Circ0001470 Acts as a miR-140-3p Sponge to Facilitate the Progression of Embryonic Development through Regulating PTGFR Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Sources and RNA Sequencing
2.2. Identification and Verification of circRNAs
2.3. Real-Time Quantitative PCR (qRT-PCR) for Gene Expression, RNase R Treatment, and Actinomycin D Assay
2.4. Vector Construction and Small Fragment Synthesis
2.5. EECs Isolation, Culture, and Transfection
2.6. Luciferase Assay
2.7. Protein Extraction and Western Blot Analysis
2.8. RNA FISH and RNA Nuclear-Cytoplasmic Fractionation
2.9. Immunohistochemistry (IHC)
2.10. RNA Immunoprecipitation (RIP)
2.11. Biotin-Coupled Probe RNA Pull-Down Assay
2.12. CCK-8 Assay
2.13. 5-Ethynyl-20-Deoxyuridine (EdU) Incorporation Assay
2.14. Cell Cycle Assay
2.15. Cell Apoptosis Assay
2.16. Lentiviral Vector Construction
2.17. Uterine Horn Injection of Mice
2.18. Statistical Analysis
3. Results
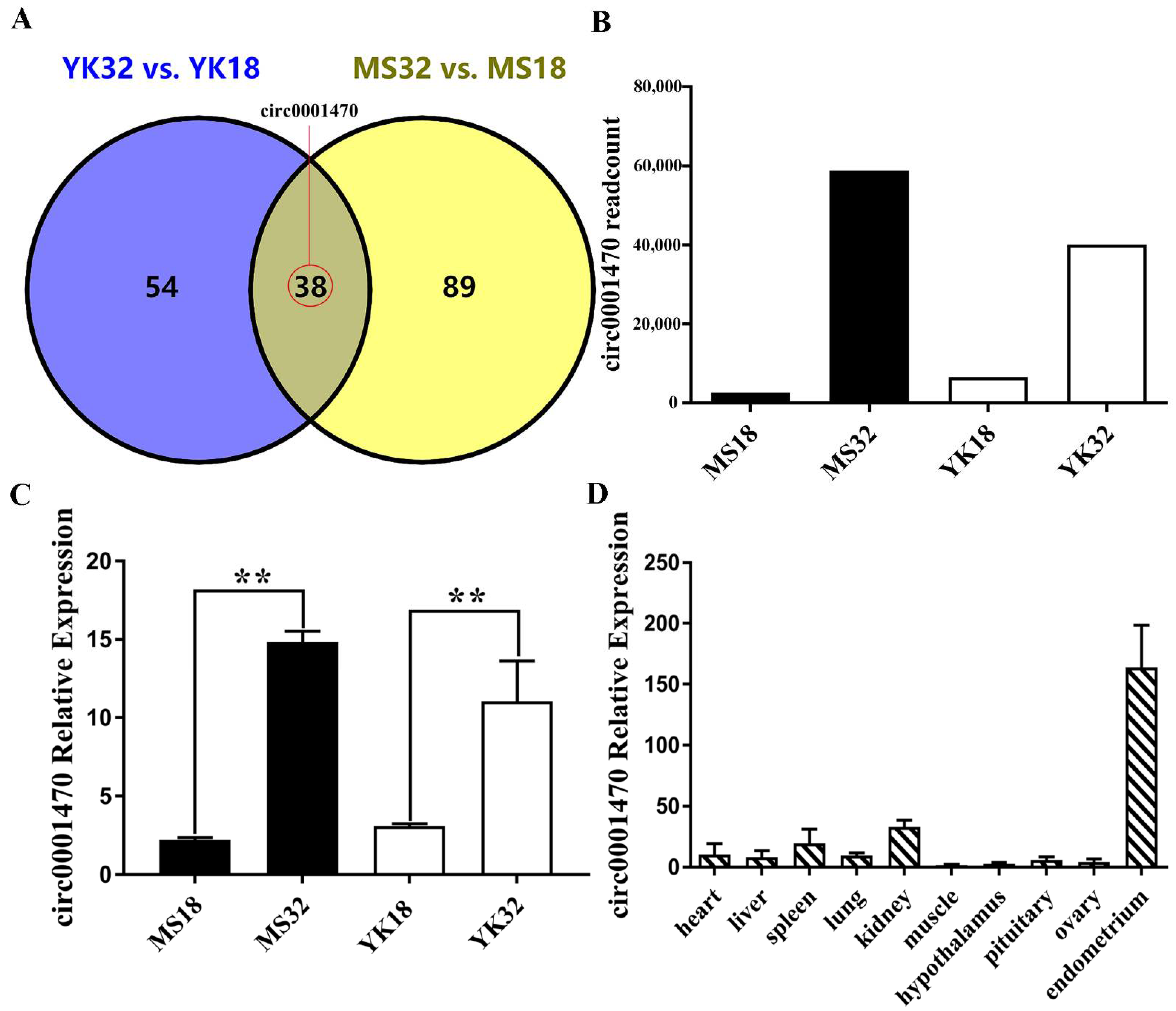
3.1. Circ0001470 Is Upregulated in Endometrial Tissue during Embryonic Development
3.2. The Characteristics of circ0001470 in EECs
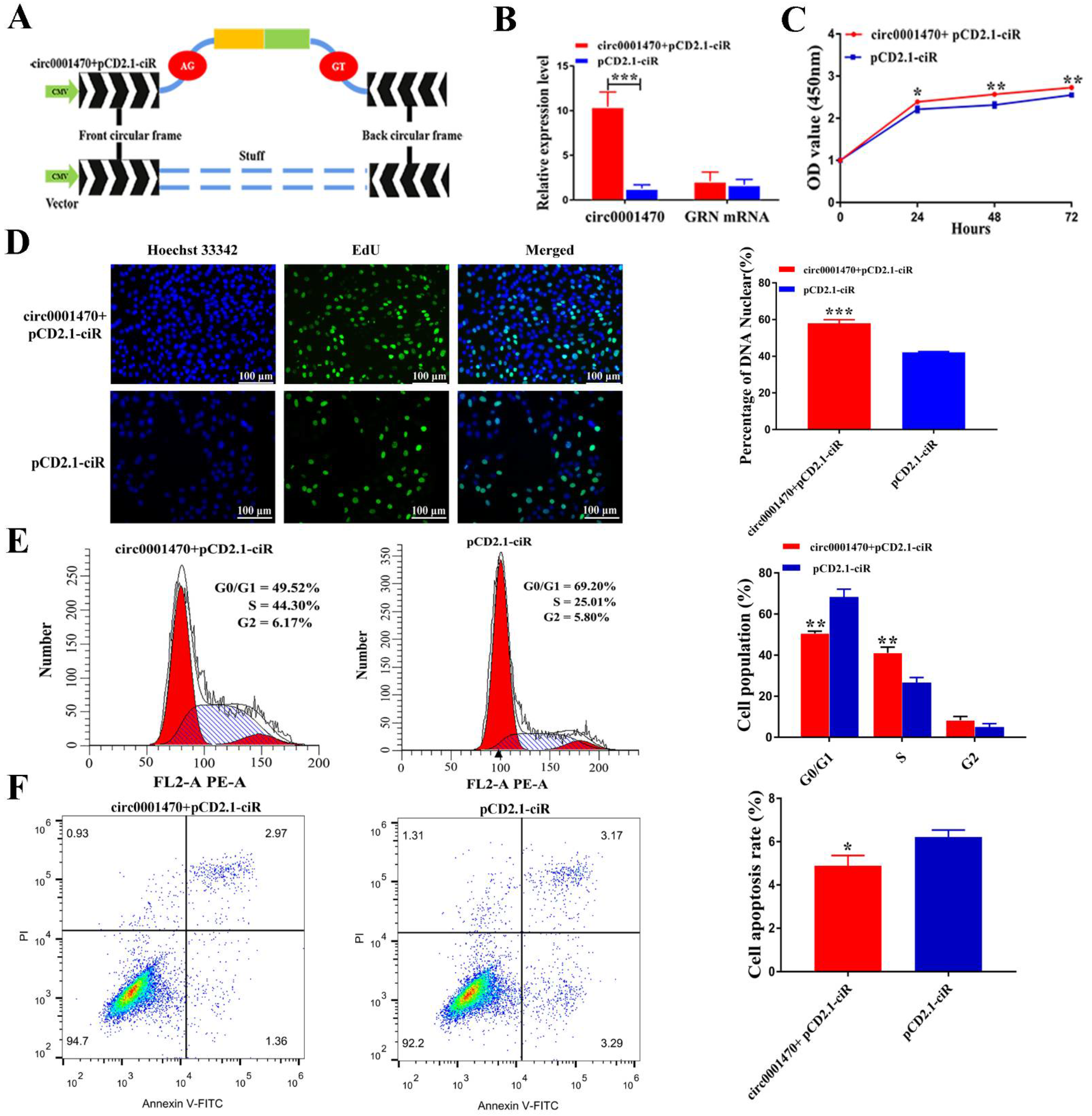
3.3. Circ0001470 Promotes EECs Proliferation, Cycle Transition, and Suppresses Apoptosis
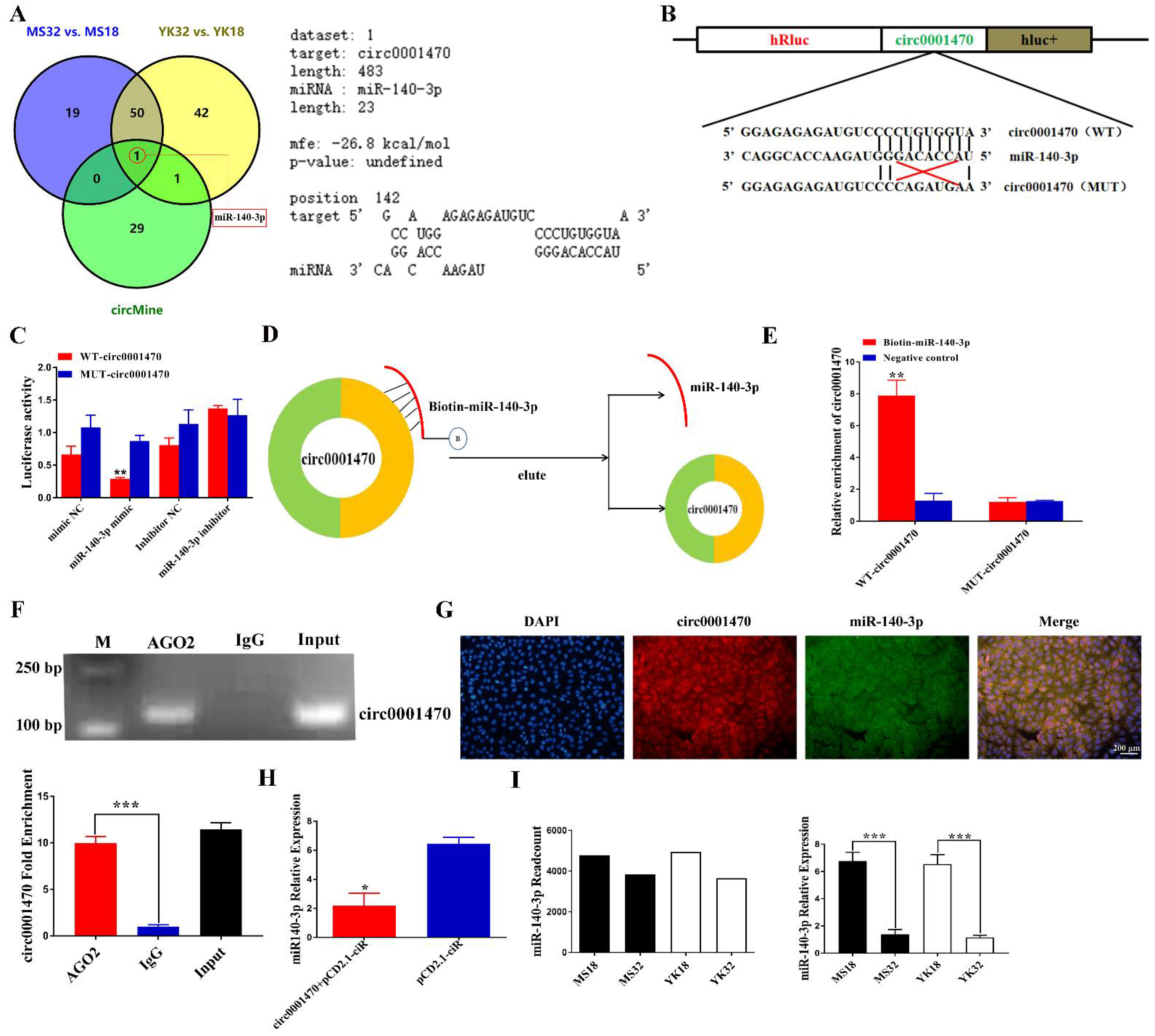
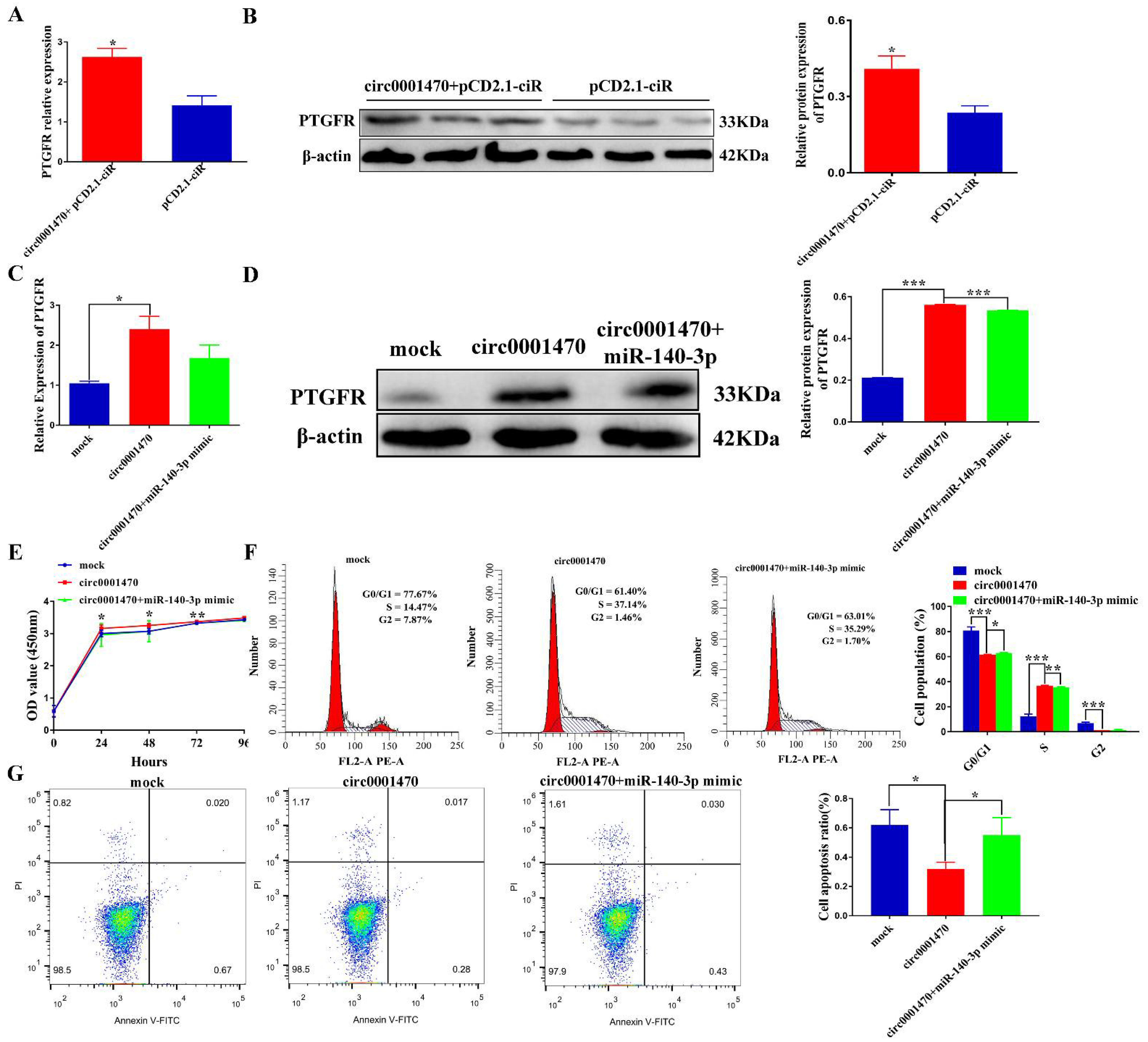
3.4. Circ0001470 Directly Targeted miR-140-3p
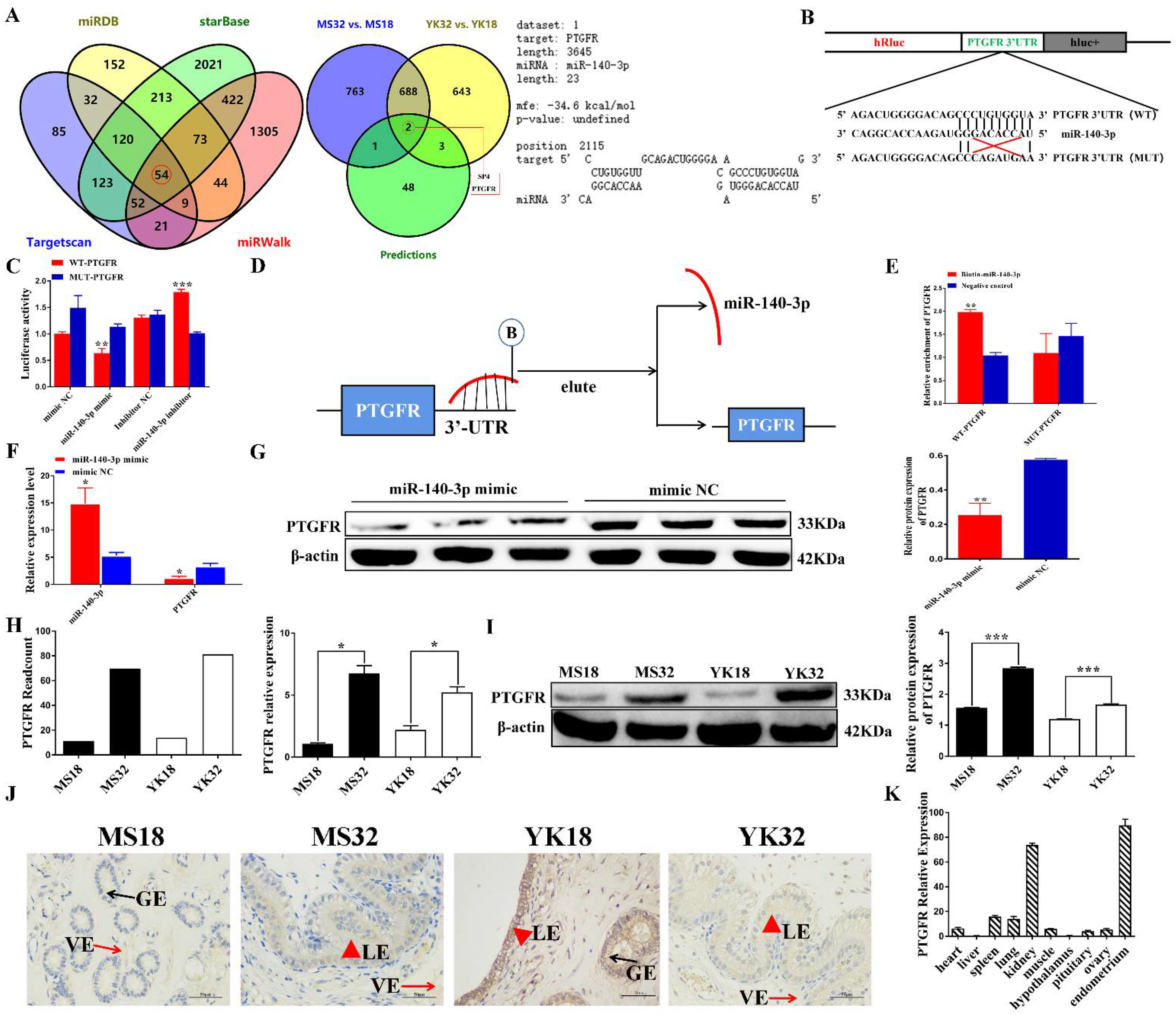
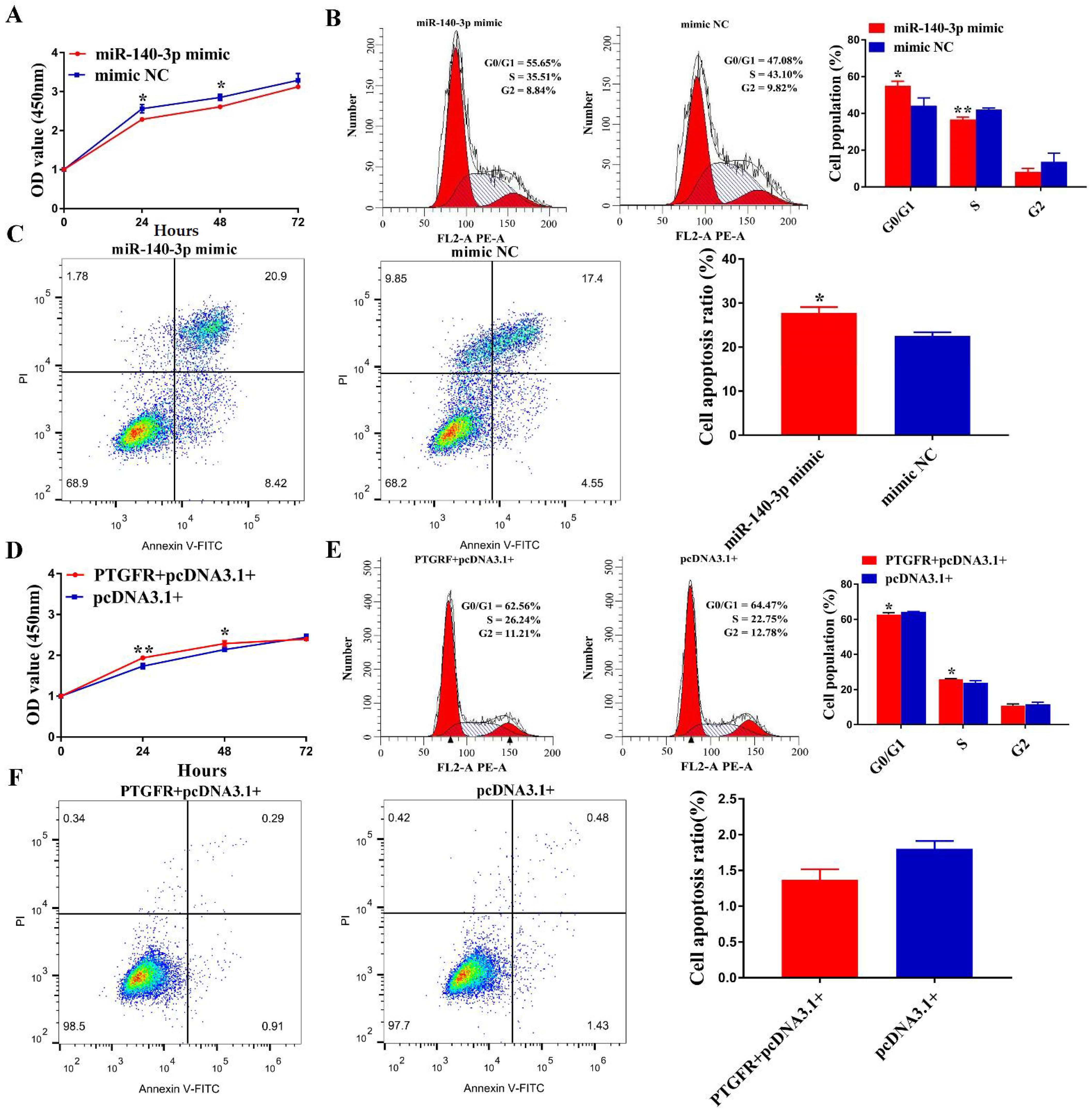
3.5. miR-140-3p Directly Targets PTGFR in EECs
3.6. MiR-140-3p Suppresses EECs Proliferation, Cycle Transition and Promotes Apoptosis by Targeting PTGFR
