-Binding Activities Enhances the Birnavirus Capsid Mechanical Stability. Sci. Rep. 2015, 5, 13486. [Google Scholar] [CrossRef] [Green Version]
- Ortega-Esteban, A.; Pérez-Berná, A.J.; Menéndez-Conejero, R.; Flint, S.J.; Martín, C.S.; de Pablo, P.J. Monitoring Dynamics of Human Adenovirus Disassembly Induced by Mechanical Fatigue. Sci. Rep. 2013, 3, 1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, H.-B.; Hevroni, L.; Kol, N.; Eckert, D.M.; Tsvitov, M.; Kay, M.S.; Rousso, I. Virion Stiffness Regulates Immature HIV-1 Entry. Retrovirology 2013, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- van Rosmalen, M.G.M.; Nemerow, G.R.; Wuite, G.J.L.; Roos, W.H. A Single Point Mutation in Precursor Protein VI Doubles the Mechanical Strength of Human Adenovirus. J. Biol. Phys. 2018, 44, 119–132. [Google Scholar] [CrossRef] [Green Version]
- Martín-González, N.; Vieira Gonçalves, L.; Condezo, G.N.; San Martín, C.; Rubiano, M.; Fallis, I.; Rubino, J.R.; Ijaz, M.K.; Maillard, J.-Y.; De Pablo, P.J. Virucidal Action Mechanism of Alcohol and Divalent Cations Against Human Adenovirus. Front. Mol. Biosci. 2020, 7, 426. [Google Scholar] [CrossRef]
- McClurkin, A.W.; Norman, J.O. Studies on Transmissible Gastroenteritis of Swine. II. Selected Characteristics of a Cytopathogenic Virus Common to Five Isolates from Transmissible Gastroenteritis. Can. J. Comp. Med. Vet. Sci. 1966, 30, 190–198. [Google Scholar]
- Sánchez, C.M.; Jiménez, G.; Laviada, M.D.; Correa, I.; Suñé, C.; Bullido, M.J.; Gebauer, F.; Smerdou, C.; Callebaut, P.; Escribano, J.; et al. Antigenic Homology among Coronaviruses Related to Transmissible Gastroenteritis Virus. Virology 1990, 174, 410–417. [Google Scholar] [CrossRef]
- Jiménez, G.; Correa, I.; Melgosa, M.P.; Bullido, M.J.; Enjuanes, L. Critical Epitopes in Transmissible Gastroenteritis Virus Neutralization. J. Virol. 1986, 60, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turoňová, B.; Schur, F.K.M.; Wan, W.; Briggs, J.A.G. Efficient 3D-CTF Correction for Cryo-Electron Tomography Using NovaCTF Improves Subtomogram Averaging Resolution to 3.4Å. J. Struct. Biol. 2017, 199, 187–195. [Google Scholar] [CrossRef]
- Sader, J.E.; Chon, J.W.M.; Mulvaney, P. Calibration of Rectangular Atomic Force Microscope Cantilevers. Rev. Sci. Instrum. 1999, 70, 3967–3969. [Google Scholar] [CrossRef] [Green Version]
- Ortega-Esteban, A.; Horcas, I.; Hernando-Pérez, M.; Ares, P.; Pérez-Berná, A.J.; San Martín, C.; Carrascosa, J.L.; de Pablo, P.J.; Gómez-Herrero, J. Minimizing Tip–Sample Forces in Jumping Mode Atomic Force Microscopy in Liquid. Ultramicroscopy 2012, 114, 56–61. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]
- Castellanos-Gonzalez, A.; Shelite, T.R.; Lloyd, N.; Sadiqova, A.; Ping, R.; Williams-Bouyer, N.; Melby, P.C.; Travi, B.L. Direct RT-PCR Amplification of SARS-CoV-2 from Clinical Samples Using a Concentrated Viral Lysis-Amplification Buffer Prepared with IGEPAL-630. Sci. Rep. 2021, 11, 14204. [Google Scholar] [CrossRef]
- Lin, S.; Lee, C.-K.; Lee, S.-Y.; Kao, C.-L.; Lin, C.-W.; Wang, A.-B.; Hsu, S.-M.; Huang, L.-S. Surface Ultrastructure of SARS Coronavirus Revealed by Atomic Force Microscopy. Cell. Microbiol. 2005, 7, 1763–1770. [Google Scholar] [CrossRef] [Green Version]
- Villarrubia, J. Algorithms for Scanned Probe Microscope Image Simulation, Surface Reconstruction, and Tip Estimation. J. Res. Natl. Inst. Stand. Technol. 1997, 102, 425. [Google Scholar] [CrossRef]
- Zeng, C.; Hernando-Pérez, M.; Dragnea, B.; Ma, X.; van der Schoot, P.; Zandi, R. Contact Mechanics of a Small Icosahedral Virus. Phys. Rev. Lett. 2017, 119, 038102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Available online: https://www.nejm.org/doi/10.1056/NEJMoa030781 (accessed on 11 October 2021).