3.7. Circ0001470 Promotes EECs Proliferation and Suppresses EECs Apoptosis through circ0001470/miR-140-3p/PTGFR Axis
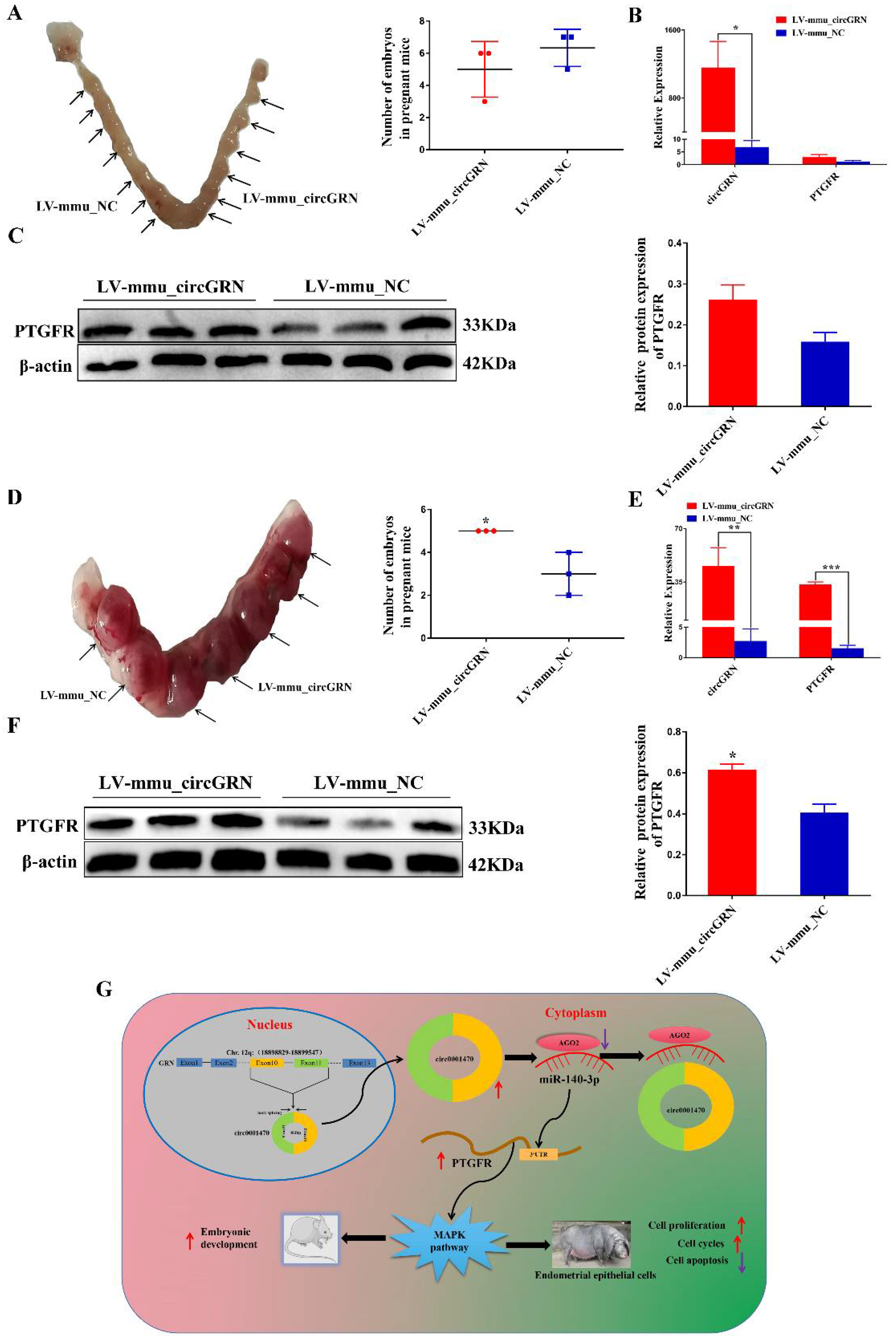
3.8. CircGRN Promotes Embryo Development In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furutani, A.; Sekiguchi, S.; Sueyoshi, M.; Sasaki, Y. Effect of intervention practices to control the porcine epidemic diarrhea (ped) outbreak during the first epidemic year (2013–2014) on time to absence of clinical signs and the number of dead piglets per sow in japan. Prev. Vet. Med. 2019, 169, 104710. [Google Scholar] [CrossRef]
- King, R.L.; Baxter, E.M.; Matheson, S.M.; Edwards, S.A. Consistency is key: Interactions of current and previous farrowing system on litter size and piglet mortality. Animal 2019, 13, 180–188. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Wang, J.; Li, Y.; Zhu, K.; Xu, Z.; Song, Y.; Song, Y.; Liu, G. Melatonin-related genes expressed in the mouse uterus during early gestation promote embryo implantation. J. Pineal Res. 2015, 58, 300–309. [Google Scholar] [CrossRef]
- Zheng, L.L.; Tan, X.W.; Cui, X.Z.; Yuan, H.J.; Li, H.; Jiao, G.Z.; Ji, C.L.; Tan, J.H. Preimplantation maternal stress impairs embryo development by inducing oviductal apoptosis with activation of the fas system. Mol. Hum. Reprod. 2016, 22, 778–790. [Google Scholar] [CrossRef] [Green Version]
- Bidarimath, M.; Tayade, C. Pregnancy and spontaneous fetal loss: A pig perspective. Mol. Reprod. Dev. 2017, 84, 856–869. [Google Scholar] [CrossRef] [Green Version]
- Wessels, J.M.; Linton, N.F.; Croy, B.A.; Tayade, C. A review of molecular contrasts between arresting and viable porcine attachment sites. Am. J. Reprod. Immunol. 2007, 58, 470–480. [Google Scholar] [CrossRef]
- Revel, A.; Achache, H.; Stevens, J.; Smith, Y.; Reich, R. Micrornas are associated with human embryo implantation defects. Hum. Reprod. 2011, 26, 2830–2840. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, H.; Kong, S.; Wang, S.; Wang, H.; Wang, H.; Armant, D.R. Physiological and molecular determinants of embryo implantation. Mol. Asp. Med. 2013, 34, 939–980. [Google Scholar] [CrossRef] [Green Version]
- Deglincerti, A.; Croft, G.F.; Pietila, L.N.; Zernicka-Goetz, M.; Siggia, E.D.; Brivanlou, A.H. Self-organization of the in vitro attached human embryo. Nature 2016, 533, 251–254. [Google Scholar] [CrossRef]
- Gu, T.; Zhu, M.J.; Schroyen, M.; Qu, L.; Nettleton, D.; Kuhar, D.; Lunney, J.K.; Ross, J.W.; Zhao, S.H.; Tuggle, C.K. Endometrial gene expression profiling in pregnant meishan and yorkshire pigs on day 12 of gestation. BMC Genom. 2014, 15, 156. [Google Scholar] [CrossRef] [Green Version]
- Kiewisz, J.; Krawczynski, K.; Lisowski, P.; Blitek, A.; Zwierzchowski, L.; Ziecik, A.J.; Kaczmarek, M.M. Global gene expression profiling of porcine endometria on days 12 and 16 of the estrous cycle and pregnancy. Theriogenology 2014, 82, 897–909. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, T.; Wu, L.; Liu, X.; Xue, S.; Lei, M. Identification of non-coding and coding rnas in porcine endometrium. Genomics 2017, 109, 43–50. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular rnas. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular rnas in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Zhang, L.; Liu, X.; Niu, M.; Cui, J.; Che, S.; Liu, Y.; An, X.; Cao, B. Analyses of circrna profiling during the development from pre-receptive to receptive phases in the goat endometrium. J. Anim. Sci. Biotechnol. 2019, 10, 34. [Google Scholar] [CrossRef]
- Dang, Y.; Yan, L.; Hu, B.; Fan, X.; Ren, Y.; Li, R.; Lian, Y.; Yan, J.; Li, Q.; Zhang, Y.; et al. Tracing the expression of circular rnas in human pre-implantation embryos. Genome Biol. 2016, 17, 130. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Ding, Y.; He, J.; Zhang, J.; Liu, X.; Chen, X.; Su, Y.; Wang, Y.; Gao, R. Altered expression patterns of circular rnas between implantation sites and interimplantation sites in early pregnant mice. J. Cell. Physiol. 2019, 234, 9862–9872. [Google Scholar] [CrossRef]
- Kartha, R.V.; Subramanian, S. Competing endogenous rnas (cernas): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular rnas are a large class of animal rnas with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Elnour, I.E.; Wang, X.G.; Zhansaya, T.; Akhatayeva, Z.; Khan, R.; Cheng, J.; Hung, Y.Z.; Lan, X.Y.; Lei, C.Z.; Chen, H. Circular rna circmyl1 inhibit proliferation and promote differentiation of myoblasts by sponging mir-2400. Cells 2021, 10, 176. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular rna profiling reveals an abundant circhipk3 that regulates cell growth by sponging multiple mirnas. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Cheng, Z.; Yu, C.; Cui, S.; Wang, H.; Jin, H.; Wang, C.; Li, B.; Qin, M.; Yang, C.; He, J.; et al. Circtp63 functions as a cerna to promote lung squamous cell carcinoma progression by upregulating foxm1. Nat. Commun. 2019, 10, 3200. [Google Scholar] [CrossRef] [Green Version]
- Su, T.; Yu, H.; Luo, G.; Wang, M.; Zhou, C.; Zhang, L.; Hou, B.; Zhang, C.; Liu, M.; Xu, D. The interaction of lncrna xloc-2222497, akr1c1, and progesterone in porcine endometrium and pregnancy. Int. J. Mol. Sci. 2020, 21, 3232. [Google Scholar] [CrossRef]
- Lal, A.; Thomas, M.P.; Altschuler, G.; Navarro, F.; O'Day, E.; Li, X.L.; Concepcion, C.; Han, Y.C.; Thiery, J.; Rajani, D.K.; et al. Capture of microrna-bound mrnas identifies the tumor suppressor mir-34a as a regulator of growth factor signaling. PLoS Genet. 2011, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Hua, R.; Zhang, X.; Li, W.; Lian, W.; Liu, Q.; Gao, D.; Wang, Y.; Lei, M. Ssc-mir-21-5p regulates endometrial epithelial cell proliferation, apoptosis and migration via the pdcd4/akt pathway. J. Cell Sci. 2020, 133, jcs248898. [Google Scholar] [CrossRef]
- Gurung, S.; Greening, D.W.; Catt, S.; Salamonsen, L.; Evans, J. Exosomes and soluble secretome from hormone-treated endometrial epithelial cells direct embryo implantation. Mol. Hum. Reprod. 2020, 26, 510–520. [Google Scholar] [CrossRef]