- Martín-González, N.; Darvas, S.M.G.; Durana, A.; Marti, G.A.; Guérin, D.M.A.; de Pablo, P.J. Exploring the Role of Genome and Structural Ions in Preventing Viral Capsid Collapse during Dehydration. J. Phys. Condens. Matter 2018, 30, 104001. [Google Scholar] [CrossRef] [Green Version]
- Denning, D.; Bennett, S.; Mullen, T.; Moyer, C.; Vorselen, D.; Wuite, G.J.L.; Nemerow, G.; Roos, W.H. Maturation of Adenovirus Primes the Protein Nano-Shell for Successful Endosomal Escape. Nanoscale 2019, 11, 4015–4024. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. Efficacy of Ethanol against Viruses in Hand Disinfection. J. Hosp. Infect. 2018, 98, 331–338. [Google Scholar] [CrossRef]
- Celik, U.; Celik, K.; Celik, S.; Abayli, H.; Sahna, K.C.; Tonbak, Ş.; Toraman, Z.A.; Oral, A. Interpretation of SARS-CoV-2 Behaviour on Different Substrates and Denaturation of Virions Using Ethanol: An Atomic Force Microscopy Study. RSC Adv. 2020, 10, 44079–44086. [Google Scholar] [CrossRef]
- Jørgensen, R.L.; Pedersen, M.S.; Chauhan, A.S.; Andreasson, L.M.; Kristiansen, G.Q.; Lisby, J.G.; Rosenstierne, M.W.; Schønning, K. An In-Well Direct Lysis Method for Rapid Detection of SARS-CoV-2 by Real Time RT-PCR in ESwab Specimens. J. Virol. Methods 2021, 289, 114062. [Google Scholar] [CrossRef]
- Srivatsan, S.; Heidl, S.; Pfau, B.; Martin, B.K.; Han, P.D.; Zhong, W.; van Raay, K.; McDermot, E.; Opsahl, J.; Gamboa, L.; et al. SwabExpress: An End-to-End Protocol for Extraction-Free COVID-19 Testing. Clin. Chem. 2021, 68, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Coronavirus Survival on Healthcare Personal Protective Equipment. Infect. Control Hosp. Epidemiol. 2010, 31, 560–561. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.H.; Peiris, J.S.M.; Lam, S.Y.; Poon, L.L.M.; Yuen, K.Y.; Seto, W.H. The Effects of Temperature and Relative Humidity on the Viability of the SARS Coronavirus. Adv. Virol. 2011, 2011, e734690. [Google Scholar] [CrossRef]
- Llauró, A.; Guerra, P.; Kant, R.; Bothner, B.; Verdaguer, N.; de Pablo, P.J. Decrease in PH Destabilizes Individual Vault Nanocages by Weakening the Inter-Protein Lateral Interaction. Sci. Rep. 2016, 6, 34143. [Google Scholar] [CrossRef] [Green Version]
- Martín-González, N.; Ibáñez-Freire, P.; Ortega-Esteban, Á.; Laguna-Castro, M.; San Martín, C.; Valbuena, A.; Delgado-Buscalioni, R.; de Pablo, P.J. Long-Range Cooperative Disassembly and Aging During Adenovirus Uncoating. Phys. Rev. X 2021, 11, 021025. [Google Scholar] [CrossRef]
- Turoňová, B.; Sikora, M.; Schürmann, C.; Hagen, W.J.H.; Welsch, S.; Blanc, F.E.C.; von Bülow, S.; Gecht, M.; Bagola, K.; Hörner, C.; et al. In Situ Structural Analysis of SARS-CoV-2 Spike Reveals Flexibility Mediated by Three Hinges. Science 2020, 370, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Chen, C.-J.; Maeda, J.; Makino, S. Nucleocapsid-Independent Specific Viral RNA Packaging via Viral Envelope Protein and Viral RNA Signal. J. Virol. 2003, 77, 2922–2927. [Google Scholar] [CrossRef] [Green Version]
- Greatorex, J.S.; Digard, P.; Curran, M.D.; Moynihan, R.; Wensley, H.; Wreghitt, T.; Varsani, H.; Garcia, F.; Enstone, J.; Nguyen-Van-Tam, J.S. Survival of Influenza A(H1N1) on Materials Found in Households: Implications for Infection Control. PLoS ONE 2011, 6, e27932. [Google Scholar] [CrossRef]