- Zhang, K.; Ge, L.; Dong, S.; Liu, Y.; Wang, D.; Zhou, C.; Ma, C.; Wang, Y.; Su, F.; Jiang, Y. Global mirna, lncrna, and mrna transcriptome profiling of endometrial epithelial cells reveals genes related to porcine reproductive failure caused by porcine reproductive and respiratory syndrome virus. Front. Immunol. 2019, 10, 1221. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of long non-coding rna and mrna profiling using rna sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef]
- Zhao, Y.; Hou, Y.; Xu, Y.; Luan, Y.; Zhou, H.; Qi, X.; Hu, M.; Wang, D.; Wang, Z.; Fu, Y.; et al. A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome. Nat. Commun. 2021, 12, 2217. [Google Scholar] [CrossRef]
- Akbar, R.; Ullah, K.; Rahman, T.U.; Cheng, Y.; Pang, H.Y.; Jin, L.Y.; Wang, Q.J.; Huang, H.F.; Sheng, J.Z. Mir-183-5p regulates uterine receptivity and enhances embryo implantation. J. Mol. Endocrinol. 2020, 64, 43–52. [Google Scholar] [CrossRef]
- Lu, J.; Huang, J.; Zhao, S.; Xu, W.; Chen, Y.; Li, Y.; Wang, Z.; Dong, Y.; You, R.; Cao, J.; et al. Foxo1 is a critical switch molecule for autophagy and apoptosis of sow endometrial epithelial cells caused by oxidative stress. Oxid. Med. Cell Longev. 2021, 2021, 1172273. [Google Scholar] [CrossRef]
- Greenbaum, L.E. Cell cycle regulation and hepatocarcinogenesis. Cancer Biol. Ther. 2004, 3, 1200–1207. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Li, H.; Yang, J.; Hao, D.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Lin, F.; et al. Circular rna profiling reveals an abundant circlmo7 that regulates myoblasts differentiation and survival by sponging mir-378a-3p. Cell Death Dis. 2017, 8, e3153. [Google Scholar] [CrossRef]
- Bian, Q.; Chen, B.; Weng, B.; Chu, D.; Tang, X.W.; Yan, S.N.; Yin, Y.F.; Ran, M.L. Circbtbd7 promotes immature porcine sertoli cell growth through modulating mir-24-3p/mapk7 axis to inactivate p38 mapk signaling pathway. Int. J. Mol. Sci. 2021, 22, 385. [Google Scholar] [CrossRef]
- Su, L.; Liu, R.; Cheng, W.; Zhu, M.; Li, X.; Zhao, S.; Yu, M. Expression patterns of micrornas in porcine endometrium and their potential roles in embryo implantation and placentation. PLoS ONE 2014, 9, e87867. [Google Scholar] [CrossRef]
- Li, W.; Xi, Y.; Xue, S.; Wang, Y.; Wu, L.; Liu, H.; Lei, M. Sequence analysis of micrornas during pre-implantation between meishan and yorkshire pigs. Gene 2018, 646, 20–27. [Google Scholar] [CrossRef]
- Li, H.; Xu, J.D.; Fang, X.H.; Zhu, J.N.; Yang, J.; Pan, R.; Yuan, S.J.; Zeng, N.; Yang, Z.Z.; Yang, H.; et al. Circular rna circrna_000203 aggravates cardiac hypertrophy via suppressing mir-26b-5p and mir-140-3p binding to gata4. Cardiovasc. Res. 2020, 116, 1323–1334. [Google Scholar] [CrossRef]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X.; Zhao, S. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the mir-140-3p/ska2 axis. Mol. Oncol. 2021, 15, 697–709. [Google Scholar] [CrossRef]
- Fu, C.; Mao, W.; Gao, R.; Deng, Y.; Gao, L.; Wu, J.; Zhang, S.; Shen, Y.; Liu, K.; Li, Q.; et al. Prostaglandin f-ptgfr signaling promotes proliferation of endometrial epithelial cells of cattle through cell cycle regulation. Anim. Reprod. Sci. 2020, 213, 106276. [Google Scholar] [CrossRef]