- Gui, M.; Liu, X.; Guo, D.; Zhang, Z.; Yin, C.-C.; Chen, Y.; Xiang, Y. Electron Microscopy Studies of the Coronavirus Ribonucleoprotein Complex. Protein Cell 2017, 8, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- de Haan, C.A.M.; Vennema, H.; Rottier, P.J.M. Assembly of the Coronavirus Envelope: Homotypic Interactions between the M Proteins. J. Virol. 2000, 74, 4967–4978. [Google Scholar] [CrossRef]
- Hernando-Pérez, M.; Zeng, C.; Miguel, M.C.; Dragnea, B. Intermittency of Deformation and the Elastic Limit of an Icosahedral Virus under Compression. ACS Nano 2019, 13, 7842–7849. [Google Scholar] [CrossRef]
- Llauró, A.; Guerra, P.; Irigoyen, N.; Rodríguez, J.F.; Verdaguer, N.; de Pablo, P.J. Mechanical Stability and Reversible Fracture of Vault Particles. Biophys. J. 2014, 106, 687–695. [Google Scholar] [CrossRef] [Green Version]
- de Pablo, P.J.; Hernando-Pérez, M.; Carrasco, C.; Carrascosa, J.L. Direct Visualization of Single Virus Restoration after Damage in Real Time. J. Biol. Phys. 2018, 44, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Esteban, A.; Condezo, G.N.; Pérez-Berná, A.J.; Chillón, M.; Flint, S.J.; Reguera, D.; San Martín, C.; de Pablo, P.J. Mechanics of Viral Chromatin Reveals the Pressurization of Human Adenovirus. ACS Nano 2015, 9, 10826–10833. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Boz, M.B.; Harvey, S.C. The Conformation of Double-Stranded DNA inside Bacteriophages Depends on Capsid Size and Shape. J. Struct. Biol. 2007, 160, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Berná, A.J.; Ortega-Esteban, A.; Menéndez-Conejero, R.; Winkler, D.C.; Menéndez, M.; Steven, A.C.; Flint, S.J.; de Pablo, P.J.; Martín, C.S. The Role of Capsid Maturation on Adenovirus Priming for Sequential Uncoating *. J. Biol. Chem. 2012, 287, 31582–31595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enjuanes, L.; Siddell, S.; Spaan, W. (Eds.) Coronaviruses and Arteriviruses; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 1998; ISBN 978-0-306-45910-8. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantero, M.; Carlero, D.; Chichón, F.J.; Martín-Benito, J.; De Pablo, P.J. Monitoring SARS-CoV-2 Surrogate TGEV Individual Virions Structure Survival under Harsh Physicochemical Environments. Cells 2022, 11, 1759. https://doi.org/10.3390/cells11111759
Cantero M, Carlero D, Chichón FJ, Martín-Benito J, De Pablo PJ. Monitoring SARS-CoV-2 Surrogate TGEV Individual Virions Structure Survival under Harsh Physicochemical Environments. Cells. 2022; 11(11):1759. https://doi.org/10.3390/cells11111759
Chicago/Turabian StyleCantero, Miguel, Diego Carlero, Francisco Javier Chichón, Jaime Martín-Benito, and Pedro José De Pablo. 2022. "Monitoring SARS-CoV-2 Surrogate TGEV Individual Virions Structure Survival under Harsh Physicochemical Environments" Cells 11, no. 11: 1759. https://doi.org/10.3390/cells11111759
APA StyleCantero, M., Carlero, D., Chichón, F. J., Martín-Benito, J., & De Pablo, P. J. (2022). Monitoring SARS-CoV-2 Surrogate TGEV Individual Virions Structure Survival under Harsh Physicochemical Environments. Cells, 11(11), 1759. https://doi.org/10.3390/cells11111759