- Gao, L.; Gao, R.; Mao, W.; Liu, B.; Zhang, S.; Tahala, D.; Fu, C.; Shen, Y.; Wu, J.; Deng, Y.; et al. Ptgfr activation promotes the expression of ptgs-2 and growth factors via activation of the pkc signaling pathway in bovine endometrial epithelial cells. Anim. Reprod. Sci. 2018, 199, 30–39. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhou, C.; Jiang, X.; Huang, S.; Li, Y.; Su, T.; Wang, G.; Zhou, Y.; Liu, M.; Xu, D. Circ0001470 Acts as a miR-140-3p Sponge to Facilitate the Progression of Embryonic Development through Regulating PTGFR Expression. Cells 2022, 11, 1746. https://doi.org/10.3390/cells11111746
Zhang L, Zhou C, Jiang X, Huang S, Li Y, Su T, Wang G, Zhou Y, Liu M, Xu D. Circ0001470 Acts as a miR-140-3p Sponge to Facilitate the Progression of Embryonic Development through Regulating PTGFR Expression. Cells. 2022; 11(11):1746. https://doi.org/10.3390/cells11111746
Chicago/Turabian StyleZhang, Long, Changfan Zhou, Xiaoyu Jiang, Shuntao Huang, Yiheng Li, Tao Su, Guowei Wang, You Zhou, Min Liu, and Dequan Xu. 2022. "Circ0001470 Acts as a miR-140-3p Sponge to Facilitate the Progression of Embryonic Development through Regulating PTGFR Expression" Cells 11, no. 11: 1746. https://doi.org/10.3390/cells11111746
APA StyleZhang, L., Zhou, C., Jiang, X., Huang, S., Li, Y., Su, T., Wang, G., Zhou, Y., Liu, M., & Xu, D. (2022). Circ0001470 Acts as a miR-140-3p Sponge to Facilitate the Progression of Embryonic Development through Regulating PTGFR Expression. Cells, 11(11), 1746. https://doi.org/10.3390/cells11111746






